Abstract
The validity for assigning disorder risk to an autism spectrum disorder (ASD) candidate gene comes from convergent genetic, clinical, and developmental neurobiology data. Here, we review these lines of evidence from multiple human genetic studies, and non-human primate and mouse experiments that support the conclusion that the MET receptor tyrosine kinase (RTK) functions to influence synapse development in circuits relevant to certain core behavioral domains of ASD. There is association of both common functional alleles and rare copy number variants that impact levels of MET expression in the human cortex. The timing of Met expression is linked to axon terminal outgrowth and synaptogenesis in the developing rodent and primate forebrain, and both in vitro and in vivo studies implicate this RTK in dendritic branching, spine maturation, and excitatory connectivity in the neocortex. This impact can occur in a cell-nonautonomous fashion, emphasizing the unique role that Met plays in specific circuits relevant to ASD.
Keywords: Autism spectrum disorder, Synaptogenesis, Forebrain, Hepatocyte growth factor, Human genetics
Dysfunctional connectivity and autism spectrum disorder
Virtually all neurodevelopmental disorders are defined by deficits in brain circuitry and function that unfold as an individual develops. Presumably, differential ontogeny of neural connectivity—that is, when, how, and which specific brain circuits are affected—dictates the presentation of clinical phenotypes that characterize each neurodevelopmental disorder. While individuals with autism spectrum disorder (ASD) often present with complex behavioral and medical phenotypes (Geschwind 2009), connectivity-based etiological hypotheses regarding the social and communication components of the diagnosis are supported by numerous functional and structural imaging studies.
Functional magnetic resonance imaging (fMRI) studies have demonstrated that distributed cortical areas are activated less synchronously in individuals with ASD during the performance of a variety of social and cognitive tasks. These observations are consistent with long-range functional connectivity in the cortex being broadly compromised in this disorder, although there is evidence that connections with frontal and temporal cortices may be especially vulnerable (see Geschwind and Levitt 2007 for review). For example, during a sentence comprehension task, there is less synchronous activation of distributed language processing areas in ASD subjects (Just et al. 2004). However, compromised synchrony is selectively observed in fronto-parietal areas during executive function (Just et al. 2007) and social processing (Kana et al. 2009) tasks. Tasks requiring face recognition (Koshino et al. 2008) or linguistic and visuospatial reasoning (Sahyoun et al. 2010) also reveal reduced functional connectivity with frontal cortical regions in individuals with ASD. In contrast, occipito-parietal connectivity is intact (Koshino et al. 2008), or even enhanced (Sahyoun et al. 2010), when performing these tasks. fMRI studies also have indicated that interhemispheric connectivity is dysfunctional in ASD (Pierce and Redcay 2008; Anderson et al. 2010).
Volumetric MRI and diffusion tensor imaging (DTI) have provided anatomical evidence supporting the alterations in functional long-range connectivity in ASD. For example, brain-wide DTI analysis of the ASD brain reveals reduced fractional anisotropy (FA) in most major long-range fiber tracts, consistent with the view of some investigators of a more global deficit in functional connectivity (Shukla et al. 2010). The structural abnormalities in the inferior fronto-occipital and superior longitudinal fasciculi that have been reported in ASD (Shukla et al. 2010; Thomas et al. 2010) are especially relevant to deficits in functional fronto-temporal and fronto-posterior connectivity discussed above. Alterations in functional interhemispheric connectivity have also been corroborated anatomically by findings of altered white matter structure within the corpus callosum (Shukla et al. 2010; Thomas et al. 2010; Alexander et al. 2007). Moreover, these studies provide evidence for heterogeneity in individual presentations of ASD. For example, Alexander et al. (2007) found reduced FA in the corpus callosum in less than one-third of their subjects with ASD, despite detecting an overall group effect for the deficit. Similarly, reduced corpus callosum volume is a variable anatomical phenotype in ASD. More significantly, the magnitude of the reduction correlates with the degree of dysfunctional connectivity and, possibly, the severity of behavioral phenotypes (Just et al. 2007, but also see Anderson et al. 2010). If similar structure–function correlations manifest independently of each other across multiple key cortical regions, this could, at least in part, underlie the well-known heterogeneity in the severity of core behavioral deficits in ASD.
In contrast to the functional hypoconnectivity of long-range connections in ASD, it has been postulated that local connectivity may be spared or hyperfunctional in ASD (Geschwind and Levitt 2007; Casanova et al. 2002; Courchesne and Pierce 2005; Levitt et al. 2004; Rubenstein 2010; Rubenstein and Merzenich 2003). This hypothesis originally arose from observations in post-mortem tissue, in particular the atypical minicolumn structure seen in several regions of the cortex in brains from ASD individuals (Casanova et al. 2002; Casanova et al. 2010). Unfortunately, current imaging techniques do not provide the structural resolution to examine local cortico-cortical connectivity. However, DTI does provide sufficient resolution to study short-range association (i.e., U-association) fiber systems, which, as recent studies indicate, are indeed compromised in the frontal, parietal, and temporal lobes of subjects with ASD (Sundaram et al. 2008; Shukla et al. 2011). In addition, definitive evidence of altered local functional connectivity in ASD is lacking, although this has not been examined as rigorously as long-range connectivity. Interestingly, recent fMRI studies have demonstrated that increased local frontal lobe connectivity is associated with an ASD risk allele that may predispose carriers to language and implicit learning endophenotypes (Scott-Van Zeeland et al. 2010).
ASD-associated genes: understanding the heterogeneity of dysfunctional connectivity
Heritability and heterogeneity are two defining characteristics of ASD. Strong genetic heritability in ASD is evident from traditional twin studies, which have demonstrated concordance values between 50% and 91% for monozygotic twins and 0% for dizygotic twins (Folstein and Rutter 1977; Steffenburg et al. 1989). However, bivariate concordance among individual traits of the ASD triad (e.g., social impairment and communication impairment or social impairment and restrictive and repetitive behaviors and interests) has proven to be modest, indicating that distinct groups of genes may influence each trait (Ronald et al. 2006a; b). This is consistent with heterogeneity in both the severity of core deficits and the comorbidity of disorders such as mental retardation and seizures in ASD (Geschwind and Levitt 2007).
Behavioral heterogeneity in ASD is the readout of connectivity of brain circuits; given the clinical complexity of individuals with the disorder, many circuits must be involved, perhaps to different degrees in each individual. In turn, the dysfunctional connectivities that underlie heterogeneous ASD subtypes are presumably owed to developmental interactions among a variety of possible ASD risk genes and environmental influences. Risk genes may selectively confer vulnerability to developing circuitry that relates to a specific ASD core deficit. Alternatively, through epistatic relationships, risk genes may interact to disrupt expression patterns and/or levels of proteins that function in specific signaling pathways that globally impact ASD-relevant circuitry. In either case, heterogeneity results not only from the particular combination of risk genes expressed, but also from their pleiotropic functions in various other pathways (Bill and Geschwind 2009). In addition, while not particularly well-understood, we speculate that disorder heterogeneity also could be impacted by co-occurrence of exacerbating environmental factors that might otherwise more broadly impact connectivity. Even further, heterogeneity can occur as a function of the timing of the genetic insult. Here, two risk genes may specifically impact circuits related to social interaction but do so during distinct developmental epochs. If the expression of one gene is related to cell migration and expression of the other is related to a later process, such as synaptogenesis, the qualitative nature of their impact on that circuitry may be quite different. For example, during hippocampal development, disruption of Disc1 function alters the migration of granule neurons (Meyer and Morris 2009) and it has been suggested that this would subsequently affect the integration of these neurons into hippocampal circuitry. In contrast, disruption of TrkB signaling in these same neurons has no effect on the migration of granule neurons, but alters their maturation such that there is a reduction in the number of synaptic contacts, as well as synaptic vesicles, made by these cells (Otal et al. 2005). Considering the immense complexity of genetic and environmental interactions over development, significant progress in understanding the etiology of ASD will likely occur with the characterization of ASD risk alleles, individually, in the context of understanding their role in mediating histogenic events that ultimately impact circuit development.
Rare mutations in genes such as NLGN3, NLGN4, NRXN1, NRXN3, and SHANK3 are enriched in ASD compared to the general population, and are intriguing because of their clearly established roles in synaptogenesis and the maintenance of synaptic connectivity (Varoqueaux et al. 2006; Tabuchi et al. 2007; Hung et al. 2008; Gibson et al. 2009). However, to date, expression studies of these genes (Varoqueaux et al. 2006; Lim et al. 1999) have not provided the resolution required to understand their potential relationship to the development of forebrain circuits affected in ASD. Further confusing the issue is the fact that, in some cases, these gene mutations have been associated with both mental retardation and ASD in the same family (Laumonnier et al. 2004). Moreover, these ASD-enriched mutations are exceedingly rare, suggesting that their contribution to ASD, which affects 1 in 110 children, at a population level is minimal. In contrast, an example of a strong link between an ASD risk allele and potential etiological mechanisms has come from studies of the PTEN locus. In 2005, Butler and colleagues found an enrichment of rare loss-of-function mutations in the PTEN gene within a subset of individuals with ASD and comorbid macrocephaly (Butler et al. 2005). Remarkably, subsequent studies in a mouse model of postnatal, forebrain-specific PTEN loss-of-function revealed ethologically relevant deficits in social behavior, as well as macrocephaly that were due to increased dendritic and axonal arborization (Kwon et al. 2006). Excitement concerning this finding, however, is tempered by the fact that germ-line PTEN mutations are rare (<1 in 10,000) and relevant to only a few ASD cases. Insight into etiological mechanisms that are more broadly relevant to idiopathic cases of ASD may come from studies of common ASD risk alleles, though this has its own challenges because of effect sizes that typically are small (Nestler and Hyman 2010). In a few recent examples, common and rare copy number variants of the same genes that are enriched in ASD have emerged, including contactin-associated protein-like 2 (Alarcon et al. 2008; Bakkaloglu et al. 2008) and the MET receptor tyrosine kinase. With respect to the MET gene, multiple laboratories have established variants in the promoter region, as well as rare copy number variations, as risk alleles for ASD. Moreover, converging data from both human and animal studies are providing insights into the biological consequences of disrupted MET signaling, both in the brain and peripheral organs. Thus, beyond the statistical correlations, neurobiological studies are providing testable hypotheses for links between disrupted MET gene expression and specific endophenotypes.
The emergence of the Met receptor tyrosine kinase as an ASD risk gene
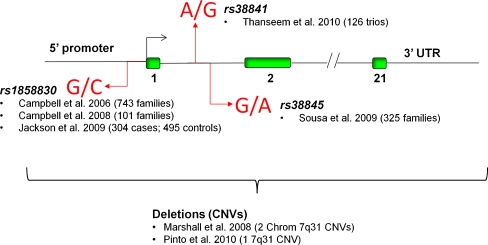
Over the past 5 years, five independent studies, using non-overlapping family and case–control cohorts, have demonstrated the association of three allelic variants of the gene encoding the MET receptor tyrosine kinase (MET) with ASD (Campbell et al. 2008; Campbell et al. 2006; Sousa et al. 2009; Jackson et al. 2009; Thanseem et al. 2010). The variants are all located in the putative 5′ regulatory region and have been associated with increased risk (Fig. 1). It is important to emphasize that genetic risk for any disorder, including ASD, requires translation into an altered biology. At least one MET variant, the rs1858830 ‘C’ allele, results in altered biological function, as shown in gel shift and transcriptional assays (Campbell et al. 2006). Specifically, the ‘C’ allele results in reduced transcriptional efficiency in vitro, a finding that is consistent with the twofold decrease in temporal neocortical MET protein expression observed in ASD post-mortem cases (Campbell et al. 2007). Furthermore, the ‘C’ allele, independent of diagnosis, correlates with levels of MET expression. With a calculated relative risk of 2.29 for ASD, the combined data strongly support the conclusion that the rs1858830 ‘C’ allele is functional, yielding reduced expression of a protein that impacts circuit development and function (see below).
Fig. 1.
Diagram illustrating the three allelic variants of the gene encoding the MET receptor tyrosine kinase (MET) that have been associated with an increased risk for ASD in multiple independent studies. In addition, rare copy number variants (CNVs) that interrupt the MET gene have been associated with ASD. See reference list for citations noted in the figure
Additional genetic studies indicate that MET and other genes encoding proteins in the MET receptor signaling pathway serve as risk factors for ASD. First, two studies report rare copy number variants that interrupt the MET gene preferentially in ASD but not in control populations (Marshall et al. 2008; Pinto et al. 2010). Second, mutations in genes that cause rare syndromes or macrocephaly, including tuberous sclerosis, neurofibromatosis, and PTEN, have high co-diagnosis of ASD. These genes encode proteins that are critical for MET receptor signaling (Levitt and Campbell 2009). Third, there is an enrichment of the rs1858830 risk variant in a stratified subpopulation of children with ASD that is characterized by co-occurring gastrointestinal (GI) disorders (Campbell et al. 2009a), which are over abundantly represented in ASD (Campbell et al. 2009a; Buie et al. 2010; Ibrahim et al. 2009). The analysis of MET variation in ASD-GI comorbid disorders was prompted by reports that MET positively regulates the development and repair of gut epithelial cells (Kitamura et al. 2000; Matsubara et al. 1998; Tahara et al. 2003). Moreover, the enrichment speaks to the relevance of connecting allelic variation to specific pathophysiological processes. Yet, despite this preponderance of convergent genetic evidence, general acceptance of MET as an ASD risk gene, based on common variation and rare copy number variations (CNVs), will be gained only insofar as its developmental neurobiological functions can be related specifically to ASD pathogenesis.
Met functions in neural development
The MET receptor was first identified as a proto-oncogene (Cooper et al. 1984), and the context in which MET function is best understood is in cancer biology (Trusolino et al. 2010). Thus, the recognition of MET serving as a key signaling component in specific neurodevelopmental events is relatively new. MET signaling commences upon the binding of its polypeptide ligand, hepatocyte growth factor (HGF). HGF binding induces MET receptor oligomerization and the autophosphorylation of intracellular tyrosine residues that serve as multi-substrate docking sites for signaling adaptor proteins (Trusolino et al. 2010; Ponzetto et al. 1994). These adaptor proteins in turn facilitate the activation of several major signaling cascades including the PI3K, ERK, and p38 systems (Trusolino et al. 2010; Stefan et al. 2001). Like other members of the receptor tyrosine kinase family, the nature of MET signaling is context- and cell-type-dependent, reflecting pleiotropic roles in cellular development.
Met signaling in cells grown in vitro has been shown to potentiate virtually every aspect of development involved in establishing neural connections. Several in vitro models have been used to show that Met activation regulates dendritic outgrowth of neocortical and hippocampal pyramidal neurons (Gutierrez et al. 2004; Lim and Walikonis 2008). Axon outgrowth can be potentiated by Met signaling, as shown in dissociated and explant culture systems (David et al. 2008; Powell et al. 2003). Further in vitro studies demonstrate that Met influences the formation, molecular differentiation and function (Akimoto et al. 2004; Tyndall et al. 2007; Tyndall and Walikonis 2006; 2007) of synaptic connections between forebrain neurons. With the exception of the putative role for Met in motor axon outgrowth (Ebens et al. 1996), however, these findings have yet to be extended to in vivo contexts. Moreover, because Met expression is readily modified by environment, one must be cautious in assuming that Met signaling functions defined in vitro will necessarily reflect in vivo physiology. For example, HGF-mediated activation of Met enhances the motility of cells derived from the fetal ganglionic eminence (Powell et al. 2001), which go on to form neocortical interneurons. Yet, recent studies from our laboratory demonstrate that fetal ganglionic eminence cells and neocortical interneurons do not express transcript or protein in vivo and, thus, genetic deletion of Met in vivo fails to alter cortical interneuron development (Judson et al. 2009; Eagleson et al. 2010). The apparent contradictory in vivo and in vitro data were reconciled by the demonstration that Met protein expression is induced in fetal ganglionic eminence neurons in culture (Eagleson et al. 2010). Thus, as with other ASD risk genes, connecting experimental data to neurodevelopmental perturbations that are relevant to ASD is challenging.
Met—a new player at the developing synapse
Recent transcript and protein mapping studies in the developing mouse forebrain have provided a clearer picture of cell-type specific and spatio-temporal features of Met expression (Judson et al. 2009; Eagleson et al. 2010), and have helped to identify testable, putative neurodevelopmental roles for the receptor in vivo. First, it was found that Met mRNA exhibits limited expression in the mouse forebrain (defined as pallial- and subpallial-derived cortical and subcortical structures) prenatally, with detectable transcript in the neocortical plate, hippocampus, amygdala, and septum by embryonic day (E) 17.5. Expression increases dramatically postnatally in these same regions, coinciding temporally with the rapid growth of neuropil elements that form synapses. In the mouse, the rapid phase of synaptogenesis extends over a 2-week period [postnatal days (P) 7–21], followed by pruning that extends through the third and fourth week. Met expression in the forebrain corresponds to phases of rapid axon terminal growth and early synaptogenesis, as shown by increases through P7, followed by a steady decline through the next week, and then a rapid and robust decrease in transcript by puberty. Importantly, Met transcript in the neocortex co-localizes with markers of excitatory projection neurons during development (Eagleson et al. 2010), but not inhibitory neurons, astrocytes, or oligodendrocytes.
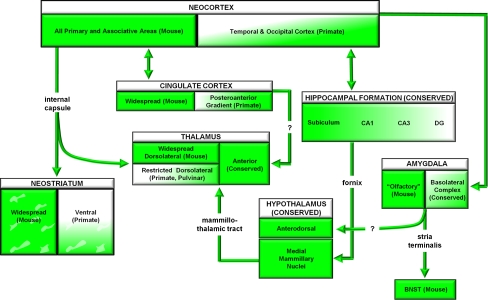
Transcript mapping reveals origins of protein translation, but immunocytochemistry reveals sites of protein localization and, perhaps, functional activity (Fig. 2). Analysis of anti-Met immunolocalization reveals a temporal pattern of protein expression that is identical to that found for Met mRNA. Spatially, however, there are substantive differences. Whereas the neocortex, amygdala, hippocampus, and septum all express Met protein (as well as transcript), additional forebrain areas that do not express the Met transcript are immunopositive. In particular, the striatum and dorsal thalamus, two major targets of corticofugal projections, express Met protein during the P0–14 period of intensive terminal axon growth and neuropil development. Importantly, a dorsal pallial promoter-driven functional deletion of Met eliminates immunostaining from all but a few structures in the forebrain (Judson et al. 2009). Thus, Met expression in the striatum and dorsal thalamus originates from excitatory projection neurons in the neocortex, with the protein transported in developing axons during axonal growth and synapse formation. In fact, intense immunolocalization of Met is observed in the internal capsule, fornix, corpus callosum, and anterior commissure, all of which carry axons from the neocortex to other cortical and subcortical destinations. Whereas the localization in subcortical structures that receive cortical input is clearly presynaptic, the subcellular distribution of Met in the neocortex proper may be both pre- and postsynaptic. This awaits ultrastructural validation in vivo.
Fig. 2.
Summary of MET/Met expression in the developing primate and mouse forebrain. Green shading indicates immunolocalization of the receptor protein in cortical and Subcortical regions and axon tracts. Regions denoted do not express the Met transcript (based on analyses in the mouse), but have detectable protein because each structure receives Met-immunoreactive axons. See (Judson et al. 2009; Judson et al. 2011) for more details
Descriptive mapping data from the developing mouse forebrain demonstrate a spatio-temporal pattern of gene and protein expression that is consistent with a role in development. What specific histogenic events then are perturbed due to the elimination of Met signaling? Laminar and cytoarchitectural organization of the neocortex and subcortical regions that express the Met transcript are normal. Detailed morphometric studies of post-pubertal, kinase-dead, conditional Met mutant mice indicate alterations in neocortical pyramidal neuron dendritic morphology (Fig. 3; Judson et al. 2010). In these conditional Met mutant mice, there are differential, compartment-specific changes in layer 2/3 pyramidal cell dendritic branching. Specifically, increases in branching in the basal dendrites are offset by accompanying reductions in branching in apical dendrites to the apical arbor. Consequently, overall dendritic length in this neuron population is unchanged relative to wild-type controls. However, because distal dendritic length is preferentially lost, the cortical volume coverage of the dendritic tree is, on average, significantly reduced. These compartment-specific changes are suggestive of presynaptic consequences of Met signaling dysfunction that are driven by distinct afferent inputs to the pyramidal cell dendritic arbor. A potential presynaptic mechanism of Met signaling dysfunction is strongly supported by the observation that dendritic branching is also altered in medium spiny neurons (MSN) in conditional Met mutant mice. Notably, unlike cortical pyramidal neurons, MSNs themselves do not express Met during development, but receive heavily Met-expressing corticostriatal afferents.
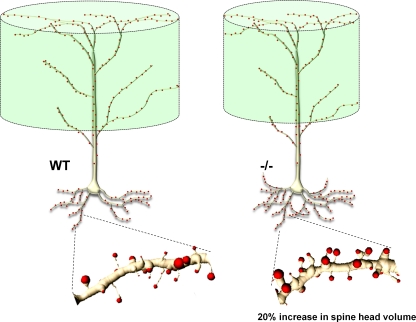
Fig. 3.
Conditional Met deletion by Emx1Cre (−/−) increases proximal and reduces distal apical dendritic branching of neocortical pyramidal neurons in post-pubertal animals. The overall impact results in a reduced volume of cortical tissue sampled by the apical dendrites. Analysis of spine number and size on basal dendrites revealed an approximately 22% increase in spine head volume, but no change in density of spines. See (Judson et al. 2010) for details
Our investigation of the conditional Met mutant mice revealed abnormal dendritic spine morphology. Spine volume on basal dendrites is increased by approximately 22%, without changes in spine numbers. Changes of this magnitude, while seemingly modest, have been shown to translate into a linear enhancement of Ca++ influx, which on a per spine basis would increase excitatory synaptic transmission (Matsuzaki et al. 2001), consistent with our recent electrophysiological data (see below). Analysis of MSN spine morphology also reveals an increase in spine volume. Because the analysis was done in post-pubertal animals, these changes are likely to be permanent.
How do these alterations in dendritic and spine morphology compare to studies examining the impact of single gene alterations for syndromic neurodevelopmental disorders? Table 1 summarizes numerous studies in mice for genes implicated in syndromic disorders for which an ASD diagnosis is common. The extent of reported dendritic and spine alterations are generally comparable to those that occur when Met is deleted functionally in conditional Met mutant mice. Of particular interest is that many of the syndromic disorders involve genes that encode proteins in the same intracellular signaling pathways (ERK/PI3 Kinase) through which Met and other receptor tyrosine kinases signal. It is unclear, however, how ubiquitously expressed signaling pathways can differentially impact specific circuits, but one possibility is that phenotypic heterogeneity may in part be owed to diverse upstream signaling disruptions that converge on these conserved downstream pathways (Bill and Geschwind 2009; Levitt and Campbell 2009). The functional implications of different syndromic disorder mutations examined in mice are varied. Different behavioral tasks have been used to detect pronounced cognitive impairments, altered social behavior and physiological changes that reflect altered synaptic function (Lee and Silva 2009; Crawley 2008).
Table 1.
Summary of in vivo analyses of morphological changes in mice carrying mutations of ASD-related syndromic disorders compared to Met deletion
| Syndrome/gene | Gene mutation | Developmental time of analyses | Morphological changes on neocortical pyramidal neurons | References |
|---|---|---|---|---|
| Met Receptor | Metfx/fx/Emx1Cre | Post-pubertal (P40) | ∼22% ↑ in ↓ spine volume; 20% in total dendritic tree volume; no change in total dendrite length | (Judson et al. 2010. JCN) |
| Fragile X | Fmr-1 null | Adult | No changes in dendrites/spines | (Galvez and Greenough 2005; Nimchinsky et al. 2001) |
| Fmr-1 null | P7 | ∼15–30% ↑ in immature spine #s; 20–28% ↑ in spine length | (Nimchinsky et al. 2001; Comery et al. 1997; Irwin et al. 2002) | |
| Neurofibromatosis-1 | NF-1 null | None | None | None |
| AngelmanSydnrome | Maternal Ube3a null | Adult | 30% ↓ in spine density; 10% ↓ in spine length | (Dindot et al. 2007) |
| Juvenile (V1 critical period) | 25% ↓ in spine density on basal dendrites in layer 2/3 | (Yashiro et al. 2009) | ||
| Juvenile (V1 critical period) | 15% ↓ in spine density on layer 5 basal dendrites but no change in apical density; no change in dendritic branching | (Sato and Stryker 2010) | ||
| Rett Syndrome | Mecp2 A140V knock-in | Adult | Segment-restricted 50% ↓ in dendritic branching | (Jentarra et al. 2010) |
| Mecp2 Hemizygous | Adult | 25% ↓ in spine number; 40% in dendrite diameter | (Fukuda et al. 2005) | |
| Mecp2 Hemizygous | Adult | 20-40% ↓ in dendritic branching; no difference in spine number | (Kishi and Macklis 2010) | |
| Tuberous Sclerosis | Tsc1 null | P20 | 15% ↑ in spine head width; 20% ↑ in spine length; 25% ↓ in spine density | (Tavazoie et al. 2005) |
Our recent efforts have focused on providing a physiological translation of the structural changes that manifest when Met is conditionally deleted from cortical excitatory neurons. Using a combination of local circuit electrophysiological mapping by laser scanning photostimulation and paired recordings, we revealed that local excitatory connections from cortical layer 2–3 pyramidal neurons onto specific subpopulations of layer 5 pyramidal neurons are enhanced twofold (Qiu et al. 2011). Moreover, layer 5 neurons that do not express Met failed to exhibit this enhanced local circuit input. These experimental data are consistent with human imaging evidence suggesting that subsets of local cortical connections in ASD may be atypically enhanced (Scott-Van Zeeland et al. 2010). We also have preliminary data showing reduced sustainability of long-term potentiation and poorer cognitive performance in behavioral tasks. As these studies emerge, the relevance of Met signaling in the establishment of proper circuit function will become more evident.
MET in ASD risk—clinical and animal studies converge
The original study of MET in two large family cohorts with ASD subjects, with a calculated relative risk of approximately 2.3, suggests that the functional ‘C’ promoter allele generates modest risk for non-syndromic, idiopathic ASD. This initial conclusion is supported by subsequent replication in additional family and case–control studies, the association of two other promoter alleles that yield smaller risk effects and CNVs (see above). Yet ASD is decidedly heterogeneous, suggesting that no single gene will increase risk for all subjects diagnosed with the disorder. In our initial analyses of more than 700 families, the MET functional allele was overrepresented in those subjects identified as being part of multi-incident families (more than one child with ASD), but not single-incident families (Campbell et al. 2006). This is consistent with the idea that common genetic predisposing factors for complex polygenic disorders, such as the ‘C’ MET promoter allele for ASD, are more likely to be enriched in multi-incident families (Risch 2001). These data indicate that further stratification of populations may yield additional insight regarding the role of MET in ASD risk.
Most children with neurodevelopmental disorders exhibit a variety of medical and psychiatric conditions (Batshaw 2002; Reber 2002), and this indeed occurs in ASD (Geschwind 2009). This likely reflects the pleiotropic nature of many of the proteins encoded by genes that increase ASD risk. A recent study from our laboratory indicates that analysis of genetic risk across functional groupings in large cohorts may provide insight into biological influences that yield such clinical heterogeneity. Specifically, we find that in families in which a child is diagnosed with ASD and has a reported gastrointestinal condition, the MET functional ‘C’ allele is even further enriched when compared to children with ASD alone (Campbell et al. 2009a; b). Why is there a connection? MET is expressed in the developing gut in humans (Kermorgant et al. 1997) and mice (Gorrindo et al. unpublished data). In addition, HGF-Met signaling in experimental models of colitis improves epithelial repair (Tahara et al. 2003). Thus, it would be predicted, based on animal studies, that alterations in Met signaling would influence circuit formation in the brain, as well as gut function. Our findings of subgroup enrichment of the functional MET risk allele are consistent with this prediction and provide insight into how genetic risk may translate into biological influences that yield relevant clinical phenotypes.
There have been significant challenges in identifying how specific risk genes may impact the core social and communication functions that are used diagnostically to identify children with ASD. Expression databases and mapping in mice (e.g., Allen Institute for Brain Science, http://www.brain-map.org/) indicate that many rodent orthologues of ASD risk genes, particularly those that carry rare mutations, can be expressed relatively broadly in the brain, and when mutated, result in significant cognitive impairments in mice (in addition to disruption of social and emotional regulation). These impairments are analogous to the generalized intellectual disability that occurs in children with ASD and those with syndromic disorders in which ASD is often co-diagnosed. Yet in instances in which the primate orthologous genes/proteins are mapped, there may be noted species differences in expression. For example, expression of CNTNAP2 is enriched at mid-gestation in frontal lobe regions of the human (Alarcon et al. 2008), but is ubiquitous in the mouse brain. To gain insight into potential species-specific patterns of MET/Met expression, we recently compared immunohistochemical staining patterns of orthologous receptors in the rhesus monkey (MET) and mouse (Met; Judson et al. 2011). We focused on late pre- and early postnatal periods of development (Fig. 2), when axonal terminal growth and synaptogenesis progress rapidly (Bourgeois 1997). The temporal expression of MET/Met in relation to axonal targeting and early phases of synapse formation is highly conserved between species, peaking near P7 in the mouse and around gestational day 150 in the monkey (Judson et al. 2011). Moreover, subcortical forebrain expression is similar, with immunostaining evident in the neuropil of many of the same amygdala subnuclei, the subiculum and CA1 of the hippocampal formation, and the medial mammillary nuclei, among other structures. Expression of MET/Met in growing axons is highly conserved across species, but in patterns within fiber tracts that reflect major differences in cortical distributions. Whereas there is widespread expression throughout the mouse neocortex, MET protein distribution in the developing monkey brain is highly restricted to temporal and occipital lobe regions. Consequently, MET immunoreactivity is detected only in the anterior commissure, the sublenticular internal capsule and the posterior corpus callosum, which carry axons between these select cortical areas. Thus, given the collective data that Met can influence synaptic development, we suggest that MET may influence the wiring of select circuits in the primate brain, particularly those that are relevant to the processing of complex visual stimuli, including faces (Pinsk et al. 2009; Pinsk et al. 2005; Moeller et al. 2008; Tsao and Livingstone 2008). This can be tested empirically, similar to recent correlations between CNTNAP2 and frontal connectivity in humans (Scott-Van Zeeland et al. 2010).
The developmental profile of MET expression provides insight into how alterations in MET expression during development may contribute to some of the endophenotypes observed in ASD. The temporal pattern of MET expression in both rodents and primates is consistent with the proposed role of this protein in influencing the wiring of forebrain circuits, most likely through combined effects on axon and dendritic growth and synapse formation and maturation. The spatial expression patterns identify the specific circuits that are likely to be affected by alterations in MET expression. Most intriguingly with respect to ASD, it appears that cortical circuits important for processing socially relevant cues (e.g., auditory and visual circuits in the primate), would be most impacted by disruptions in MET signaling.
Concluding remarks
Functional data in children and adults with ASD suggest that it is a neurodevelopmental disorder that has its roots in the disruption of developing connectivity (Geschwind and Levitt 2007; Minshew et al. 1997). It therefore should not come as a surprise that many of the most compelling candidate genes for rare/syndromic and idiopathic forms of ASD are involved in aspects of brain wiring and synaptic function (Bourgeron 2009). However, there are practical limitations to providing definitive evidence beyond correlational human and animal studies that a particular gene is a risk factor for ASD. Excitement regarding claims of developmental neurobiological relevance or function at the forming synapse is muted by the fact that there are thousands of pre- and postsynaptic gene products (Ryan and Grant 2009), and a large part of the genome is expressed at some time in primate brain development (Johnson et al. 2009). Thus, there is a strong likelihood that any statistically identified ASD candidate gene can reasonably be proposed to have biological relevance to the disorder. The challenge, therefore, is to gather convergent lines of evidence that show consistency in clinical, genetic, and neurobiological phenomena that relate to ASD risk. Here, recent published and preliminary findings were summarized that, as we suggest, make a strong case for a new player that contributes to ASD vulnerability at the developing synapse—the MET receptor tyrosine kinase.
References
- Akimoto M, Baba A, Ikeda-Matsuo Y, Yamada MK, Itamura R, Nishiyama N, et al. Hepatocyte growth factor as an enhancer of nmda currents and synaptic plasticity in the hippocampus. Neuroscience. 2004;128(1):155–162. doi: 10.1016/j.neuroscience.2004.06.031. [DOI] [PubMed] [Google Scholar]
- Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander AL, Lee JE, Lazar M, Boudos R, DuBray MB, Oakes TR, et al. Diffusion tensor imaging of the corpus callosum in Autism. Neuroimage. 2007;34(1):61–73. doi: 10.1016/j.neuroimage.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Anderson JS, Druzgal TJ, Froehlich A, Dubray MB, Lange N, Alexander AL, et al. Decreased Interhemispheric Functional Connectivity in Autism. Cereb Cortex. 2010 Oct 12. [DOI] [PMC free article] [PubMed]
- Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82(1):165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batshaw ML. Children with disabilities. 5th ed. Batshaw ML, editor. Baltimore: Brookes; 2002.
- Bill BR, Geschwind DH. Genetic advances in autism: heterogeneity and convergence on shared pathways. Curr Opin Genet Dev. 2009;19(3):271–278. doi: 10.1016/j.gde.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois J. Synaptogenesis, heterochrony and epigenesis in the mammalian neocortex. Acta Paediatr Suppl. 1997;422:27–33. doi: 10.1111/j.1651-2227.1997.tb18340.x. [DOI] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19(2):231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Buie T, Campbell DB, Fuchs GJ, 3rd, Furuta GT, Levy J, Vandewater J, et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics. 2010;125(Suppl 1):S1–S18. doi: 10.1542/peds.2009-1878C. [DOI] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42(4):318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci USA. 2006;103(45):16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, D'Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, et al. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann Neurol. 2007;62(3):243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- Campbell DB, Li C, Sutcliffe JS, Persico AM, Levitt P. Genetic evidence implicating multiple genes in the MET receptor tyrosine kinase pathway in autism spectrum disorder. Autism Res. 2008;1(3):159–168. doi: 10.1002/aur.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, Buie TM, Winter H, Bauman M, Sutcliffe JS, Perrin JM, et al. Distinct genetic risk based on association of MET in families with co-occurring autism and gastrointestinal conditions. Pediatrics. 2009;123(3):1018–1024. doi: 10.1542/peds.2008-0819. [DOI] [PubMed] [Google Scholar]
- Campbell DB, Warren D, Sutcliffe JS, Lee EB, Levitt P. Association of MET with social and communication phenotypes in individuals with autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2009 Jun 22. [DOI] [PMC free article] [PubMed]
- Casanova MF, Buxhoeveden DP, Switala AE, Roy E. Minicolumnar pathology in autism. Neurology. 2002;58(3):428–432. doi: 10.1212/wnl.58.3.428. [DOI] [PubMed] [Google Scholar]
- Casanova MF, El-Baz A, Vanbogaert E, Narahari P, Switala A. A topographic study of minicolumnar core width by lamina comparison between autistic subjects and controls: possible minicolumnar disruption due to an anatomical element in-common to multiple laminae. Brain Pathol. 2010;20(2):451–458. doi: 10.1111/j.1750-3639.2009.00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, et al. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proceedings of the National Academy of Sciences of the United States of America. 1997 May 13;94(10):5401-4. [DOI] [PMC free article] [PubMed]
- Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984 Sep 6–11;311(5981):29–33. [DOI] [PubMed]
- Courchesne E, Pierce K. Why the frontal cortex in autism might be talking only to itself: local over-connectivity but long-distance disconnection. Curr Opin Neurobiol. 2005;15(2):225–230. doi: 10.1016/j.conb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57(6):809–818. doi: 10.1016/j.neuron.2008.03.001. [DOI] [PubMed] [Google Scholar]
- David MD, Yeramian A, Duñach M, Llovera M, Cantí C, Herreros AG, et al. Signalling by neurotrophins and hepatocyte growth factor regulates axon morphogenesis by differential beta-catenin phosphorylation. J Cell Sci. 2008;121(Pt 16):2718–2730. doi: 10.1242/jcs.029660. [DOI] [PubMed] [Google Scholar]
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Human Molecular Genetics. 2007 Sep 27;17(1):111–8. [DOI] [PubMed]
- Eagleson KL, Campbell DB, Thompson BL, Bergman MY, Levitt P. The autism risk genes MET and PLAUR differentially impact cortical development. Autism Research. 2010;3:1–16. doi: 10.1002/aur.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebens A, Brose K, Leonardo ED, Hanson MG, Bladt F, Birchmeier C, et al. Hepatocyte growth factor/scatter factor is an axonal chemoattractant and a neurotrophic factor for spinal motor neurons. Neuron. 1996;17(6):1157–1172. doi: 10.1016/S0896-6273(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18(4):297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Itoh M, Ichikawa T, Washiyama K, Goto Y-i. Delayed maturation of neuronal architecture and synaptogenesis in cerebral cortex of Mecp2-deficient mice. J Neuropathol Exp Neurol. 2005 Jun 1;64(6):537–44. [DOI] [PubMed]
- Galvez R, Greenough WT. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am J Med Genet. 2005 Jun 1;135A(2):155–60. [DOI] [PubMed]
- Geschwind DH. Advances in autism. Annu Rev Med. 2009;60:367–380. doi: 10.1146/annurev.med.60.053107.121225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17(1):103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Gibson JR, Huber KM, Sudhof TC. Neuroligin-2 deletion selectively decreases inhibitory synaptic transmission originating from fast-spiking but not from somatostatin-positive interneurons. J Neurosci. 2009;29(44):13883–13897. doi: 10.1523/JNEUROSCI.2457-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez H, Dolcet X, Tolcos M, Davies A. HGF regulates the development of cortical pyramidal dendrites. Development. 2004;131(15):3717–3726. doi: 10.1242/dev.01209. [DOI] [PubMed] [Google Scholar]
- Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28(7):1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Idupulapati M, Gilbert ME, Harris JB, Chakravarti AB, Rogers EJ, et al. Dendritic spine and dendritic field characteristics of layer V pyramidal neurons in the visual cortex of fragile-X knockout mice. Am J Med Genet. 2002 Jul 29;111(2):140–6. [DOI] [PubMed]
- Ibrahim SH, Voigt RG, Katusic SK, Weaver AL, Barbaresi WJ. Incidence of gastrointestinal symptoms in children with autism: a population-based study. Pediatrics. 2009;124(2):680–686. doi: 10.1542/peds.2008-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PB, Boccuto L, Skinner C, Collins JS, Neri G, Gurrieri F, et al. Further evidence that the rs1858830 C variant in the promoter region of the MET gene is associated with autistic disorder. Autism Res. 2009;2(4):232–236. doi: 10.1002/aur.87. [DOI] [PubMed] [Google Scholar]
- Jentarra GM, Olfers SL, Rice SG, Srivastava N, Homanics GE, Blue M, et al. Abnormalities of cell packing density and dendritic complexity in the MeCP2 A140V mouse model of Rett syndrome/X-linked mental retardation. Mar. 2010;9:1–15. doi: 10.1186/1471-2202-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Ž, Coppola G, Bogdanović D, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62(4):494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson MC, Bergman MY, Campbell DB, Eagleson KL, Levitt P. Dynamic gene and protein expression patterns of the autism-associated met receptor tyrosine kinase in the developing mouse forebrain. J Comp Neurol. 2009;513(5):511–531. doi: 10.1002/cne.21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson M, Eagleson K, Wang L, Levitt P. Evidence of cell-nonautonomous changes in dendrite and dendritic spine morphology in the met-signaling-deficient mouse forebrain. J Comp Neurol. 2010;518:4463–4478. doi: 10.1002/cne.22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson MC, Amaral DG, Levitt P. Conserved Subcortical and Divergent Cortical Expression of Proteins Encoded by Orthologs of the Autism Risk Gene MET. Cerebral Cortex. 2011 Dec 1;in press. [DOI] [PMC free article] [PubMed]
- Just MA, Cherkassky VL, Keller TA, Minshew NJ. Cortical activation and synchronization during sentence comprehension in high-functioning autism: evidence of underconnectivity. Brain. 2004;127(Pt 8):1811–1821. doi: 10.1093/brain/awh199. [DOI] [PubMed] [Google Scholar]
- Just MA, Cherkassky VL, Keller TA, Kana RK, Minshew NJ. Functional and anatomical cortical underconnectivity in autism: evidence from an FMRI study of an executive function task and corpus callosum morphometry. Cereb Cortex. 2007;17(4):951–961. doi: 10.1093/cercor/bhl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana RK, Keller TA, Cherkassky VL, Minshew NJ, Just MA. Atypical frontal-posterior synchronization of Theory of Mind regions in autism during mental state attribution. Soc Neurosci. 2009;4(2):135–152. doi: 10.1080/17470910802198510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermorgant S, Walker F, Hormi K, Dessirier V, Lewin MJ, Lehy T. Developmental expression and functionality of hepatocyte growth factor and c-Met in human fetal digestive tissues. Gastroenterology. 1997;112(5):1635–1647. doi: 10.1016/S0016-5085(97)70046-5. [DOI] [PubMed] [Google Scholar]
- Kishi N, Macklis JD. MeCP2 functions largely cell-autonomously, but also non-cell-autonomously, in neuronal maturation and dendritic arborization of cortical pyramidal neurons. Exp Neurol. 2010 Feb 11;222(1):51–8. [DOI] [PMC free article] [PubMed]
- Kitamura S, Kondo S, Shinomura Y, Isozaki K, Kanayama S, Higashimoto Y, et al. Expression of hepatocyte growth factor and c-met in ulcerative colitis. Inflamm Res. 2000;49(7):320–324. doi: 10.1007/PL00000212. [DOI] [PubMed] [Google Scholar]
- Koshino H, Kana RK, Keller TA, Cherkassky VL, Minshew NJ, Just MA. fMRI investigation of working memory for faces in autism: visual coding and underconnectivity with frontal areas. Cereb Cortex. 2008;18(2):289–300. doi: 10.1093/cercor/bhm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50(3):377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74(3):552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-S, Silva AJ. The molecular and cellular biology of enhanced cognition. Nat Rev Neurosci. 2009;10(2):126–140. doi: 10.1038/nrn2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Campbell DB. The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J Clin Invest. 2009;119(4):747–754. doi: 10.1172/JCI37934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Eagleson KL, Powell EM. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci. 2004;27(7):400–406. doi: 10.1016/j.tins.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Lim CS, Walikonis RS. Hepatocyte growth factor and c-Met promote dendritic maturation during hippocampal neuron differentiation via the Akt pathway. Cell Signal. 2008;20(5):825–835. doi: 10.1016/j.cellsig.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, et al. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274(41):29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara Y, Ichinose M, Yahagi N, Tsukada S, Oka M, Miki K, et al. Hepatocyte growth factor activator: a possible regulator of morphogenesis during fetal development of the rat gastrointestinal tract. Biochem Biophys Res Commun. 1998;253(2):477–484. doi: 10.1006/bbrc.1998.9808. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Ellis-Davies GC, Nemoto T, Miyashita Y, Iino M, Kasai H. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2001;4(11):1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Morris JA. Disc1 regulates granule cell migration in the developing hippocampus. Hum Mol Genet. 2009;18(17):3286–3297. doi: 10.1093/hmg/ddp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshew NJ, Goldstein G, Siegel DJ. Neuropsychologic functioning in autism: profile of a complex information processing disorder. J Int Neuropsychol Soc. 1997;3(4):303–316. [PubMed] [Google Scholar]
- Moeller S, Freiwald WA, Tsao DY. Patches with links: a unified system for processing faces in the macaque temporal lobe. Science. 2008;320(5881):1355–1359. doi: 10.1126/science.1157436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13(10):1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001 Jul 15;21(14):5139–46. [DOI] [PMC free article] [PubMed]
- Otal R, Martinez A, Soriano E. Lack of TrkB and TrkC signaling alters the synaptogenesis and maturation of mossy fiber terminals in the hippocampus. Cell Tissue Res. 2005;319(3):349–358. doi: 10.1007/s00441-004-1020-5. [DOI] [PubMed] [Google Scholar]
- Pierce K, Redcay E. Fusiform function in children with an autism spectrum disorder is a matter of "who". Biol Psychiatry. 2008;64(7):552–560. doi: 10.1016/j.biopsych.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsk MA, DeSimone K, Moore T, Gross CG, Kastner S. Representations of faces and body parts in macaque temporal cortex: a functional MRI study. Proc Natl Acad Sci USA. 2005;102(19):6996–7001. doi: 10.1073/pnas.0502605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsk MA, Arcaro M, Weiner KS, Kalkus JF, Inati SJ, Gross CG, et al. Neural representations of faces and body parts in macaque and human cortex: a comparative fMRI study. J Neurophysiol. 2009;101(5):2581–2600. doi: 10.1152/jn.91198.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzetto C, Bardelli A, Zhen Z, Maina F, dalla Zonca P, Giordano S, et al. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell. 1994;77(2):261–271. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- Powell EM, Mars WM, Levitt P. Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron. 2001;30(1):79–89. doi: 10.1016/S0896-6273(01)00264-1. [DOI] [PubMed] [Google Scholar]
- Powell EM, Mühfriedele S, Bolz JUR, Levitt P. Differential regulation of thalamic and cortical axonal growth by hepatocyte growth factor/scatter factor. Dev Neurosci. 2003;25(2–4):197–206. doi: 10.1159/000072268. [DOI] [PubMed] [Google Scholar]
- Qiu S, Anderson CT, Levitt P, Shepherd GMG. Circuit-specific intracortical hyperconnectivity in mice with deletion of the autism-associated Met receptor tyrosine kinase. J Neurosci. 2011;31:5855–5864. doi: 10.1523/JNEUROSCI.6569-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber M. Dual diagnosis: mental retardation and psychiatric disorders. 5th ed. Batshaw ML, editor. Baltimore: Brookes; 2002.
- Risch N. Implications of multilocus inheritance for gene-disease association studies. Theor Popul Biol. 2001;60(3):215–220. doi: 10.1006/tpbi.2001.1538. [DOI] [PubMed] [Google Scholar]
- Ronald A, Happe F, Bolton P, Butcher LM, Price TS, Wheelwright S, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45(6):691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- Ronald A, Happe F, Price TS, Baron-Cohen S, Plomin R. Phenotypic and genetic overlap between autistic traits at the extremes of the general population. J Am Acad Child Adolesc Psychiatry. 2006;45(10):1206–1214. doi: 10.1097/01.chi.0000230165.54117.41. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL. Three hypotheses for developmental defects that may underlie some forms of autism spectrum disorder. Curr Opin Neurol. 2010;23(2):118–123. doi: 10.1097/WCO.0b013e328336eb13. [DOI] [PubMed] [Google Scholar]
- Rubenstein JLR, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2(5):255–267. doi: 10.1034/j.1601-183X.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan TJ, Grant SGN. The origin and evolution of synapses. Nat Rev Neurosci. 2009;10(10):701–712. doi: 10.1038/nrn2717. [DOI] [PubMed] [Google Scholar]
- Sahyoun CP, Belliveau JW, Soulieres I, Schwartz S, Mody M. Neuroimaging of the functional and structural networks underlying visuospatial vs. linguistic reasoning in high-functioning autism. Neuropsychologia. 2010;48(1):86–95. doi: 10.1016/j.neuropsychologia.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proceedings of the National Academy of Sciences. 2010 Mar 23;107(12):5611–6. [DOI] [PMC free article] [PubMed]
- Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010 Nov 3;2(56):56ra80. [DOI] [PMC free article] [PubMed]
- Shukla DK, Keehn B, Muller RA. Tract-specific analyses of diffusion tensor imaging show widespread white matter compromise in autism spectrum disorder. J Child Psychol Psychiatry. 2010;52(3):286–295. doi: 10.1111/j.1469-7610.2010.02342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla DK, Keehn B, Smylie DM, Muller RA. Microstructural abnormalities of short-distance white matter fiber tracts in autism spectrum disorder. Neuropsychologia. 2011 Feb 17. [DOI] [PMC free article] [PubMed]
- Sousa I, Clark TG, Toma C, Kobayashi K, Choma M, Holt R, et al. MET and autism susceptibility: family and case-control studies. Eur J Hum Genet. 2009;17(6):749–758. doi: 10.1038/ejhg.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan M, Koch A, Mancini A, Mohr A, Weidner KM, Niemann H, et al. Src homology 2-containing inositol 5-phosphatase 1 binds to the multifunctional docking site of c-Met and potentiates hepatocyte growth factor-induced branching tubulogenesis. J Biol Chem. 2001;276(5):3017–3023. doi: 10.1074/jbc.M009333200. [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30(3):405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- Sundaram SK, Kumar A, Makki MI, Behen ME, Chugani HT, Chugani DC. Diffusion tensor imaging of frontal lobe in autism spectrum disorder. Cereb Cortex. 2008;18(11):2659–2665. doi: 10.1093/cercor/bhn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318(5847):71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara Y, Ido A, Yamamoto S, Miyata Y, Uto H, Hori T, et al. Hepatocyte growth factor facilitates colonic mucosal repair in experimental ulcerative colitis in rats. J Pharmacol Exp Ther. 2003;307(1):146–151. doi: 10.1124/jpet.103.054106. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005 Dec 6;8(12):1727–34. [DOI] [PubMed]
- Thanseem I, Nakamura K, Miyachi T, Toyota T, Yamada S, Tsujii M, et al. Further evidence for the role of MET in autism susceptibility. Neurosci Res. 2010;68(2):137–141. doi: 10.1016/j.neures.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Thomas C, Humphreys K, Jung KJ, Minshew N, Behrmann M. The anatomy of the callosal and visual-association pathways in high-functioning autism: A DTI tractography study. Cortex. 2010 Aug 3. [DOI] [PMC free article] [PubMed]
- Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11(12):834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- Tsao DY, Livingstone M. Mechanisms of face perception. Ann Rev Neurosci. 2008;31:411–437. doi: 10.1146/annurev.neuro.30.051606.094238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndall SJ, Walikonis RS. The receptor tyrosine kinase Met and its ligand hepatocyte growth factor are clustered at excitatory synapses and can enhance clustering of synaptic proteins. Cell Cycle. 2006;5(14):1560–1568. doi: 10.4161/cc.5.14.2918. [DOI] [PubMed] [Google Scholar]
- Tyndall SJ, Walikonis RS. Signaling by hepatocyte growth factor in neurons is induced by pharmacological stimulation of synaptic activity. Synapse. 2007;61(4):199–204. doi: 10.1002/syn.20362. [DOI] [PubMed] [Google Scholar]
- Tyndall SJ, Patel SJ, Walikonis RS. Hepatocyte growth factor-induced enhancement of dendritic branching is blocked by inhibitors of N-methyl-d-aspartate receptors and calcium/calmodulin-dependent kinases. J Neurosci Res. 2007;85(11):2343–2351. doi: 10.1002/jnr.21390. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51(6):741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Riday T, Condon K, Roberts A. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009 Jan 1. [DOI] [PMC free article] [PubMed]