Abstract
Amino-acid selective isotope labeling of proteins offers numerous advantages in mechanistic studies by revealing structural and functional information unattainable from a crystallographic approach. However, efficient labeling of proteins with selected amino acids necessitates auxotrophic hosts, which are often not available. We have constructed a set of auxotrophs in a commonly used Escherichia coli expression strain C43(DE3), a derivative of E. coli BL21(DE3), which can be used for isotopic labeling of individual amino acids or sets of amino acids. These strains have general applicability to either soluble or membrane proteins that can be expressed in E. coli. We present examples in which proteins are selectively labeled with 13C- and 15N-amino acids and studied using magic-angle spinning solid-state NMR and pulsed EPR, demonstrating the utility of these strains for biophysical characterization of membrane proteins, radical-generating enzymes and metalloproteins.
Keywords: isotope, auxotroph, E. coli, protein expression, NMR, EPR
1. Introduction
Amino-acid selective isotope labeling of proteins represents a general approach in which a defined set of amino acid types in proteins is enriched with stable isotopes such as 2H, 13C and 15N. Such selectively labeled samples are extremely useful in structural and mechanistic studies using a number of spectroscopic methods. For example, selective incorporation of isotopically labeled amino acids into large membrane proteins results in great simplification of the NMR spectra from magic-angle spinning (MAS)1 solid-state NMR (SSNMR) techniques, and is essential to apply this very powerful approach to structural studies of large membrane protein complexes[1]. Selective incorporation of isotopically labeled amino acids is also an essential tool in studying proteins using pulsed EPR methods[2], which can provide detailed information about the electronic structure of paramagnetic centers and their interactions with the surrounding protein. Such information, including the number, strength and orientation of hydrogen bonds, is particularly important to understand reactivity and mechanism, and provides critical details beyond the information available from X-ray crystallography. Vibrational spectroscopies, i.e., FTIR, Raman and resonance Raman, also greatly benefit from selective isotope labeling for making assignments (isotope editing)[3]. These techniques can provide detailed information about protein-protein and protein-ligand interactions and dynamics. In the current work, we describe a set of auxotrophic strains of E. coli designed to facilitate the labeling of either membrane proteins or soluble proteins with selected amino acid types enriched with stable isotopes. Efficient and unscrambled labeling of target proteins is demonstrated, and the samples of the isotopically labeled proteins are evaluated using MAS SSNMR and pulsed EPR techniques.
A variety of selective isotope labeling approaches have been developed within the last decade. For example, proteins can be enriched with 13C at specific sites of all amino acid residues with the addition of 13C-labeled carbon sources such as glycerol, glucose or succinic acid in the expression medium[4–6]. However, it is advantageous to selectively incorporate isotope labels at selected amino acid residue types for relatively large proteins[7]. Auxotrophic bacterial strains are often used as expression hosts[8,9] in order to achieve the efficient labeling of the selected residues without dilution of isotopes by the endogenous amino acid metabolic and catabolic processes, which result in scrambling the labels to other residue types. Residue-specific labeling of protein samples can sometimes be performed without the use of auxotrophic hosts by the addition of excess non-labeled amino acids or enzyme inhibitors that block the interconversions between different amino acids. Although the isotopic dilution and scrambling cannot be completely eliminated, labeling specificity can be markedly improved with such an approach[10]. In recent years, in vitro protein synthesis has advanced significantly and, accordingly, there have been numerous NMR studies on samples prepared in vitro[11–13]. Sample preparations by cell-free protein synthesis have several advantages over in vivo biosynthesis but also have limitations. In particular, cell-free expression remains limited to small and medium size proteins with relatively simple assembly processes and are not yet suitable for the preparation of large, multisubunit membrane proteins, for example. For such proteins, in vivo expression in biological hosts such as E. coli remains the only practical option for isotopic labeling.
A major obstacle to selective isotopic enrichment of proteins with rare stable isotopes is the shortage of suitable auxotrophic strains that are compatible with commonly used expression vectors. E. coli BL21(DE3) incorporates an inducible T7 RNA polymerase gene and is one of the most popular hosts for protein production[14]. However, BL21(DE3) is not optimal for high-yield production of membrane proteins (or some soluble proteins) since overexpression often results in toxicity[15]. The C43(DE3) strain, available from Lucigen Inc. (Middleton, WI), is a derivative of the BL21(DE3) strain that is optimized for the successful overproduction of membrane proteins[16,17]. This strain also grows well in defined growth medium without compromising the yield of recombinant protein and is, therefore, well suited for isotopic labeling of both soluble proteins as well as membrane proteins with selected amino acids. In this study, auxotrophic C43(DE3) strains, requiring the addition of selected amino acids in the growth medium, were generated by genomic insertion/deletion mutagenesis. Genes were also targeted which encode enzymes, such as deaminases, which would otherwise result in interconverting amino acids and scrambling the labels[9]. Each auxotrophic strain is designed for the selective labeling of one or more residue types within a defined set of amino acids. These strains can be used for cost effective, high-yield production of any recombinant soluble or membrane protein that can be expressed in E. coli.
Using the newly engineered C43(DE3) auxotrophs, we demonstrate their utility using two proteins: 1) the cytochrome bo3 (cyt bo3) ubiquinol oxidase[18] (CyoABCD) from E. coli, a 144 kDa membrane protein containing 4 transmembrane subunits, two heme prosthetic groups, one ubiquinone and one copper; 2) a small (13 kDa), water-soluble [2Fe-2S] ferredoxin[19] (FdxB) from Pseudomonas putida thought to be involved in the assembly of Fe-S clusters. Three cyt bo3 samples were prepared, in which Ile/Leu, Ile/Tyr and His/Tyr amino acid pairs were, respectively, enriched with 13C and 15N. The MAS SSNMR experiments performed with these samples indicated clean and nearly complete 13C and 15N isotopic enrichment of the selected amino acid types. In addition, selectively 15N labeled cyt bo3 samples were used in pulsed EPR experiments to determine the amino acids interacting with the stabilized ubi-semiquinone species bound at the high affinity quinone binding site (QH) of the enzyme. Successful application of these auxotrophs for the heterologous overproduction of a selectively 15N labeled, foreign metalloprotein, FdxB, is also demonstrated. 2D pulsed EPR was used to resolve for the first time an N-H…S interaction between a glutamine side-chain and a terminal cysteine sulfur ligand within the reduced [2Fe-2S] cluster.
2. Materials and Methods
2.1. Materials
All the amino acid isotopes used in this study were purchased from Cambridge Isotope Laboratories (Andover, MA). n-dodecyl β-D-maltoside (DDM) was bought from Anatrace (Maumee, OH), and isopropyl β-D-thiogalactopyranoside (IPTG) from Anatrace (Maumee, OH) and Wako Pure Chemical Industries (Tokyo, Japan). Oligonucleotide primers were ordered from Integrated DNA Technologies (Coralville, IA) and Operon Biotechnologies (Tokyo, Japan). Other chemicals used in the bacterial growth and sample buffer were from Sigma-Aldrich (St. Louis, MO) unless stated otherwise.
2.2. Construction of E. coli C43(DE3) auxotrophs
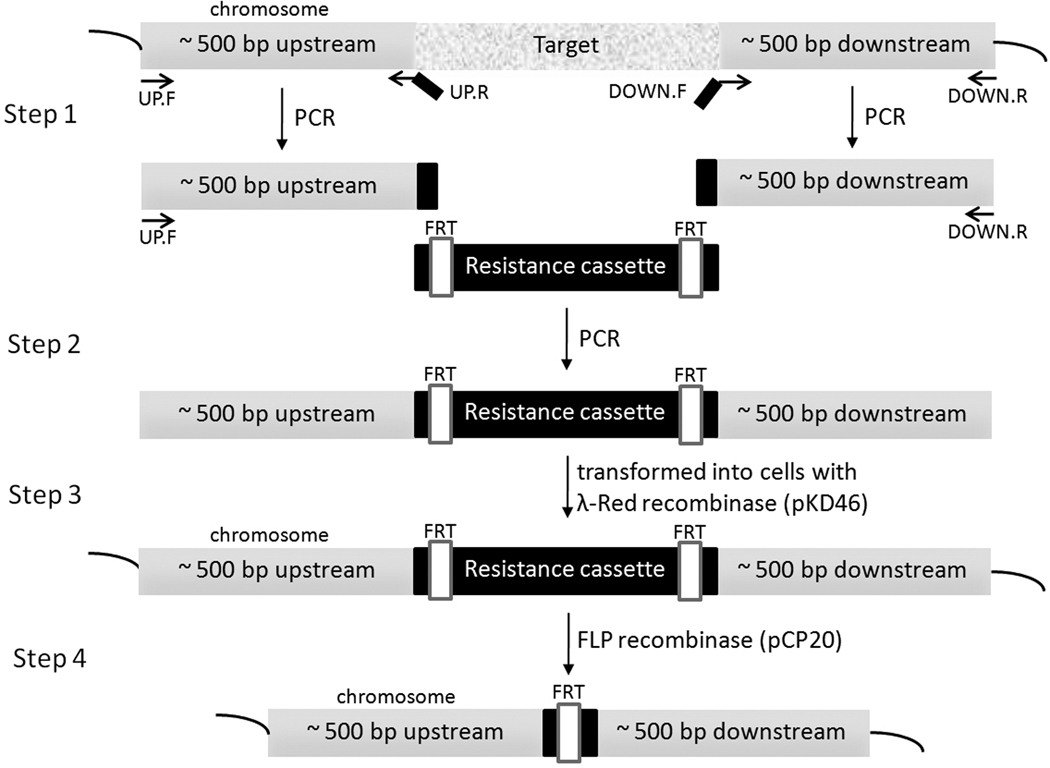
Each target gene was deleted from the chromosome of C43 (DE3) E. coli strain using λ-Red recombination system[20]. The procedure utilizes a linear double stranded DNA generated by PCR to contain an antibiotic resistance marker flanked by about 45 bp sequences homologous to the upstream and downstream regions of the target gene. The DNA was then transformed into E. coli cells expressing λ-Red recombinase from pKD46. The knock-out strain that has undergone cross-over can then be readily selected by the resistance marker. We found that unlike E. coli K-12 strain, the efficiency of the homologous recombination by λ-Red recombinase in the C43(DE3) strain was not good enough to reliably delete the target gene if only 45 bp long homologous regions were used. Thus, we generated the linear double-stranded DNA with approximately 500 bp long upstream and downstream homologous regions flanking a resistance cassette by PCR (Fig. 1)[21]. Using the longer homologous regions dramatically improved the recombination rate. After each knock-out strain was selected by the resistance cassette, the deletion was confirmed by analyzing the DNA segment amplified from the target region of the chromosome, and if possible, also by the requirement of an amino acid for growth in minimal medium. An extremely convenient feature incorporated by Datsenko and Wanner[20] was that the antibiotic cassettes were bordered by FLP recognition tags (FRT), which allowed us to remove the cassettes and reuse the same markers for subsequent deletion of different targets (Fig. 1). Table 1 summarizes the properties of the C43(DE3) auxotrophs generated in this work.
Figure 1.
Schematic showing the deletion of a chromosomal gene with the λ-Red recombination system. Step 1 – Approximately 500 bp long upstream and downstream regions of the target gene were amplified from the E. coli chromosome by PCR. Note that each product has an overlap (black) with the resistance cassette. Step 2 – The linear double-stranded DNA was generated by PCR using the two DNA products from step 1 and the resistance cassette. Step 3 – The linear DNA was transformed into cells expressing λ-Red recombination system from pKD46. The new knock-out strain was selected used its resistance marker. Step 4 – If necessary, the resistance cassette was removed by FLP recombinase expressed from pCP20 vector[20].
Table 1.
C43(DE3) amino-acid auxotrophs constructed with the λ-Red recombination system
| Strain | Genes deleted | Amino acids required for growth |
|---|---|---|
| ML2 | ilvE | Ile, Leua |
| ML3 | hisG | His |
| ML4 | avtA | Val |
| ML5 | metA | Met |
| ML6 | ilvE avtA | Ile, Val, Leua |
| ML8 | argH | Arg |
| ML12 | ilvE avtA aspC | Ile, Val, Leua, Tyrb |
| ML14 | tyrA | Tyr |
| ML17 | glnA | Gln |
| ML21 | hisG tyrA | His, Tyr |
| ML22 | ilvE avtA aspC hisG | Ile, Val, His, Leua, Tyrb |
| ML26 | ilvE avtA aspC hisG argH | Ile, Val, His, Arg, Leua, Tyrb |
| ML30 | ilvE avtA aspC hisG argH glnA | Ile, Val, His, Arg, Gln, Leua, Tyrb |
| ML31 | ilvE avtA aspC hisG argH metA | Ile, Val, His, Arg, Met, Leua, Tyrb |
| ML32 | argH glnA | Arg, Gln |
| ML34 | argH glnA hisG | Arg, Gln, His |
| ML36 | ilvE avtA aspC hisG metA | Ile, Val, His, Met, Leua, Tyrb |
| ML40 | ilvE avtA aspC hisG argH metA lysA | Ile, Val, His, Arg, Met, Lys, Leua, Tyrb |
| ML41 | ilvE avtA aspC hisG argH metA lysA thrC | Ile, Val, His, Arg, Met, Lys, Thr, Leua, Tyrb |
| C43_cysE | cysE | Cys |
In the presence of 0.4–1 mM Tyr, tyrB is repressed and Leu is required for growth in minimal medium.
Addition of 0.4–1 mM Tyr represses tyrB, allowing these strains to be used for isotope labeling of Tyr.
2.3. Expression of amino-acid specific isotopically labeled cyt bo3 samples
Cyt bo3 was expressed from the inducible pET17b vector using an M63 minimal medium as previously reported[22]. Each liter of minimal medium contained 3 g KH2PO4, 7 g K2HPO4, 2 g glucose, 2 g NH4Cl, 10 mg thiamine, 100 mg ampicillin, 50 mg kanamycin, 10 µM CuSO4, 30 µM FeSO4 and 1 mM MgSO4. Supplementary Table 1 and Table 2 list the C43(DE3) auxotrophs used for the protein expression along with the amino acids supplemented in the minimal growth medium. For each sample, a 2 mL LB culture was grown from a glycerol stock for 10 h at 37 °C. 20 µL of the LB culture was then transferred to 10 mL M63 minimal medium with additional amino acids. After about 12 h of growth at 37 °C, the culture was then enlarged to the final volume (either 3 L or 4 L). When the cell density reached an OD600 of ~ 0.6, 0.5 mM IPTG was introduced into the culture, which was then harvested 4–6 h later. For the Ile, Tyr labeled and Tyr, His labeled samples, the culture volume in each 2L-flask was reduced from 1 L to 0.3 L and the duration of induction was increased from 4 h to 6 h. The 6xHis-tagged cyt bo3 was purified from the E. coli membranes solubilized with 1 % DDM using Ni-NTA affinity chromatography as previously reported[22]. The procedure consistently gave high yields of the enzymes (Supplementary Table 1).
2.4. Preparation of cyt bo3 samples for SSNMR
The purified cyt bo3 samples contained about 0.05 – 0.1 % DDM which was removed by dialysis with 12 L 25 mM Tris-HCl pH 8 solution for 3 days. The detergent-depleted samples were then pelleted in an ultracentrifuge at 180,000 × g for 12–16 h and then transferred into 3.2 mm diameter thin wall solid-state NMR rotors with a microspatula.
2.5. SSNMR related methods
CP-MAS 1D 15N and 13C and 2D 13C-13C experiments were performed on a 14.1 Tesla (600 MHz 1H frequency) Varian Infinity Plus spectrometer equipped with Varian T3 1H-13C-15N 3.2-mm probe. The NCC experiments were performed on an 11.7 Tesla (500 MHz 1H frequency) Varian Infinity Plus spectrometer and a 17.6 Tesla (750 MHz 1H frequency) Varian Unity Inova spectrometer equipped with Varian Balun™ 1H-13C-15N 3.2-mm probes. Samples were packed into 3.2-mm limited speed rotors (Varian, now part of Agilent Technologies, Santa Clara, CA). All 1D and CC 2D data sets were acquired at −20.4 ± 0.5 °C actual sample temperature with 100 scfh flow (determined by ethylene glycol calibration)[23]. The NCC experiments were acquired with the variable temperature gas maintained at −50 °C on the 500 MHz spectrometer and −45 °C on the 750 MHz spectrometer with 100 scfh flow with 1H decoupling applied during acquisition and evolution periods on average at ~70 kHz. As we have previously shown, the overall spectral intensity improves at lower temperature, but the relative peak intensities among different spin systems does not change; this effect is due to a combination of enhanced Boltzmann polarization, improved probe performance, and slight increases in the CP enhancement as the temperature is lowered from −20 C to −50 C[22]. More recently, we improved instrumentation for collecting data closer to the phase transition and determined that membrane protein spectral resolution is superior at −20 C [24]. The relative intensity among spin systems does not change over this temperature range. Hard π/2 pulse widths were ~2.5 µs for 1H and 13C. MAS rates were 10~13.3 kHz. Spectra were processed with nmrPipe[25], employing zero filling and Lorentzian-to-Gaussian line broadening for each dimension before Fourier transformation. Back linear prediction and polynomial baseline correction were applied to the frequency domain in the direct dimension. Chemical shifts were referenced externally with adamantane (assuming the downfield peak to resonate at 40.48 ppm)[26]. Additional experimental details are listed in the figure captions. Peak picking and assignments were performed with Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco).
2.6. Generation of the QH semiquinone in cyt bo3 for pulsed EPR
The cyt bo3 samples were first dialyzed against 50 mM potassium phosphate at pH 8.3, 0.05 % DDM, 10 mM EDTA and 5 % glycerol and concentrated to 200–400 µM in a final volume of ~ 150 µl. Each sample was then reduced under argon by a 500-fold excess of sodium ascorbate and incubated at 4 °C for 3 h. The sample was then transferred to an argon-flushed EPR tube and rapidly frozen in liquid nitrogen.
2.7. Preparation of FdxB samples for pulsed EPR
The fdxB gene coding for the ISC-like [2Fe-2S] ferredoxin (FdxB) of P. putida JCM 20004 (formerly P. ovalis IAM 1002) has been cloned and sequenced as a part of its isc gene cluster (DDBJ/EMBL/GenBank code AB109467) and heterologously overexpressed in E. coli BL21-CodonPlus(DE3)-RIL strain (Stratagene) using a pET28aFDXB-SG vector (based on a pET28a His-tag expression vector, Novagen), purified, and crystallized[19]. The structure of the recombinant FdxB has been determined and refined at 1.90-Å resolution (the final R-factor, 0.182; free-R factor, 0.216), and the coordinates and structural factors have been deposited in the Protein Data Bank (PDB ID code: 3AH7)[19].
For preparation of the uniformly 15N-labeled FdxB (U-FdxB) sample, the pET28aFDXB-SG vector was transformed into the host strain, E. coli CodonPlus(DE3)-RIL (Stratagene), and the transformants were grown overnight at 25 °C in the CHL-15N (<97 atm%) medium (Chlorella Industry Co. Ltd., Fukuoka, Japan) containing 50 mg/L kanamycin and 0.2 mM FeCl3, and the recombinant holoprotein was overproduced with 0.5 g/L Algal 15N(98.7–99.2%)-Amino Acid Mix (Chlorella Industry Co. Ltd., Fukuoka, Japan) and 1 mM IPTG for ~18–22 h at 25 °C[27]. This system was suitable for heterologous overproduction of the uniformly 15N-labeled iron-sulfur holoproteins by employing the combination of a pET28a vector (Novagen) plus E. coli CodonPlus(DE3)-RIL host strain (Stratagene) system, because our previous overproduction procedures using the combinations of the pTrc99A vector/E. coli CodonPlus(DE3)-RIL host strain/M9 salt-based synthetic medium system[28,29] were much less successful with the P. putida fdxB gene, giving negligible amount of a recombinant holo-protein (insufficient for high-resolution pulsed EPR analysis of iron-sulfur proteins typically requiring ~0.4 mM or more), for reasons that are not clear. The cells (from 1L culture medium in 2L-flask) were pelleted by centrifugation, and the U-FdxB holoprotein was purified as reported previously for the nonlabeled protein[19]. The purified protein was concentrated with Centriprep-10 and Microcon-YM10 apparatus (Amicon) to ~3 mM, rapidly frozen in liquid nitrogen, and stored at −80 °C until use.
For preparation of the selectively Gln 15Nε-labeled FdxB sample [on the 14N(N/A)-protein background], the E. coli C43(DE3) auxotroph strain ML17 (Table 1) having the deletion of glnA gene was used as parent cells for heterologous overexpression of the P. putida fdxB gene. The ML17 strain lacking the glnA gene (see [9]) is useful for selecive labeling of Gln 15Nε, but minor diffusion and dilution of the input (not severe as compared to Glu and Asp, which are located at the center of the E. coli metabolic pathways) can occur when used in the absence of appropriate non-labeled amino acids because Gln Nα preferentially diffuses into Glu.
Because the pET28aFDXB-SG vector carries the kanamycin-resistance marker gene[19], the resistance cassette in the host strain ML17 was removed by FLP recombinase expressed from pCP20 vector[20] (Step 4 in Fig. 1), and the deletion of the cognate glnA gene was subsequently verified by PCR. The pET28aFDXB-SG vector was transformed into the resulting host strain ML17, and the transformants were grown overnight at 25 °C in 1L culture (in 2L-flask) of the nonlabeled CHL medium (Chlorella Industry Co. Ltd., Fukuoka, Japan) containing 50 mg/L kanamycin, 0.2 mM FeCl3, 0.5 g/L 14N(N/A) L-glutamine, and 0.5 g/L nonlabeled Algal Amino Acid Mix (Chlorella Industry Co. Ltd., Fukuoka, Japan [note that the nonlabeled Algal Amino Acid Mix is the commercial amino acid mixture derived from the acid extract of Chlorella and therefore does not contain Gln and Asn]). The cells were harvested by centrifugation, and the resulting cell pellet was subsequently inoculated into a total of 2 L culture (using two 2L-flasks, each containing 1L culture medium) of the freshly prepared, nonlabeled CHL medium (Chlorella Industry Co. Ltd., Fukuoka, Japan) containing 50 mg/L kanamycin, 0.2 mM FeCl3, 0.5 g/L nonlabeled Algal Amino Acid Mix (Chlorella Industry Co. Ltd., Fukuoka, Japan), and ~0.45 g/L L-glutamine labeled at the 15Nε2 position (98%+) (Cambridge Isotope Laboratories, Inc., Andover, MA) for 30 min at 37 °C. Then the recombinant FdxB holoprotein was overproduced in this culture with 1 mM IPTG for 6 h at 30 °C. The cells were pelleted by centrifugation, and the Gln 15Nε-labeled FdxB holoprotein was purified as described above for the U-FdxB holoprotein.
For high-resolution pulsed EPR analysis, the FdxB samples (~3 mM, ~40–50 µL) were reduced with sodium dithionite under continuous flow of dry argon gas inside suprasil quartz EPR tubes (Wilmad) prior sealing, rapidly frozen in liquid nitrogen, and shipped in the frozen state in dry ice by international priority delivery service from Tokyo, Japan, to Urbana, U.S.A.
2.8. Pulsed EPR related methods
The pulsed EPR experiments were carried out at 70 K (cyt bo3) and 20 K (FdxB) using an X-band Bruker ELEXSYS E580 spectrometer equipped with Oxford CF 935 cryostats. The 2D, four-pulse experiment (π/2-τ-π/2-t1-π-t2-π/2-τ-echo, also called HYSCORE)[30], was employed with appropriate phase-cycling schemes to eliminate unwanted features from the experimental echo envelopes. The intensity of the echo after the fourth pulse was measured with t2 and t1 varied and constantτ. The length of a π/2 pulse was nominally 16 ns and a π pulse 32 ns. HYSCORE data were collected in the form of 2D time-domain patterns containing 256×256 points with steps of 20 or 32 ns. Spectral processing was performed using Bruker WIN-EPR software.
3. Results
3.1. Construction and use of E. coli C43(DE3) auxotrophs
Of the amino acids chosen for isotope labeling in this study, Ile, Leu, and Tyr have biosynthetic pathways involving transaminases, which are known to be responsible for the scrambling of isotope labels intended for single residue types (Supplementary Fig. 1). These transaminases are encoded by the ilvE, avtA, aspC and tyrB genes[9]. Deletion of the ilvE, avtA and aspC genes from the chromosome resulted in an auxotroph for Ile and Val. To extend the auxotrophy to include Leu, the tyrB locus also needed to be inactivated. Multiple attempts to replace the tyrB region with an antibiotic resistance cassette were not successful, for reasons that are not clear. However, the expression of tyrB has been shown to be repressed by the presence of Tyr or Phe in the growth medium[31]. Hence, in the presence of Tyr, some auxotroph strains in Table 1 behave like Leu auxotrophs (Supplementary Fig. 2), allowing us to label selected pairs of amino acids for SSNMR experiments.
3.2. Magic-angle spinning solid-state NMR analysis of pair-wise labeled cyt bo3 samples
The three cyt bo3 samples prepared in this study for MAS SSNMR experiments have Ile-Leu, Ile-Tyr and His-Tyr amino acids labeled with 13C and 15N, and will be denoted as IL-bo3, IY-bo3 and HY-bo3 respectively (Supplementary Table 1). The lack of transaminase activities in the auxotroph strains increased the doubling time of the E. coli culture in the minimal medium by a factor of 1.5. However, this had only minor effects on the expression level of cyt bo3, yielding 5–9 mg per liter of media. The spectra collected from these samples were compared to those of the uniformly 13C, 15N labeled sample (U-bo3) described in our previous work[22]. We used 13C one-dimensional (1D) spectra to assess the overall sensitivity, resolution, and sample integrity (Supplementary Fig. 3). These 13C 1D spectra were acquired at a sample temperature of −20.4°C, just below the lipid phase transition, for optimal cross polarization (CP) transfer and resolution. To quantify the amount of the isotopically labeled material in the MAS SSNMR rotor, we performed 13C direct polarization (DP) experiments for each sample. Integrated intensities from the DP spectra were compared to signal intensity obtained from a DP experiment performed on a known amount of 13C-labeled adamantane (within an error of ~10%). Through explicit calculation of DP signal intensities per 13C label expected for each sample, relative to the known standard, it was concluded that the U-bo3, IL-bo3, IY-bo3, and HY-bo3 samples contain 11.0, 9.7, 9.5, and 11.1 mg of protein in 3.2-mm thin wall MAS rotor, respectively. The 13C CP spectra (Supplementary Fig. 3b–d) demonstrate typical chemical shift patterns for Ile, Leu, Tyr, and His residues (according to statistics available at the BioMagResBank, http://www.bmrb.wisc.edu). The resolution compared to the uniformly labeled sample (Supplementary Fig. 3a) is greatly enhanced.
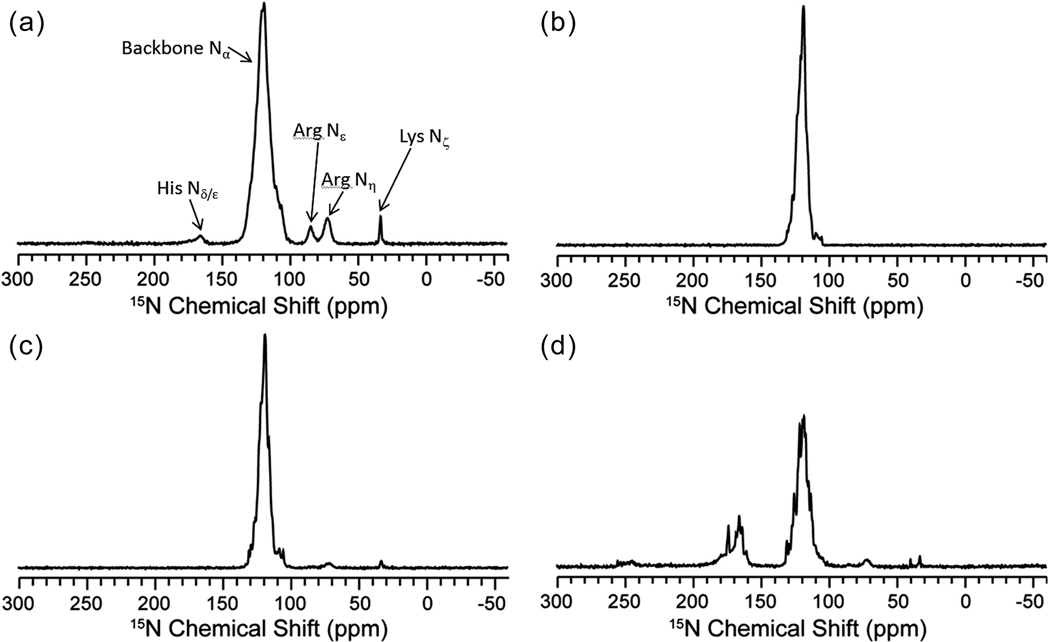
Following 13C 1D experiments, we quantified the 15N labeling. Chemical shifts of 15N have a large dispersion due to sensitivity to local interactions, including such factors as protonation, hydrogen-bonding and dynamics, especially for the His side-chains. The 15N 1D spectra of the pair-wise labeled samples (Fig. 2b–d) show little or no 15N scrambling to other unwanted amino acid types. The 15N 1D of the U-bo3 sample (Fig. 2a) contains amide nitrogen resonances between 100 and 135 ppm, which is consistent with the range of labeled amino acids in the sample. Additionally, the U-bo3 spectrum demonstrates intensities for 15N side-chain resonances, such as His 15Nδ/ε (~165 ppm), Arg 15Nε (~85 ppm), Arg 15Nε (~70 ppm), and Lys 15Nδ (~35 ppm). In contrast, the IL-bo3 spectrum (Fig. 2b) has no observable intensity at these side-chain resonances. Similar ratios of the His 15Nδ/ε signal intensities to the normalized amide 15N signal intensities for the U-bo3 and HY-bo3 samples are consistent with virtually complete 15N labeling at the selected residues in the HY-bo3 sample. The IY-bo3 (Fig. 2c) and HY-bo3 (Fig. 2d) spectra displayed small intensities at ~70 ppm and ~35 ppm corresponding to the Arg and Lys side-chain resonances. Evaluation of these side-chain signal intensities in comparison to the amide region indicated that the unintended incorporation of the 15N isotope represents less than 3% of the overall populations. This is consistent with the lack of scrambling we observed in the 13C-15N 2D experiments described below. Note that the intensity of all spectra are scaled relative to signal to noise. Thus, although the peaks ranging from 160–180 ppm appear to be significantly less intense in the U-bo3 (Fig. 2a) compared to Fig. 2d, this is just because of the relative signal strength of the amide region. Inherently, there was less signal in the HY-bo3 sample since there are significantly less isotopically labeled amide nitrogens.
Figure 2.
15N 1D of (a) U-bo3 (2048 scans), (b) IL-bo3 (40960 scans), (c) IY-bo3 (49152 scans), and (d) HY-bo3 (106496 scans) acquired on a 600 MHz spectrometer (1H frequency).
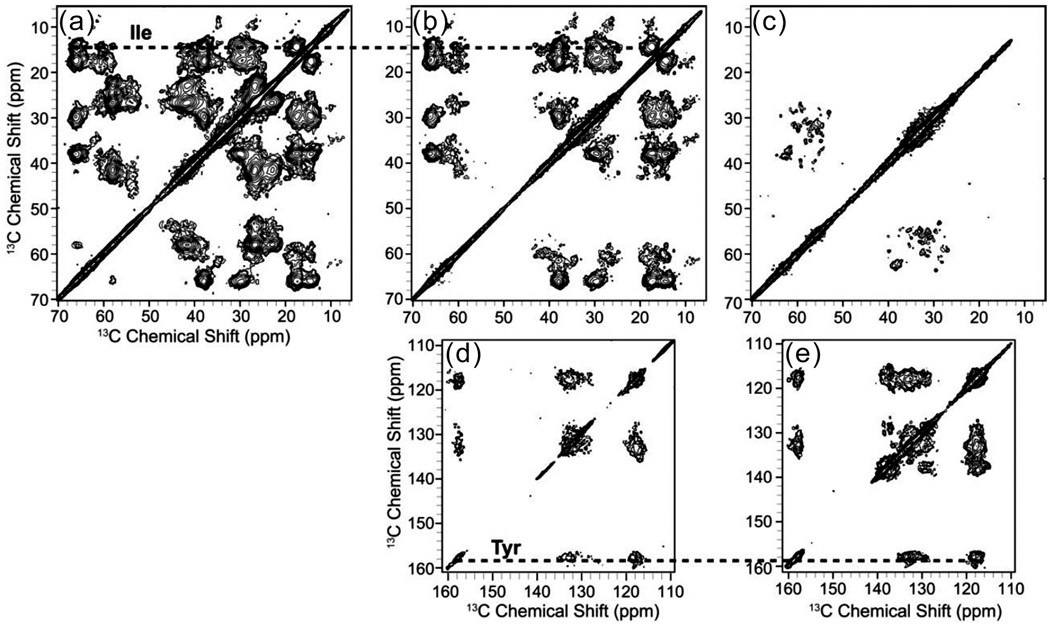
To further evaluate the labeling pattern and the improvements to spectral resolution of the pair-wise labeled cyt bo3 samples, 13C-13C 2D correlation experiments were acquired with 50 ms of DARR (dipolar-assisted rotational resonance) mixing[32] to observe the intraresidue (methyl, aliphatic, carbonyl, and aromatic chemical shift) correlations for each spin system. The aliphatic regions of IL-bo3 and IY-bo3 have resonances that show clean Ile spin system labeling (13Cα~66 ppm, 13Cβ~39 ppm, 13Cγ1~28 ppm, 13Cγ2~18 ppm, 13Cδ1~14 ppm) (Fig. 3a–b). Similarly, clean Tyr spin systems (13Cγ~130 ppm, 13Cδ~133 ppm 13Cε~118 ppm, 13Cζ~158 ppm) can be identified in the aromatic regions of both the IY-bo3 and HY-bo3 spectra (Fig. 3d–e). Notably Cα-Cα correlations between adjacent Ile and Leu pairs are also visible in the IL-bo3 spectrum (Fig. 3a). The significant chemical shift dispersions of Ile and His signals indicate that those residues have subtle differences in the side chain conformations which can be observed under the improved resolution from the selectively labeled samples. The individual His Cα and Cβ linewidths are approximately 0.4 ppm, indicating a homogeneous sample preparation, similar to the U-bo3 sample (Fig. 3d).
Figure 3.
Expansions of the aliphatic region of (a) IL-bo3, (b) IY-bo3 and (c) HY-bo3, and aromatic region of (d) IY-bo3 and (e) HY-bo3. 13C-13C 2D spectra were collected on a 600 MHz spectrometer (1H frequency) with 50 ms DARR mixing. Data were acquired for 160, 96 and 120 hrs for the IL-, IY-, and HY-bo3 samples, respectively, and processed with 40 Hz (F1) and 40 Hz (F2) net Gaussian-to-Lorentzian line broadening. Dashed lines highlight the spin systems of residues in common between the samples (Ile and Tyr).
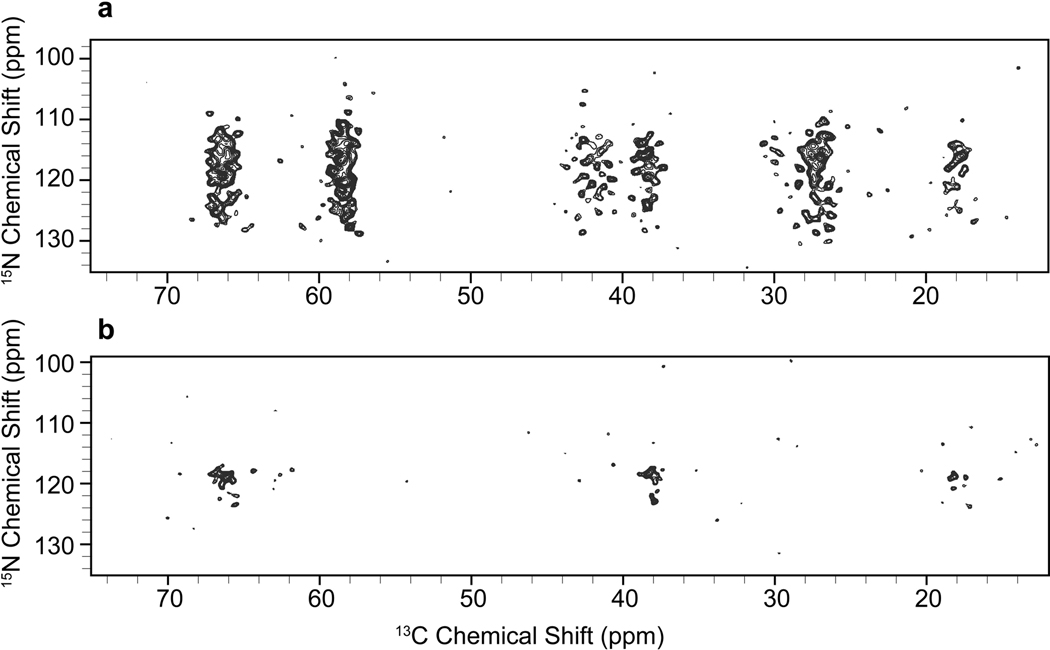
Heteronuclear backbone correlation experiments for chemical shifts assignments also benefit from selective isotope labeling. Intraresidue N(CA)CX 2D experiments were performed on the IL-bo3 and IY-bo3 samples (Supplementary Fig. 4). These experiments again confirmed the clean labeling pattern and demonstrated the good amide 15N dispersion within each amino acid type. Interresidue N(CO)CX correlation experiments were also performed on the IL-bo3 and IY-bo3 samples (Fig. 4) in order to assign pairs of labeled amino acids (i.e.- IL, II, LL, IY or YY pairs). There are 19 interresidue correlations expected (11 I-I, 1 Y-Y, 4 I-Y, and 3 Y-I) in the IY-bo3 spectrum, and 19 correlations are observed. The high level of 15N dispersion for these amino acids means that nearly every pair can be resolved in these two dimensions, whereas 3D or 4D experiments would be required for a U-bo3 sample. In the IL-bo3 spectrum, there are 48 expected pairs (11 I-I, 7 L-L, 17 I-L, and 13 L-I) but 73 labeled pairs are observed. We attribute the additional signals to intraresidue polarization transfer peaks that are observed as a consequence of the higher sensitivity of data collection in this case.
Figure 4.
15N-(13CO)-13CX 2D spectra of (a) IL-bo3 collected on a 500 MHz spectrometer (1H frequency) with 30 ms DARR mixing for 35 hrs, processed with 20 Hz (F1, 15N) and 15 Hz (F2, 13C) net Gaussian-to-Lorentzian line broadening and (b) IY-bo3 collected on a 750 MHz spectrometer (1H frequency) with 50 ms DARR mixing for 75 hrs, processed with 35 Hz (F1, 15N) and 20 Hz (F2, 13C) net Gaussian-to-Lorentzian line broadening.
3.3. Identification of the nitrogen donors hydrogen bonded with the semiquinone in the QH-site of cyt bo3 by pulsed EPR studies on selectively 15N labeled samples
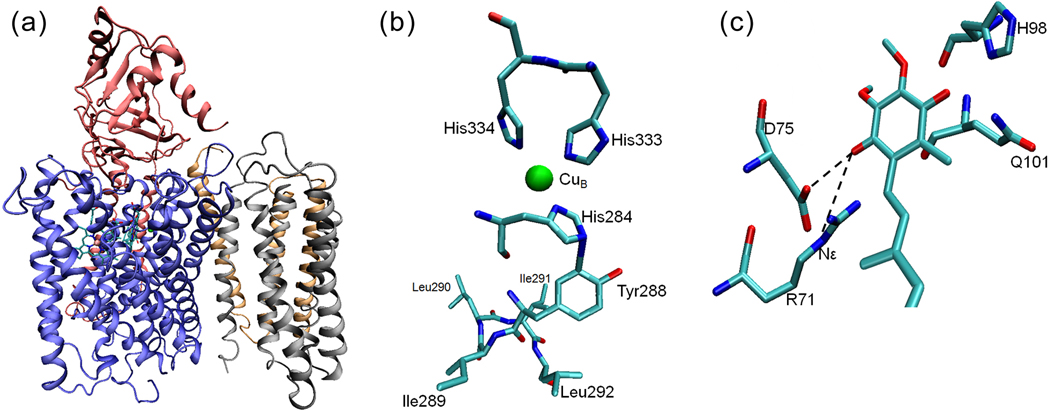
During the turnover of cyt bo3, a semiquinone (SQ) intermediate is generated at a high affinity quinone binding site (QH). The X-ray structure of cyt bo3 does not contain any bound quinone[18], but site-directed mutagenesis studies suggest roles for residues R71, D75, H98, and Q101at the QH site[18,33]. Selective 15N isotope labeling with auxotrophs was used to identify the nitrogen involved in the formation of the strongest H-bond with the QH SQ in the cyt bo3. The auxotroph strains were used to label the protein with 15N as follows (ref. [2] and this work): 1) Arginine: a) uniform labeling; b) the two Nε positions; c) the peptide position; 2) Histidine: a) uniform labeling; b) the two ring positions (Nδ and Nε); c) the Nδ position only; 3) Glutamine with 15N in the Nε position; 4) uniformly labeled with 15N except for Arg, Gln and His (Supplementary Table 2). In our previously published study [2], 2D ESEEM spectra of the SQ in these proteins clearly identified the Nε of R71 as the H-bond donor carrying the most transferred unpaired spin density, giving rise to the model shown in Fig. 5c.
Figure 5.
The X-ray crystal structure of cytochrome bo3 from E. coli[18]. (a) A cartoon representation of the four subunits of cyt bo3 with heme b, heme o3 and CuB. (b) The residues near the CuB site. His333, His334 and His284 are ligands to CuB. The proposed cross-link between His284 and Tyr288 is immediately followed by a unique quartet of ILIL. All residues shown are from subunit I. (c) A cartoon model of the high-affinity ubiquinone binding site (QH) of the wide-type cyt bo3 showing the strong H-bonds between the ubisemiquinone and the Nε of R71 as well as the side-chain carboxylic group of D75.
The pulsed EPR approach was extended to the D75H mutant, which is of interest because it is spectroscopically identical to the wild type enzyme, but lacks catalytic function[33,34]. Using the D75H mutant samples with selective Histidine isotopic labels, it was determined that the Nε from a histidine residue, presumably H75, carries most of the unpaired spin density instead of the Nε of R71 in the wild type cyt bo3 (Supplementary Fig. 5).
3.4. Pulsed EPR characterization of a weakly coupled N-H…S hydrogen bond interaction between a Gln side chain and the [2Fe-2S] center of the recombinant ISC-like ferredoxin from P. putida
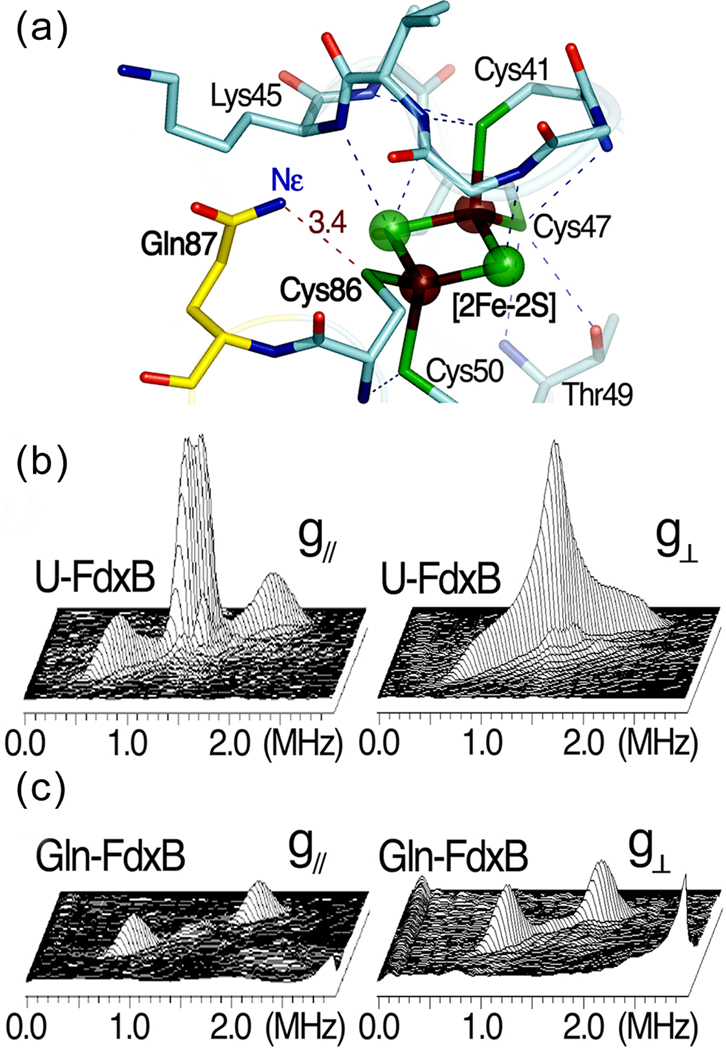
The newly developed E. coli auxotrophs described above can also be used for the heterologous overproduction of specific isotope labeled (foreign) proteins for biophysical studies. In this example, a selectively 15N labeled, adrenodoxin-like [2Fe-2S] ferredoxin (FdxB) from P. putida[19], was used for pulsed EPR analysis. The 1.90-Å structure of FdxB (PDB code 3AH7) shows an extensive (N/O)-H…S hydrogen bond network around the [2Fe-2S] cluster, with nearly the same pattern as found in other closely related homologs such as adrenodoxin and putidaredoxin, including a conserved N-H…S hydrogen bond between the Nε of Gln87 and the Cys86 Sγ ligand at the Nε-Sγ distance of 3.4 Å in FdxB[19] (Fig. 6a). The extensive (N/O)-H…S hydrogen bond network around the biological Fe-S clusters is one of the most important themes in modulating their redox properties and potential electron transfer pathways in biological electron transfer conduits[35]. Paramagnetic relaxation due to the reduced (antiferromagnetically coupled Fe3+-Fe2+) cluster severely broadens signals from nuclei in the vicinity (~8 Å) of the paramagnetic centers, making it difficult to fully assign hyperfine interactions (hfi) with the paramagnetic metal center by multidimensional heteronuclear NMR[36]. Pulsed EPR (2D ESEEM) can also be used to characterize the nuclear spin interactions with the paramagnetic center. Uniformly 15N-labeled FdxB (denoted as U-FdxB) was prepared, but the 2D ESEEM spectra did not resolve individual nuclei, showing only two broad (aggregate) 15N cross-features with apparent hfi splittings of ~1.2 and ~0.2 MHz, respectively (Fig. 6b). Therefore, FdxB was prepared with glutamine specifically 15N-labeled at the Nε position (denoted as Gln-FdxB). The spectra collected from Gln-FdxB clearly resolve, for the first time, the sharp cross-peaks assigned to a single N-H…S hydrogen bond interaction between the 15Nε of Gln87 and the terminal Cys86 Sγ ligand of the reduced cluster in Gln-FdxB, with the isotropic hfi splitting of ~0.9 MHz (Fig. 6c).
Figure 6.
The X-ray crystal structure and 2D pulsed EPR studies on the [2Fe-2S](Cys)4 cluster binding site of P. putida FdxB. (a) The hydrogen bond network around the [2Fe-2S](Cys)4 cluster binding site in the 1.90-Å structure of P. putida FdxB (PDB code: 3AH7) [19], (b) the 2D ESEEM characterization of weakly coupled 15N nuclei in uniformly 15N-labeled FdxB (U-FdxB) and (c) selectively Gln15Nε-labeled FdxB (Gln-FdxB) in the reduced state. The 2D ESEEM spectra in 3D stacked presentation (b,c) were measured at the low-field edge of the EPR line near g// (342.5 mT, left) and the intermediate position at g⊥ (357 mT, right). The cross-peaks for 15Nε of Gln87, which is involved in the N-H…S hydrogen bond with Cys86 Sγ (a), were clearly resolved with the hfi splitting of ~0.9 MHz ((2.01, 0.92) MHz and 15A=1.09 MHz near g// (left); (1.95, 1.13) MHz and 15A=0.82 MHz at g⊥ (right)) in the Gln-FdxB spectra (c).
4. Discussion
Selective labeling of amino acids with stable isotopes, applied in conjunction with a number of spectroscopic techniques such as NMR, EPR and infrared spectroscopies, is undoubtedly a powerful method in deciphering the structures and mechanisms of proteins. Here we have described a set of strains designed to facilitate the selective labeling of proteins that can be expressed in E. coli. The use of these strains is illustrated by examples using MAS SSNMR and pulsed EPR studies of selectively labeled samples. The approach presented here is not limited to amino acid type-specific isotope labeling. Cofactors such as hemes and quinones can also be selectively labeled in the same manner using suitable strains as reported recently[37].
4.1. Solid-state NMR studies of cyt bo3 containing pair-wise selectively labeled amino acids
In order to assign resonances of residues in such a large complex, dramatic simplification of the spectra by the use of amino acid selective labeling is required. For initial experiments, Ile-Leu, Ile-Tyr and His-Tyr amino acid pairs were isotopically enriched in cyt bo3 because the active-site Tyr residue is immediately followed by an ILIL sequence, giving rise to a YIL triplet and an ILIL quartet, which are unique to this complex (Fig. 5b).
One- and two-dimensional SSNMR spectra of each pair-wise labeled sample demonstrated vast improvement in resolution over the 13C, 15N uniformly labeled sample. The labeling precision and the absence of isotope scrambling are critical in analyzing heteronuclear multidimensional spectra in order to accurately assign correlations for the labeled residues. The 2D spectra display overall enhancement of the resolution and superior chemical shift dispersion, despite the highly helical nature of this protein. This approach also implies a substantial improvement in sensitivity, because fewer polarization transfer steps and indirect dimensions are required, relative to 3D or 4D spectra, and thus the 2D spectra have higher sensitivity. These data justify further efforts aimed at chemical shift assignments, as well as functional studies via SSNMR. The amino acid type-specific labeling approach described here applied in conjunction with the recent development of an algorithm for optimal unique pair labeling can facilitate biomolecular NMR assignments[38].
4.2. Pulsed EPR studies of the 15N labeled nitrogen donors hydrogen bonded to the semiquinone form of the ubiquinone bound at the QH-site of cyt bo3
In this example, the low resolution structural model from X-ray crystallography and site-directed mutagenesis provided information of which amino acids might be stabilizing the ubisemiquinone at the QH site in the protein[18,33]. The enzyme stabilizes a semiquinone form of ubiquinone at this site which is an intermediate during catalytic turnover, so a detailed description of the hydrogen bonding partners and electronic structure of the ubisemiquinone is important to understand the function of the enzyme. Individual amino acid types were isotopically labeled and incorporated into cyt bo3 and these samples have allowed us to definitively conclude that the Nε of an Arg71 is a hydrogen bond donor to the semiquinone in the wild type enzyme [2]. A mutant (D75H), which is catalytically inactive, lacks this strong hydrogen bond to the semiquinone. These experiments relied on the use of the auxotrophic strains described in this work, and the labeling pattern that was both precise and clean.
4.3. Application of E. coli C43(DE3) auxotrophs as the host cells for heterologous overproduction of selectively 15N labeled foreign metalloproteins for biophysical studies
It is demonstrated that the new auxotrophs can be used for high-level, heterologous expression of a selectively 15N-labeled metalloprotein, FdxB from P. putida[19]. The single N-H…S hydrogen bond interaction between the 15Nε of Gln and the terminal sulfur ligand to the reduced [2Fe-2S] cluster was successfully resolved and characterized for the first time by 2D pulsed EPR. This is usually impossible even with the use of uniformly 15N-labeled proteins, presumably because of the significant spectral overlap of many weakly coupled 15N nuclei from the peptide backbone near the cluster[27]. The incorporation of tRNA genes for the E. coli rare codons into the C43(DE3) auxotrophs will further expand their application for heterologous, high-level expression of genes from sources other than E. coli.
Supplementary Material
Acknowledgments
We thank Dr. Jihyun F. Kim from the Systems Microbiology Research Center, KRIBB, Daejeon 305-806, South Korea) for providing the genome sequence of E. coli BL21(DE3). We thank Dr. John E. Cronan and Dr. Nicholas R. De Lay from the Department of Microbiology at the University of Illinois at Urbana-Champaign for providing the pKD46, pKD3, pKD4 and pCP20 plasmids. We thank Dr. Deborah Berthold from the Department of Chemistry at the University of Illinois at Urbana-Champaign for advice on the manuscript. This work was supported by the DE-FG02-87ER13716 (RBG) and DE-FG02-08ER15960 (SAD) Grants from US Department of Energy, National Institute of Health & NIGMS Roadmap Initiative (R01GM075937), the JSPS Grant-in-aid 21659111 (TI), the International Collaborations in Chemistry Grant from JSPS (TI) and NSF (CHE-1026541 to SAD), NIH grant GM062954 (SAD), and the NIH Molecular Biophysics Training Grant (PHS 5 T32 GM008276) and the Ullyot Fellowship to LJS.
Footnotes
Abbreviations: magic-angle spinning (MAS) solid-state NMR (SSNMR); Electron spin resonance, EPR; FTIR, Fourier transform infrared; DDM, n-dodecyl β-D-maltoside; IPTG, isopropyl β-D-thiogalactopyranoside; basepairs, bp; optical density, OD; ethylene diamine tetra-acetic acid, EDTA; iron-sulfur cluster biosynthesis, ISC; Hyperfine Sub-level Correlation, HYSCORE; cross-polarization, CP; direct polarization, DP; semiquinone, SQ; hfi, hyperfine
References
- 1.McDermott A. Annu. Rev. Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 2.Lin MT, Samoilova RI, Gennis RB, Dikanov SA. J. Am. Chem. Soc. 2008;130:15768–15769. doi: 10.1021/ja805906a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haris PI. Biochem. Soc. Trans. 2010;38:940–946. doi: 10.1042/BST0380940. [DOI] [PubMed] [Google Scholar]
- 4.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 5.Hong M, Jakes K. J. Biomol. NMR. 1999;14:71–74. doi: 10.1023/a:1008334930603. [DOI] [PubMed] [Google Scholar]
- 6.Gammeren AJ, Hulsbergen FB, Hollander JG, Groot HJ. J. Biomol. NMR. 2005;31:279–293. doi: 10.1007/s10858-005-1604-8. [DOI] [PubMed] [Google Scholar]
- 7.Hiller M, Higman VA, Jehle S, van Rossum B-J, Kühlbrandt W, Oschkinat H. J. Am. Chem. Soc. 2008;130:408–409. doi: 10.1021/ja077589n. [DOI] [PubMed] [Google Scholar]
- 8.Whittaker JW. Methods Mol. Biol. 2007;389:175–188. doi: 10.1007/978-1-59745-456-8_13. [DOI] [PubMed] [Google Scholar]
- 9.Waugh DS. J. Biomol. NMR. 1996;8:184–192. doi: 10.1007/BF00211164. [DOI] [PubMed] [Google Scholar]
- 10.Tong KI, Yamamoto M, Tanaka T. J. Biomol. NMR. 2008;42:59–67. doi: 10.1007/s10858-008-9264-0. [DOI] [PubMed] [Google Scholar]
- 11.Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Mei Ono A, Guntert P. Nature. 2006;440:52–57. doi: 10.1038/nature04525. [DOI] [PubMed] [Google Scholar]
- 12.Staunton D, Schlinkert R, Zanetti G, Colebrook SA, Campbell ID. Magn. Reson. Chem. 2006;44:S2–S9. doi: 10.1002/mrc.1835. [DOI] [PubMed] [Google Scholar]
- 13.Sobhanifar S, Reckel S, Junge F, Schwarz D, Kai L, Karbyshev M, Löhr F, Bernhard F, Dötsch V. J. Biomol. NMR. 2010;46:33–43. doi: 10.1007/s10858-009-9364-5. [DOI] [PubMed] [Google Scholar]
- 14.Studier FW, Moffatt BA. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 15.Dumon-Seignovert L, Cariot G, Vuillard L. Protein Expr. Purif. 2004;37:203–206. doi: 10.1016/j.pep.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 16.Miroux B, Walker JE. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 17.Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Högbom M, van Wijk KJ, Slotboom DJ, Persson JO, de Gier JW. Proc. Natl. Acad. Sci. USA. 2008;105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek M, Laakkonen L, Puustinen A, Iwata S, Wikström M. Nature Struct. Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- 19.Iwasaki T, Kappl R, Bracic G, Shimizu N, Ohmori D, Kumasaka T. J. Biol. Inorg. Chem. 2011;16:923–935. doi: 10.1007/s00775-011-0793-8. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko KA, Wanner BL. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derbise A, Lesic B, Dacheux D, Ghigo JM, Carniel E. FEMS Immunol. Med. Microbiol. 2003;38:113–116. doi: 10.1016/S0928-8244(03)00181-0. [DOI] [PubMed] [Google Scholar]
- 22.Frericks HL, Zhou DH, Yap LL, Gennis RB, Rienstra CM. J. Biomol. NMR. 2006;36:55–71. doi: 10.1007/s10858-006-9070-5. [DOI] [PubMed] [Google Scholar]
- 23.Van Geet AL. Anal. Chem. 1968;40:2227–2229. [Google Scholar]
- 24.Tang M, Sperling LJ, Berthold DA, Nesbitt AE, Gennis RB, Rienstra CM. J. Am. Chem. Soc. 2011;133:4359–4366. doi: 10.1021/ja107775w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 26.Morcombe CR, Zilm KW. J. Magn. Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 27.Iwasaki T, Samoilova RI, Kounosu A, Ohmori D, Dikanov SA. J. Am. Chem. Soc. 2009;131:13659–13667. doi: 10.1021/ja903228w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kounosu A, Li Z, Cosper NJ, Shokes JE, Scott RA, Imai T, Urushiyama A, Iwasaki T. J. Biol. Chem. 2004;279:12519–12528. doi: 10.1074/jbc.M305923200. [DOI] [PubMed] [Google Scholar]
- 29.Iwasaki T, Kounosu A, Uzawa T, Samoilova RI, Dikanov SA. J. Am. Chem. Soc. 2004;126:13902–13903. doi: 10.1021/ja045898x. [DOI] [PubMed] [Google Scholar]
- 30.Höfer P, Grupp A, Nebenführ H, Mehring M. Chem. Phys. Lett. 1986;132:279–282. [Google Scholar]
- 31.Yang J, Camakaris H, Pittard J. Mol. Microbiol. 2002;45:1407–1419. doi: 10.1046/j.1365-2958.2002.03108.x. [DOI] [PubMed] [Google Scholar]
- 32.Takegoshi K, Nakamura S, Terao T. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 33.Hellwig P, Yano T, Ohnishi T, Gennis RB. Biochemistry. 2002;41:10675–10679. doi: 10.1021/bi012146w. [DOI] [PubMed] [Google Scholar]
- 34.Yap LL, Samoilova RI, Gennis RB, Dikanov SA. J. Biol. Chem. 2007;282:8777–8785. doi: 10.1074/jbc.M611595200. [DOI] [PubMed] [Google Scholar]
- 35.Beinert H, Holm RH, Munck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 36.Machonkin TE, Westler WM, Markley JL. Inorg. Chem. 2005;44:779–797. doi: 10.1021/ic048624j. [DOI] [PubMed] [Google Scholar]
- 37.Lin MT, Shubin AA, Samoilova RI, Narasimhulu KV, Baldansuren A, Gennis RB, Dikanov SA. J. Biol. Chem. 2011;286:10105–10114. doi: 10.1074/jbc.M110.206821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hefke F, Bagaria A, Reckel S, Ullrich SJ, Dotsch V, Glaubitz C, Guntert P. J. Biomol. NMR. 2011;49:75–84. doi: 10.1007/s10858-010-9462-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.