Abstract
The nuclear lamina is a protein meshwork that lines the nuclear envelope in metazoan cells. It is composed largely of a polymeric assembly of lamins, which comprise a distinct sequence homology class of the intermediate filament protein family. On the basis of its structural properties, the lamina originally was proposed to provide scaffolding for the nuclear envelope and to promote anchoring of chromatin and nuclear pore complexes at the nuclear surface. This viewpoint has expanded greatly during the past 25 years, with a host of surprising new insights on lamina structure, molecular composition and functional attributes. It has been established that the self-assembly properties of lamins are very similar to those of cytoplasmic intermediate filament proteins, and that the lamin polymer is physically associated with components of the cytoplasmic cytoskeleton and with a multitude of chromatin and inner nuclear membrane proteins. Cumulative evidence points to an important role for the lamina in regulating signaling and gene activity, and in mechanically coupling the cytoplasmic cytoskeleton to the nucleus. The significance of the lamina has been vaulted to the forefront by the discovery that mutations in lamins and lamina-associated polypeptides lead to an array of human diseases. A key future challenge is to understand how the lamina integrates pathways for mechanics and signaling at the molecular level. Understanding the structure of the lamina from the atomic to supramolecular levels will be essential for achieving this goal.
Keywords: nucleus, nuclear lamina, intermediate filament, nuclear envelope, human disease, cytoskeleton
Introduction
A nuclear lamina-like structure was made first described in the protozoan Amoeba proteus by thin section EM, where it appeared as an ~300 nm thick “honeycomb” layer apposed to the inner surface of the nuclear envelope (NE) (Harris and James, 1952). An analogous “fibrous lamina” subsequently was reported in some vertebrate and other higher eukaryotic cells, where it was seen as a zone of intermediate electron density up to 40–60 nm thick sandwiched between the inner nuclear membrane (INM) and the shell of peripheral nuclear heterochromatin (Fawcett, 1966). However, since a discrete lamina subjacent to the NE was not evident in the large majority of vertebrate cells by thin section EM, it seemed possible that a fibrous lamina might be an uncommon specialization of a few cell types. This question was addressed by landmark studies from the Blobel laboratory involving rat liver nuclei, which show no conspicuous lamina in thin section EM. Initially they observed that when nuclei were treated with nonionic detergent to solubilize the nuclear membrane lipids, the proteinaceous nuclear pore complexes (NPCs) remained intact and associated at their nucleoplasmic side with the membrane-denuded nuclear surface (Aaronson and Blobel, 1974). More remarkably, when NE “ghosts” isolated by nuclease digestion were treated with nonionic detergent, a shell-like structure derived from the NE persisted (Aaronson and Blobel, 1975). By EM, this material contained the detergent insoluble NPCs attached at their nucleoplasmic base to a thin fibrous lamina-like structure derived from the entire circumference of the nucleus (Aaronson and Blobel, 1975; Dwyer and Blobel, 1976). Moreover, if isolated NEs were depleted of chromatin by high salt treatment, the lamina was clearly visible as a ~15 nm fibrillar layer juxtaposed to the INM (Dwyer and Blobel, 1976). This suggested that a lamina could indeed be a widespread NE component. One of us (Larry Gerace) was a graduate student in the Blobel laboratory soon after this discovery, and undertook a characterization of the three major polypeptides present in the “pore complex-lamina” fraction (Aaronson and Blobel, 1975; Dwyer and Blobel, 1976). Using antibodies prepared to individual polypeptides, it was found by immunofluorescence microscopy that these proteins are concentrated at the nuclear periphery (Gerace et al., 1978; Krohne et al., 1978). Moreover, in immuno-EM of liver nuclei, the proteins were localized specifically to the lamina and not to NPCs (Gerace et al., 1978), and thereby came to be known as lamins A, B and C. Subsequently, the rat liver “lamin B” was designated lamin B1 when the vertebrate lamin B2 isotype was described (Vorburger et al., 1989). Immunofluorescence labeling indicated that lamins are widespread among different vertebrate cells (Gerace et al., 1978; Krohne et al., 1978). Since these proteins undergo reversible disassembly during mitosis in concert with NE disassembly/reformation (Gerace and Blobel, 1980), it was proposed that lamins form a polymeric core component of the lamina (Fig. 1).
Figure 1. Schematic diagram of the nuclear envelope.
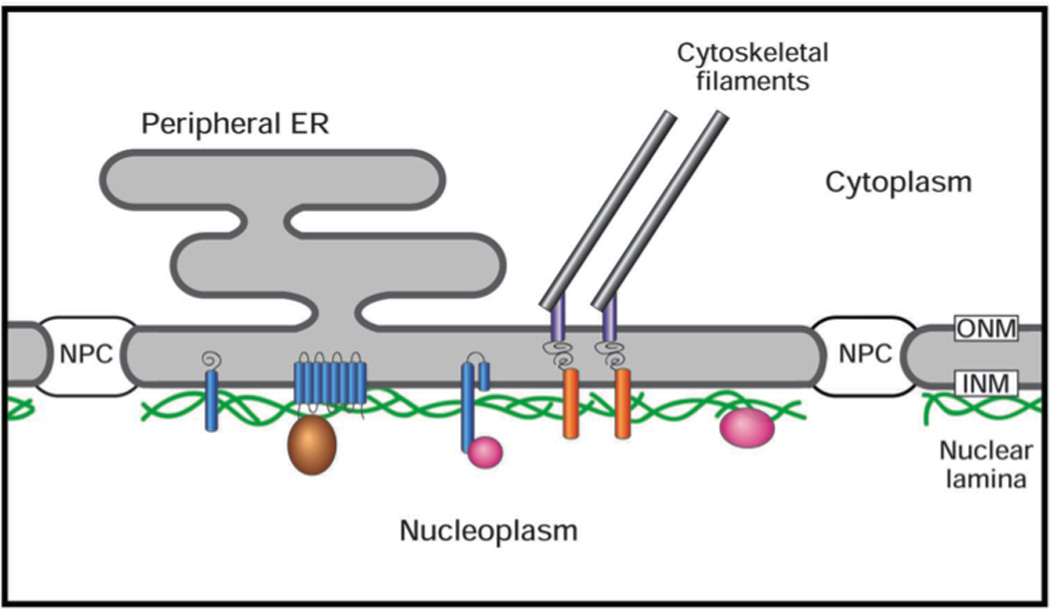
The outer nuclear membrane (ONM), which is continuous with the peripheral ER, is joined to inner nuclear membrane (INM) at the nuclear pore complex (NPC). The nuclear lamina comprises lamin filament polymers (green) and associated membrane-spanning proteins (blue), and peripheral proteins (brown and pink). The lamina is connected to the cytoplasmic cytoskeletal filaments by the LINC complex, which consists of Sun domain proteins (orange) spanning the INM that attached to nesprins (grey) that span the ONM.
Approaching the lamina at a molecular level
Key breakthroughs for understanding the organization of the nuclear lamina occurred in 1986 through a convergence of molecular biological and structural approaches. First, the Blobel (Fisher et al., 1986) and Kirschner (McKeon et al., 1986) laboratories accomplished cDNA cloning of lamins A/C. They reported that the two lamins arise from alternative splicing of the same gene, and that they share an internal ~350 amino acid region with strong sequence homology to the “rod domain” that is the hallmark feature of cytoplasmic IF proteins. The rod domain generates the IF backbone, and contains mostly heptad repeats assembled in a parallel, unstaggered α-helical coiled-coil (Herrmann et al., 2007). The rod is flanked at its N- and C-termini by non-heptad containing “end” domains that vary in sequence between different IF classes. Whereas the cDNA sequencing placed lamins squarely in the IF protein family, lamins were demonstrated to be IF proteins structurally from a collaboration between the laboratories of Ueli Aebi and Larry Gerace (Aebi et al., 1986), who at the time were faculty members at the Johns Hopkins Medical School. The Aebi group had been developing techniques to study various supramolecular assemblies by surface reconstruction, and was using the NPC of Xenopus laevis oocytes as one of their models (Buhle et al., 1985). Straightforward methods were available to isolate the huge (400 µm diameter) nucleus from this cell by micromanipulation. Moreover, NEs could be cleanly separated from the nuclear contents because the chromosomes in this meiotic prophase cell are detached from the NE, in contrast to somatic cells. Although well-preserved NPCs had been seen in oocyte NE preparations and a lamina had been suggested to exist (Scheer et al., 1976), its structure had not been fully appreciated. A key realization by the Johns Hopkins groups was that the lamina was extremely fragile after solubilization of nuclear membranes. Accordingly, isolated oocyte NEs were first whole mounted on grids and then treated in situ with a nonionic detergent to solubilize the membranes prior to examination by freeze-drying/ rotary metal shadowing. This strategy “unveiled” breathtaking en face structural views of the nuclear lamina, particularly in areas where the NPCs appeared to be removed by mechanical forces. Rather than simply having a “fibrous” character, the oocyte lamina was found to comprise a quasi-orthogonal meshwork of ~10 nm filaments (Fig. 2). Subsequent work showed that the lamina of this meiotic cell contains mainly one lamin isotype, as distinguished from the multiple isotypes expressed in somatic cells (Nigg, 1992).
Figure 2. View of the nuclear lamina from Xenopus oocyte en face.

Electron micrograph of a replica of a freeze dried/ metal shadowed Xenopus oocyte NE after treatment with Triton X-100. The lamina is revealed as a quasi-orthogonal filament meshwork in areas where NPCs have been removed by mechanical forces (inset), but the network also can be seen in NPC-attached regions (upper left). Reproduced from (Aebi et al., 1986) with permission. Bars, 1 µm.
The Gerace group had developed methods to isolate lamins from rat liver nuclei for biochemical and structural studies, and worked with the Aebi laboratory to study these molecules in detail. Examination of purified lamins A/C and lamin B by glycerol spraying/ metal shadowing revealed that the basic protomeric subunit of both lamin subtypes, consisting largely of a dimer by analytical ultracentrifugation, was a long rod with two globular masses attached at one end (Fig. 3) (Aebi et al., 1986). This made complete sense in light of the sequences of lamins A/C: the rod would correspond to the predicted α-helical coiled-coil of the dimer, and the two globular masses would represent the C-terminal alternatively spliced tail domains of lamins A/C (Fisher et al., 1986; McKeon et al., 1986), where most of the remaining protein mass (~190–250 amino acids) occurred. Subsequent work involving examination of recombinant deletion mutants of lamins confirmed this interpretation (Gieffers and Krohne, 1991; Heitlinger et al., 1992). Bolstered by these results and informed by his fortuitous interest in assembly of the cytoplasmic IF protein keratin, Ueli Aebi tested the ability of the isolated rat lamins A/C dimers to assemble into filaments. Shortly thereafter, he provided compelling EM images documenting reconstitution of ~10 nm wide filaments from isolated lamins A/C, by dialysis from a solubilizing condition into a solution of more physiological pH and ionic strength (Fig. 4) (Aebi et al., 1986). However, the 10 nm lamin filaments were only transient intermediates during in vitro assembly, and continued to laterally associate over time to form filament bundles and paracrystals with a periodic axial repeat of ~25 nm, similar to the repeat seen with oocyte lamina filaments examined in situ (Aebi et al., 1986). Thus, the cDNA sequencing results were beautifully complemented by the structural analysis of oocyte NEs and the in vitro reconstitution of purified lamins, which established that lamins form bona fide IFs.
Figure 3. Structure of the purified lamins A/C dimers by rotary shadowing.
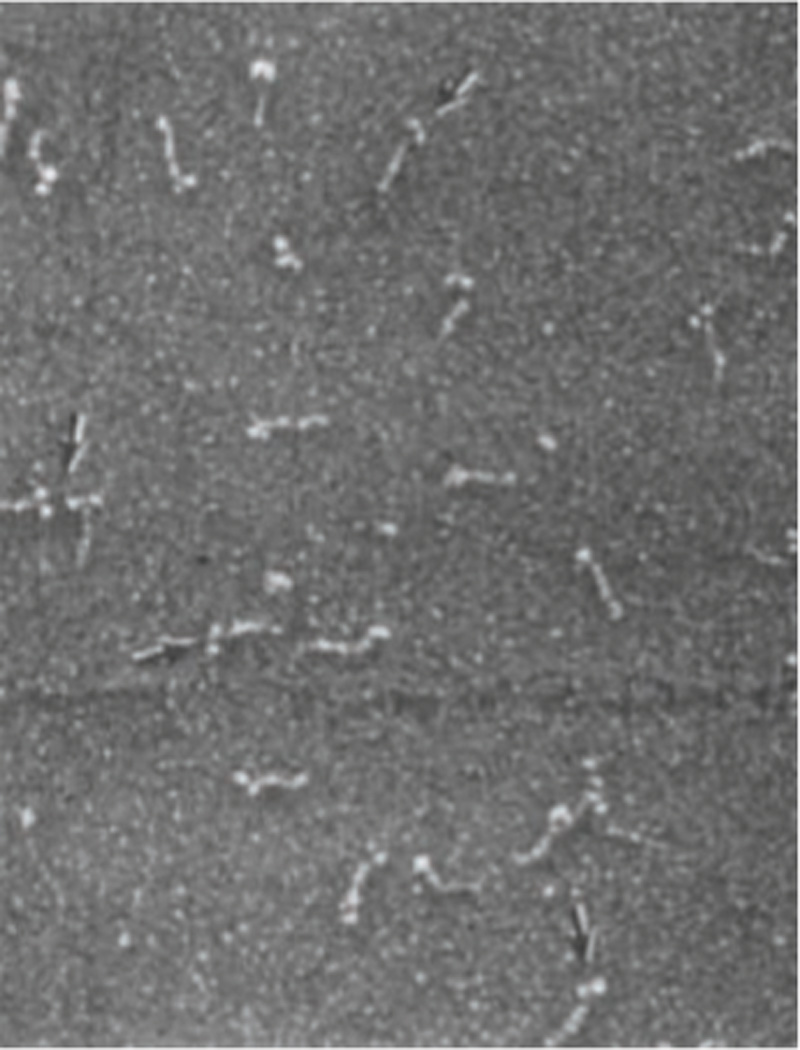
Electron micrograph of a sample of purified lamins A/C from rat liver NEs that was prepared by glycerol spraying/ rotary shadowing. The lamin dimer is seen as an ~52 nm rod attached to 2 masses (arrows). Reproduced from (Aebi et al., 1986) with permission.
Figure 4. Reconstitution of 10 nm filaments from purified lamins A/C.
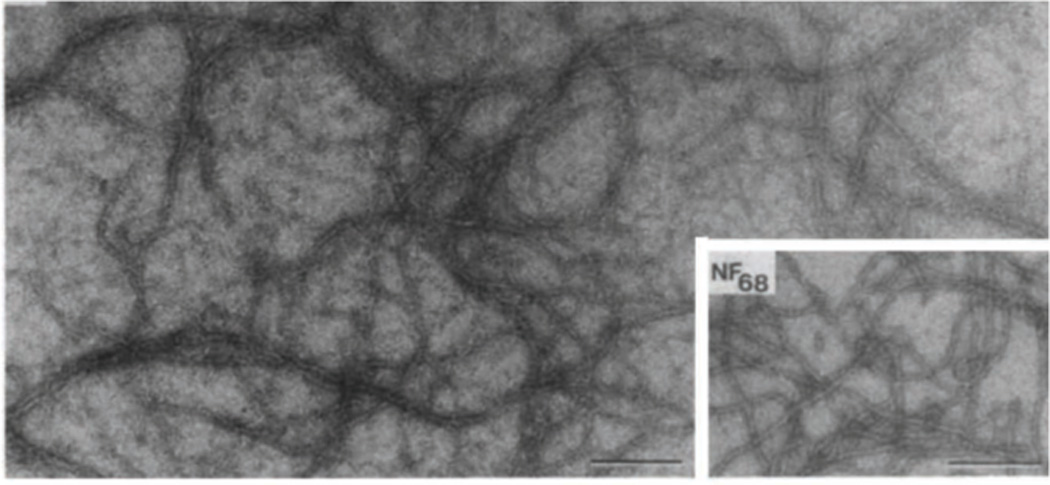
Electron micrograph of a negatively stained specimen of rat liver lamins A/C dialyzed into 25 mM MES, 200 mM NaC1 pH 6.5. Inset. For comparison, a sample of 10 nm filaments reconstituted from the purified 68 kD neurofilament protein of bovine brain is shown. Bars, 250 nm. Reproduced from (Aebi et al., 1986) with permission.
The inscrutable lamina of somatic cells
Genome and cDNA sequencing has revealed that lamin genes are ubiquitously present in metazoan organisms. Whereas C. elegans has a single lamin gene (Riemer et al., 1993), there are two lamin gene classes in more complex higher eukaryotes, A-type and B-type lamins (Dechat et al., 2010; Gruenbaum et al., 2005). Of the three lamin genes in mammals, one encodes the alternatively spliced products lamins A/C (LMNA), which are expressed mostly in differentiated cells. The other two code for lamin B1 (LMNB1) and lamin B2 (LMNB2), which are expressed at varying levels throughout development (Dechat et al., 2010; Gruenbaum et al., 2005). Correspondingly, the relative levels of the three lamin isotypes often differ in various cultured cell lines [e.g (Olins et al., 2001; Zwerger et al., 2010)]. In addition to lamins, there are many dozens of lamina-associated transmembrane proteins concentrated at the inner nuclear membrane (Fig. 1), whose expression profile is regulated in a developmental and cell type-specific manner (Korfali et al., 2010; Schirmer et al., 2005). Most of the well-characterized NE transmembrane proteins reside at the INM and bind to lamins, including emerin, lamin B receptor (LBR), lamina associated polypeptide (LAP)1, LAP2 and MAN1 (Gruenbaum et al., 2005; Schirmer and Foisner, 2007). Some of these inner membrane proteins show preferential binding to different lamin isotypes, and appear to promote the membrane association of lamins (Anderson et al., 2009; Gruenbaum et al., 2005; Schirmer and Foisner, 2007). Furthermore, lamins and some lamina associated proteins bind to DNA and/or chromatin proteins (Fig. 1) (Gruenbaum et al., 2005; Mattout et al., 2007; Taniura et al., 1995), which appears to foster the close apposition of chromatin to the NE in somatic cells. An additional group of lamina-associated proteins comprise the LINC complex, consisting of SUN domain proteins spanning the INM that are associated with nesprins spanning the outer membrane, which in turn are attached to actin filaments, microtubules and cytoplasmic IFs (Fig. 1) (Burke and Roux, 2009; Starr and Fridolfsson, 2010). Thus, the lamin polymer forms the core of a supramolecular assembly that interconnects the cytoplasmic cytoskeleton to the membranes of the NE and to chromatin.
Although 25 years have lapsed since the visualization of the Xenopus oocyte nuclear lamina, determining the supramolecular structure of the somatic cell lamina has proven elusive. Analysis has been particularly confounded by the shell of chromatin that is tenaciously associated with the NE/lamina in somatic cells. Hints that the somatic lamina may have at least a quasi-regular organization came from thin section EM of detergent-treated rat liver NEs, where the lamina appears as a chicken wire-like mesh in tangential views (Dwyer and Blobel, 1976). However, whether lamins of somatic cells form long 10 nm filaments, a network of short interconnected filaments, or some other polymeric entity remains unresolved. An equally important question is whether the A and B lamin subtypes that are expressed in higher metazoans are assembled in homotypic or heterotypic polymers. Biochemical studies have reported that isolated lamin subtypes have the capacity for both homotypic and heterotypic association in vitro (Kapinos et al., 2010; Schirmer and Gerace, 2004). Moreover, immunofluorescence microscopy has suggested that different lamin isotypes (Shimi et al., 2008) as well as lamin A vs lamin C (Kolb et al., 2011) are partially segregated from each other in cultured cells, although whether such segregation occurs in the cells of animal tissues is unresolved. These results support the notion that the lamina may contain both homotypic and heterotypic polymers of lamins, with regional structural variations at the sub-micrometer level. Fluorescence recovery after photobleaching (FRAP) analysis of cultured cells expressing GFP-tagged lamins indicates that lamins A, B1, and C all have very low diffusional mobilities at the NE of cultured cells (Broers et al., 1999; Moir et al., 2000), with little recovery of fluorescence in bleached areas after hours. Thus, lamin protomers seem to remain stably integrated in polymers after their initial assembly. Nonetheless, the NE roughly doubles in surface area during the cell cycle (Maul et al., 1972), and the lamina must rearrange to accommodate incorporation of newly synthesized lamins during this period. Whether de novo lamin insertion occurs throughout the lamina or at regional “hotspots” is not known. It may be that highly ordered arrays of lamin filaments as seen in the Xenopus oocyte lamina occur only in the nucleus of certain cell types, such as in postmitotic differentiated tissues.
The formation of lamin polymers at the NE without doubt is driven by the intrinsic self-association behavior of lamins. In addition, this process may be modulated locally by lamin-binding proteins of the INM and/or by chromatin (Schirmer and Foisner, 2007). Conversely or in addition, the organization of the lamin polymer could help to specify the lateral distribution of transmembrane proteins of the NE and chromatin packaging. FRAP analysis indicates that lamina-associated transmembrane proteins, although more mobile than lamins, still have low diffusional mobility at the NE (Ellenberg et al., 1997; Ostlund et al., 2006). This is consistent with the notion that they are physically tethered to lamins and/or other immobile nuclear components at the INM. It is conceivable that many membrane proteins of the INM are organized into discrete multi-protein complexes that cannot be resolved by conventional light microscopy, as suggested by in vitro binding studies involving emerin (Mansharamani and Wilson, 2005).
Confocal sections through the nucleus of cultured cells frequently reveal puncta with a high concentration of lamins and INM proteins that appear to occur in the interior of the nucleus, far from the NE (Ellenberg et al., 1997). These usually represent cross-sections of “nucleoplasmic reticulum,” tubular invaginations of the INM or of the entire NE that can extend microns into the nucleus (Malhas et al., 2011). Although the functions of these invaginations are unclear, they may provide a mechanism for directing NE functions to more internal nuclear regions. In addition to the nucleoplasmic reticulum, which can be viewed as an extension of the NE, a fraction of lamins occurs in a more diffuse internal nucleoplasmic localization (Moir et al., 2000; Shimi et al., 2008). Differential chemical extraction may provide a means for separating the internal vs peripheral lamins (Kolb et al., 2011). Whether either of the two pools has a unique postsynthetic modification or other biochemical signature is unknown. (FRAP) analysis indicates that the internal lamins have very low diffusional mobility, similar to those at the NE (Moir et al., 2000). Their low mobility may be due to assembly in a polymer and/or association with other immobile nuclear components. Functional insight related to internal nucleoplasmic lamins A/C has come from the analysis of mice with a null allele of the gene encoding LAP2α (Naetar et al., 2008), a non-membrane associated isoform of LAP2 that directly binds to lamins A/C. LAP2α is localized in internal regions of the nucleoplasm, and acts to regulate pRb (Dorner et al., 2006). In mouse cells/tissues with a LAP2α knockout, the internal nucleoplasmic lamins A/C, but not those at the NE, were lost (Naetar et al., 2008). The pathologies seen for the LAP2α knockout mice, which include hyperproliferation of epithelia and erythroid progenitor cells, raise the possibility that important functions of lamins may occur in internal nuclear regions as well as at the NE. Recent work has reported the association of nuclear actin with lamins (Simon et al., 2010) and these interactions could further extend the functional properties of lamins.
Advances and roadblocks to understanding the nuclear lamin polymer
Whereas the higher order structural organization of lamins in somatic cells is not clear, substantial progress has been made in understanding the molecular basis for lamin self-association into polymers. This has been accomplished by in vitro reconstitution and structural analysis of recombinant lamins, most of which has been spurred by the hand of Ueli Aebi. Examination of bacterially expressed, recombinant lamins from a wide diversity of organisms including human (Kapinos et al., 2010), chicken (Heitlinger et al., 1991; Heitlinger et al., 1992), frog (Gieffers and Krohne, 1991), fly (Sasse et al., 1997), and worm (Karabinos et al., 2003) has revealed that the basic protomer of all lamin subtypes is a dimer, which is able to self-associate into higher order structures including filament bundles and paracrystals.
Importantly, the in vitro assembly pathway of lamins was found to involve an early intermediate involving head-to-tail longitudinal polymers of lamin dimers (Fig. 3). First reported for recombinant chicken lamin B2 and lamin A (Heitlinger et al., 1991; Heitlinger et al., 1992), this behavior subsequently was found for lamins from frog (Gieffers and Krohne, 1991), fly (Sasse et al., 1997) and worm (Karabinos et al., 2003). The ~48 nm spacing of the globular domains along the longitudinal axis of the nascent lamin filaments was less than the ~52 nm length of the dimer rod, implying overlap between the highly conserved N- and C-terminal ends of the rod domain of adjacent dimers. With further assembly, the head-to-tail lamin polymers were suggested to assemble in a staggered antiparallel fashion, on the way to forming filaments and paracrystals with the characteristic ~25 nm axial repeat (Foeger et al., 2006; Heitlinger et al., 1991; Heitlinger et al., 1992; Sasse et al., 1997). The staggered, antiparallel protomer packing of lamins in higher order assemblies roughly resembles that of cytoplasmic IF proteins (Herrmann et al., 2007). By contrast, the early in vitro assembly intermediate of lamins differs from that of cytoplasmic IF proteins, which involves “unit length” 10 nm-wide structures that are based on the strong lateral interactions of tetramers (Herrmann et al., 2007). Tomographic reconstructions of 10 nm filaments assembled from the C. elegans lamin (Karabinos et al., 2003) have been generated from cryo-EM preparations (Ben-Harush et al., 2009). This work revealed that the filaments contain 3–4 tetrameric protofilaments, each of which contains 2 antiparallel, partially staggered arrays of head-to-tail dimers.
While the strong self-assembly properties of lamins have precluded atomic resolution structural determination of the full-length proteins, high resolution structures have been obtained for several lamin segments. X-ray crystallography (Dhe-Paganon et al., 2002) and solution NMR (Krimm et al., 2002) revealed that the tail domain of lamins A/C contains a 105 residue Ig-fold domain with β sheet secondary structure. The architecture of the remainder of the tail is not known, and its folding in cells may depend in significant part on interacting proteins. It has not been possible to crystallize the full length rod domain of lamins, but Aebi and collaborators have determined X-ray structures of fragments of the rod domain containing the C-terminal ~90 residues (Kapinos et al., 2011; Strelkov et al., 2004). The work suggested that this region not only can engage in parallel coiled-coil interactions, but also may partially unzip for parallel or antiparallel interactions with the rod domain of longitudinally adjacent dimers (Kapinos et al., 2010). These intriguing results could provide a molecular explanation for the overlap between the N- and C-terminal portions of the rod domain seen with the early assembly intermediate involving the head-to-tail polymer of lamin dimers. It is evident that understanding the structure of lamin polymers at the atomic level is a problem of enormous magnitude, which may require new technologies.
A functional window provided by human disease mutations
The functional relevance of the nuclear lamina has been underscored by discoveries made over the past 15 years that have linked human diseases to mutations in the genes for lamina proteins, most commonly in the LMNA gene (Cohen et al., 2008; Worman et al., 2009). These findings have provided a valuable framework to obtain molecular insight into lamina protein functions. Mutations in LMNA have been linked to at least 12 different clinical disorders (Worman and Bonne, 2007). The most prevalent of these are striated muscle diseases, the prototype of which is Emery-Dreifuss muscular dystrophy (EDMD) (Wheeler and Ellis, 2008; Worman and Bonne, 2007). EDMD and the other myopathies associated with LMNA mutations all include dilated cardiomyopathy to a variable extent, and are suggested to reflect the same basic disorder with variations in severity and specific organ involvement (Worman et al., 2009). Diseases arising from mutations in LMNA also include partial lipodystrophy syndromes, peripheral neuropathy and rare premature aging disorders, including Hutchison-Guilford Progeria Syndrome (HGPS) (Worman et al., 2009). Mice with a knockout of the Lmna gene, while indistinguishable from wild type animals at birth, die by 8 weeks of age with severe muscular dystrophy and dilated cardiomyopathy (Nikolova et al., 2004; Sullivan et al., 1999). Since this phenotype is strongly reminiscent of the striated muscule diseases caused by LMNA mutations, the Lmna null mouse has have been extensively studied as a model for EDMD, together with models containing knock-in human LMNA disease alleles (Stewart et al., 2007). Although over 300 disease alleles have been described for LMNA (Worman and Bonne, 2007), very few disease mutations have been found in either LMNB1 or LMNB2. This is understandable in light of the findings that mice with a deletion of either Lmnb1 (Vergnes et al., 2004) or Lmnb2 (Coffinier et al., 2010) show perinatal lethality.
In addition to being caused by mutations in LMNA, EDMD can be caused by mutations in the genes for several lamina-associated transmembrane proteins of the NE (Worman and Bonne, 2007). The prototype for this is the EMD gene that encodes emerin, a lamin A-binding transmembrane protein of the INM. Indeed, EMD was the first NE protein gene that was linked to human disease (Bione et al., 1994; Manilal et al., 1996). EDMD also can be caused by mutations in the genes encoding nesprin-1 and nesprin-2 (Zhang et al., 2007), components of the LINC complex that connect the lamina to cytoplasmic actin filaments via the Sun proteins (Fig. 1) (Crisp et al., 2006; Starr and Fridolfsson, 2010). The revelation that clinically identical forms of EDMD can be caused by mutations in the genes for multiple nuclear lamina proteins strongly supports the view that the lamina is a structurally and functionally integrated network of proteins.
Based on the phenotypes associated with mutations in the genes for lamin A and other lamina associated proteins, two explanations have been discussed for the primary molecular basis of the diseases. One model postulates that the diseases initially arise from aberrations in nuclear mechanics/structure, whereas a second proposes that the pathologies at first derive from abnormalities in signaling/gene expression (Cohen et al., 2008; Worman et al., 2009). The two models are not mutually exclusive, and alterations in one of these cellular features could influence the other. Indeed, perturbations in both nuclear mechanics and in signaling/gene expression have been observed in many cultured cell models for lamina-related diseases. Fibroblasts from human patients or from mouse models with mutations in the gene for lamin A frequently have dysmorphic nuclei, including NE herniations and other shape irregularities (Cohen et al., 2008; Worman et al., 2009). Moreover, biophysical studies have documented that the nucleus of fibroblasts from Lmna null mice is more deformable and fragile than that of wild type fibroblasts, and also manifests a deficiency in stress-induced signaling (Lammerding et al., 2004). Whatever the most proximal cause(s) of the lamina-associated diseases, aberrant signaling is a common denominator that likely contributes to disease phenotypes in most cases. The best illustration of this concept comes from work involving a mouse knockin model for EDMD (H222P/H222P), which showed increased ERK and JNK signaling in the heart prior to development of dilated cardiomyopathy (Muchir et al., 2007). The finding that disease onset could be retarded with MEK inhibitors supports a causal role of increased MAPK signaling in disease pathogenesis (Muchir et al., 2009).
A considerably more complex phenotype arises from the LMNA mutations that cause HGPS. These mutations interfere with removal of the farnesylated tail of lamin A that normally occurs after assembly of newly synthesized lamin A at the NE (Davies et al., 2011). Patient or mouse fibroblasts with mutant alleles that prevent removal of the farnesylated tail of lamin A have dysmorphic nuclei, impaired DNA damage repair, aberrant chromatin structure, diminished Wnt signaling, and a substantial decrease in nuclear deformability accompanied by increased fragility (Pereira et al., 2008; Worman et al., 2009). At the organismal level, mutations causing HGPS are proposed to cause a reduction in mesenchymal stem cell pools or an alteration in their differentiation pathway (Scaffidi and Misteli, 2008; Zhang et al., 2011). Mouse models indicate that retention of the farnesylated tail of lamin A is a toxic gain-of-function (Davies et al., 2011), but the “target” of the farnesylated tail at the NE, and whether it directly affects nuclear mechanics or signaling/gene expression, is unknown.
In addition to the mechanisms involving aberrant nuclear mechanics and perturbed signaling, a third type of mechanism potentially could contribute to disease phenotypes related to lamina proteins. This involves the connection of the nuclear lamina/nucleus to the cytoplasmic cytoskeleton that is mediated by the LINC complex (Fig. 1). It has been demonstrated in numerous studies of worms and flies that the LINC complex is important for nuclear positioning and migration during development (Burke and Roux, 2009; Starr and Fridolfsson, 2010). Moreover mice containing various combinations of null alleles of the genes encoding Sun1/Sun2 and nesprin-1/nesprin-2 have defective brain development due to defects in neuronal migration (Zhang et al., 2009), a process that requires nuclear movement over long distances. A similar phenotype occurs in mice with null alleles of Lmnb2 (Coffinier et al., 2010), emphasizing the importance of the lamin polymer for LINC complex functions. A role for the LINC complex in nuclear polarization of cultured mammalian cells also has been described (Luxton et al., 2011; Roux et al., 2009). Other than regulating nuclear positioning and movement, it is possible that the connection of the nucleus to the cytoplasmic cytoskeleton by the LINC complex controls intracellular stretch related signaling (Lammerding et al., 2004) that is altered when nuclear/cytoskeletal connection is perturbed. Thus, aberrations in LINC complex could be highly intertwined with other perturbations seen in lamina related diseases.
The molecular and structural details of how mutations in the lamin A protein lead to altered nuclear mechanics and signaling are not understood. With EDMD, LMNA disease mutations are commonly dominant and are distributed throughout the entire lamin protein, in the head and tail domains as well as in the rod (Worman and Bonne, 2007). Some of these mutations can alter in vitro and in vivo assembly properties of the lamin (Wiesel et al., 2008). If the mutant lamin A copolymerizes with wild type lamins, it could change the mechanical properties of lamin polymers, and alter the binding interfaces that are provided for other regulatory proteins (see below). Alternatively, non-uniform assembly of mutant lamin could promote the nuclear herniations and shape aberrations that are commonly observed (Cohen et al., 2008; Worman et al., 2009). Clearly, understanding the structure of the lamin polymeric meshwork at high resolution, and determining how this is influenced by mutant lamins, is a central question. The discovery that DNA replication (Shumaker et al., 2008) and transcription (Spann et al., 2002) can be influenced by lamins is consistent with the importance of these proteins in nuclear mechanics, signaling and gene expression.
Silence of the lamina
Numerous cytological and molecular approaches have revealed that heterochromatin and inactive genes preferentially accumulate at the NE/lamina during the course of differentiation (Guelen et al., 2008; Kalverda et al., 2008; Pickersgill et al., 2006; Towbin et al., 2009). Moreover, a common theme that has emerged over the past decade is that many signaling pathways are negatively regulated by lamina components (Heessen and Fornerod, 2007). Although NPCs are attached to the lamina and are active participants in gene expression due to their role in nucleocytoplamic trafficking, adjacent regions of the lamina between NPCs appear to serve as a nuclear subcompartment for “silence”.
The basis for heterochromatin association with the nuclear lamina very likely is due to multiple molecular interactions. One of these may involve the INM protein LBR, which binds the heterochromatin protein HP1 (Ye et al., 1997). In addition, since lamins bind chromatin via histones (Mattout et al., 2007; Taniura et al., 1995), a structural feature of the lamin polymer may provide a preferential binding site for a corresponding feature of condensed chromatin, such as found in the 30 nm chromatin fiber. Studies involving inducible tethering of a reporter gene to the nuclear lamina in mammalian cells have revealed that lamina association can lead to transcriptional inactivation (Finlan et al., 2008; Reddy et al., 2008).
In addition to it role in heterochromatin organization, the lamina provides a location where transcriptional activators are sequestered from the transcription machinery (Heessen and Fornerod, 2007) or where they are inactivated by other mechanisms. In the latter category, there are many examples illustrating negative regulation of signaling by nuclear lamina components, both in mouse models (Cohen et al., 2007; Muchir et al., 2007) and in cultured cells (Datta et al., 2009; Gonzalez et al., 2008; Huber et al., 2009; Mansharamani and Wilson, 2005; Pan et al., 2005). A number of transcriptional repressors also bind to INM proteins (Gruenbaum et al., 2005). The binding of these factors and their chromatin targets to the lamina could promote gene inactivation by mediating proximity to a gene silencing compartment. For example, the chromatin protein BAF, which is concentrated at the NE by binding to the LEM domain that is found in several INM proteins, has complex roles related to chromatin structure and compaction, NE reassembly at the end of mitosis, and activation of certain genes (Margalit et al., 2007; Segura-Totten and Wilson, 2004). Binding of BAF to LEM proteins might attenuate its functions in transcriptional activation and promote its functions in chromatin compaction. There are dozens of NE-specific transmembrane proteins that have tissue-selective expression patterns (Korfali et al., 2010; Schirmer et al., 2005), and these are likely to include important modulators of chromatin organization and transcription factors. Dissection of the physical interactions and functions of these will be an important area of study in the future.
Summary
The importance of the nuclear lamina in cell function has been clearly established by the discovery of disease-causing mutations in the genes for lamina proteins. The disease phenotypes, together with insights from analysis of cultured cells where lamina proteins have been targeted, have revealed an unexpectedly broad array of functions. Not only does the lamina provide an attachment site for chromatin and an environment that promotes gene inactivation, it also directly couples the cytoplasmic cytoskeleton to the nucleus. These functions are dependent on the core polymeric assembly of lamins, but major roles also are provided by other lamina components, including transmembrane proteins of the NE. Notwithstanding the substantial functional insight that has been gained from analysis of disease models for lamin A and other lamina components, the most proximal causes of the diseases remain unclear. Although some basic principles for lamin self-association have been established, the structures formed by higher order arrays of lamins with other membrane proteins is mostly unknown. A key future challenge is to determine the structure of the lamina as an integrated supramolecular assembly. This will be needed to fully understand how the lamina coordinates mechanical and signaling pathways and contributes to disease.
Figure 5. In vitro assembly of head-to-tail dimers chicken lamin B2.
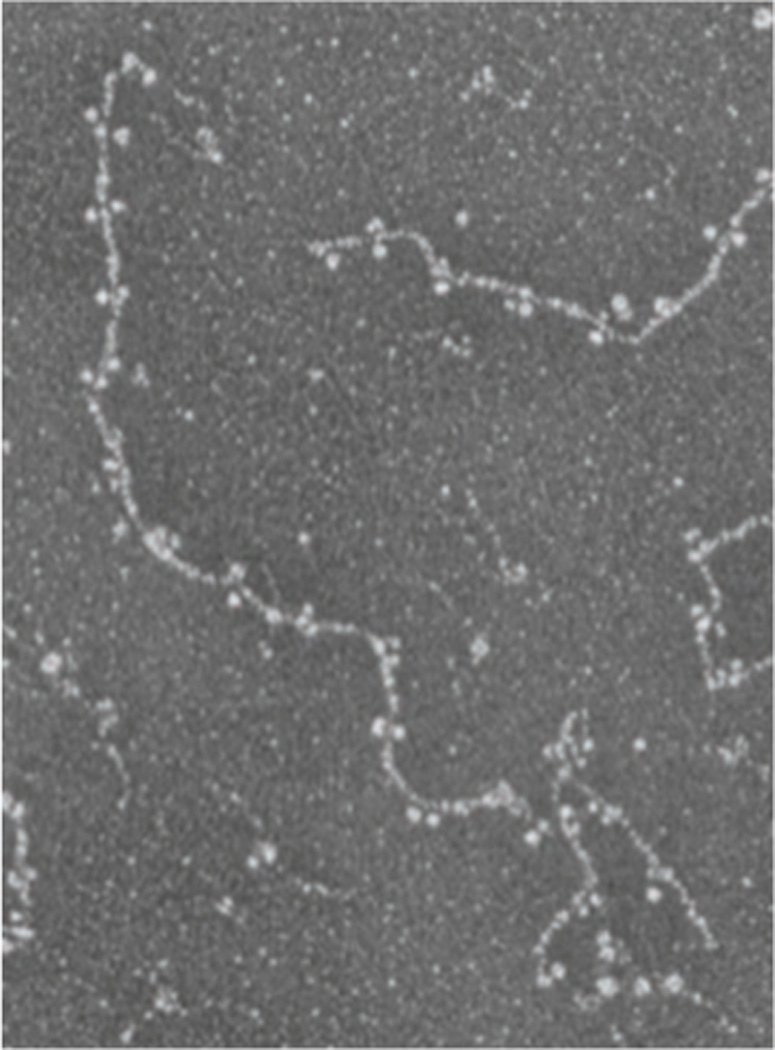
Electron micrograph of a glycerol sprayed/ rotary shadowed sample of purified recombinant chicken lamin B2 dimers dialyzed into a buffer containing 25 mM MES, 150 mM NaC1 pH 6.5. Longitudinal head-to-tail polymers of lamin dimers are shown. ©1991 Rockefeller University Press. Originally published in J. Cell Biol. 113:485–495.
Acknowledgements
The authors were supported by NIH RO1GM28521.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aaronson RP, Blobel G. On the attachment of the nuclear pore complex. J Cell Biol. 1974;62:746–754. doi: 10.1083/jcb.62.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaronson RP, Blobel G. Isolation of nuclear pore complexes in association with a lamina. Proc Natl Acad Sci U S A. 1975;72:1007–1011. doi: 10.1073/pnas.72.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol. 2009;186:183–191. doi: 10.1083/jcb.200901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Harush K, Wiesel N, Frenkiel-Krispin D, Moeller D, Soreq E, et al. The supramolecular organization of the C. elegans nuclear lamin filament. J Mol Biol. 2009;386:1392–1402. doi: 10.1016/j.jmb.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Bione S, Maestrini E, Rivella S, Mancini M, Regis S, et al. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet. 1994;8:323–327. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- Broers JL, Machiels BM, van Eys GJ, Kuijpers HJ, Manders EM, et al. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112(Pt 20):3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- Buhle EL, Jr, Aebi U, Smith PR. Correlation of surface topography of metal-shadowed specimens with their negatively stained reconstructions. Ultramicroscopy. 1985;16:436–450. doi: 10.1016/0304-3991(85)90110-x. [DOI] [PubMed] [Google Scholar]
- Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Coffinier C, Chang SY, Nobumori C, Tu Y, Farber EA, et al. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci U S A. 2010;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TV, Kosti O, Stewart CL. The nuclear envelope protein MAN1 regulates TGFbeta signaling and vasculogenesis in the embryonic yolk sac. Development. 2007;134:1385–1395. doi: 10.1242/dev.02816. [DOI] [PubMed] [Google Scholar]
- Cohen TV, Hernandez L, Stewart CL. Functions of the nuclear envelope and lamina in development and disease. Biochem Soc Trans. 2008;36:1329–1334. doi: 10.1042/BST0361329. [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Guan T, Gerace L. NET37, a nuclear envelope transmembrane protein with glycosidase homology, is involved in myoblast differentiation. J Biol Chem. 2009;284:29666–29676. doi: 10.1074/jbc.M109.034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BS, Coffinier C, Yang SH, Barnes RH, 2nd, Jung HJ, et al. Investigating the purpose of prelamin A processing. Nucleus. 2011;2:4–9. doi: 10.4161/nucl.2.1.13723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol. 2010;2:a000547. doi: 10.1101/cshperspect.a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S, Werner ED, Chi YI, Shoelson SE. Structure of the globular tail of nuclear lamin. J Biol Chem. 2002;277:17381–17384. doi: 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- Dorner D, Vlcek S, Foeger N, Gajewski A, Makolm C, et al. Lamina-associated polypeptide 2alpha regulates cell cycle progression and differentiation via the retinoblastoma-E2F pathway. J Cell Biol. 2006;173:83–93. doi: 10.1083/jcb.200511149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N, Blobel G. A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J Cell Biol. 1976;70:581–591. doi: 10.1083/jcb.70.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat. 1966;119:129–145. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc Natl Acad Sci U S A. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foeger N, Wiesel N, Lotsch D, Mucke N, Kreplak L, Aebi U, Gruenbaum Y, Herrmann H. Solubility properties and specific assembly pathways of the B-type lamin from Caenorhabditis elegans. J Struct Biol. 2006;155:340–350. doi: 10.1016/j.jsb.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Gerace L, Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieffers C, Krohne G. In vitro reconstitution of recombinant lamin A and a lamin A mutant lacking the carboxy-terminal tail. Eur J Cell Biol. 1991;55:191–199. [PubMed] [Google Scholar]
- Gonzalez JM, Navarro-Puche A, Casar B, Crespo P, Andres V. Fast regulation of AP-1 activity through interaction of lamin A/C, ERK1/2, and c-Fos at the nuclear envelope. J Cell Biol. 2008;183:653–666. doi: 10.1083/jcb.200805049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Harris P, James TW. Electron microscope study of the nuclear membrane of Amoeba proteus in thin section. Experientia. 1952;8:384–385. doi: 10.1007/BF02176196. [DOI] [PubMed] [Google Scholar]
- Heessen S, Fornerod M. The inner nuclear envelope as a transcription factor resting place. EMBO Rep. 2007;8:914–919. doi: 10.1038/sj.embor.7401075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitlinger E, Peter M, Haner M, Lustig A, Aebi U, Nigg EA. Expression of chicken lamin B2 in Escherichia coli: characterization of its structure, assembly, and molecular interactions. J Cell Biol. 1991;113:485–495. doi: 10.1083/jcb.113.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitlinger E, Peter M, Lustig A, Villiger W, Nigg EA, et al. The role of the head and tail domain in lamin structure and assembly: analysis of bacterially expressed chicken lamin A and truncated B2 lamins. J Struct Biol. 1992;108:74–89. doi: 10.1016/1047-8477(92)90009-y. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- Huber MD, Guan T, Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol Cell Biol. 2009;29:5718–5728. doi: 10.1128/MCB.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Roling MD, Fornerod M. Chromatin organization in relation to the nuclear periphery. FEBS Lett. 2008;582:2017–2022. doi: 10.1016/j.febslet.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Kapinos LE, Burkhard P, Herrmann H, Aebi U, Strelkov SV. Simultaneous formation of right- and left-handed anti-parallel coiled-coil interfaces by a coil2 fragment of human lamin A. J Mol Biol. 2011;408:135–146. doi: 10.1016/j.jmb.2011.02.037. [DOI] [PubMed] [Google Scholar]
- Kapinos LE, Schumacher J, Mucke N, Machaidze G, Burkhard P, et al. Characterization of the head-to-tail overlap complexes formed by human lamin A, B1 and B2 "half-minilamin" dimers. J Mol Biol. 2010;396:719–731. doi: 10.1016/j.jmb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Karabinos A, Schunemann J, Meyer M, Aebi U, Weber K. The single nuclear lamin of Caenorhabditis elegans forms in vitro stable intermediate filaments and paracrystals with a reduced axial periodicity. J Mol Biol. 2003;325:241–247. doi: 10.1016/s0022-2836(02)01240-8. [DOI] [PubMed] [Google Scholar]
- Kolb T, Maass K, Hergt M, Aebi U, Herrmann H. Lamin A and lamin C form homodimers and coexist in higher complex forms both in the nucleoplasmic fraction and in the lamina of cultured human cells. Nucleus. 2011;2:425–433. doi: 10.4161/nucl.2.5.17765. [DOI] [PubMed] [Google Scholar]
- Korfali N, Wilkie GS, Swanson SK, Srsen V, Batrakou DG, et al. The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture. Mol Cell Proteomics. 2010;9:2571–2585. doi: 10.1074/mcp.M110.002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm I, Ostlund C, Gilquin B, Couprie J, Hossenlopp P, et al. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 2002;10:811–823. doi: 10.1016/s0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- Krohne G, Franke WW, Ely S, D'Arcy A, Jost E. Localization of a nuclear envelope-associated protein by indirect immunofluorescence microscopy using antibodies against a major polypeptide from rat liver fractions enriched in nuclear envelope-associated material. Cytobiologie. 1978;18:22–38. [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GG, Gomes ER, Folker ES, Worman HJ, Gundersen GG. TAN lines: A novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2011;2:173–181. doi: 10.4161/nucl.2.3.16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A, Goulbourne C, Vaux DJ. The nucleoplasmic reticulum: form and function. Trends Cell Biol. 2011;21:362–373. doi: 10.1016/j.tcb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Manilal S, Nguyen TM, Sewry CA, Morris GE. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum Mol Genet. 1996;5:801–808. doi: 10.1093/hmg/5.6.801. [DOI] [PubMed] [Google Scholar]
- Mansharamani M, Wilson KL. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J Biol Chem. 2005;280:13863–13870. doi: 10.1074/jbc.M413020200. [DOI] [PubMed] [Google Scholar]
- Margalit A, Brachner A, Gotzmann J, Foisner R, Gruenbaum Y. Barrier-to-autointegration factor--a BAFfling little protein. Trends Cell Biol. 2007;17:202–208. doi: 10.1016/j.tcb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Mattout A, Goldberg M, Tzur Y, Margalit A, Gruenbaum Y. Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J Cell Sci. 2007;120:77–85. doi: 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, et al. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J Cell Biol. 1972;55:433–447. doi: 10.1083/jcb.55.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Shan J, Bonne G, Lehnart SE, Worman HJ. Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum Mol Genet. 2009;18:241–247. doi: 10.1093/hmg/ddn343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Pavlidis P, Decostre V, Herron AJ, Arimura T, et al. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J Clin Invest. 2007;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naetar N, Korbei B, Kozlov S, Kerenyi MA, Dorner D, et al. Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat Cell Biol. 2008;10:1341–1348. doi: 10.1038/ncb1793. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Assembly-disassembly of the nuclear lamina. Curr Opin Cell Biol. 1992;4:105–109. doi: 10.1016/0955-0674(92)90066-l. [DOI] [PubMed] [Google Scholar]
- Nikolova V, Leimena C, McMahon AC, Tan JC, Chandar S, et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest. 2004;113:357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins AL, Herrmann H, Lichter P, Kratzmeier M, Doenecke D, et al. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp Cell Res. 2001;268:115–127. doi: 10.1006/excr.2001.5269. [DOI] [PubMed] [Google Scholar]
- Ostlund C, Sullivan T, Stewart CL, Worman HJ. Dependence of diffusional mobility of integral inner nuclear membrane proteins on A-type lamins. Biochemistry. 2006;45:1374–1382. doi: 10.1021/bi052156n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Estevez-Salmeron LD, Stroschein SL, Zhu X, He J, et al. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signaling by the transforming growth factor-{beta} superfamily of cytokines. J Biol Chem. 2005;280:15992–16001. doi: 10.1074/jbc.M411234200. [DOI] [PubMed] [Google Scholar]
- Pereira S, Bourgeois P, Navarro C, Esteves-Vieira V, Cau P, et al. HGPS and related premature aging disorders: from genomic identification to the first therapeutic approaches. Mech Ageing Dev. 2008;129:449–459. doi: 10.1016/j.mad.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Riemer D, Dodemont H, Weber K. A nuclear lamin of the nematode Caenorhabditis elegans with unusual structural features; cDNA cloning and gene organization. Eur J Cell Biol. 1993;62:214–223. [PubMed] [Google Scholar]
- Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse B, Lustig A, Aebi U, Stuurman N. In vitro assembly of Drosophila lamin Dm0--lamin polymerization properties are conserved. Eur J Biochem. 1997;250:30–38. doi: 10.1111/j.1432-1033.1997.t01-1-00030.x. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Kartenbeck J, Trendelenburg MF, Stadler J, Franke WW. Experimental disintegration of the nuclear envelope. Evidence for pore-connecting fibrils. J Cell Biol. 1976;69:1–18. doi: 10.1083/jcb.69.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Gerace L. The stability of the nuclear lamina polymer changes with the composition of lamin subtypes according to their individual binding strengths. J Biol Chem. 2004;279:42811–42817. doi: 10.1074/jbc.M407705200. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Foisner R. Proteins that associate with lamins: many faces, many functions. Exp Cell Res. 2007;313:2167–2179. doi: 10.1016/j.yexcr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Identification of novel integral membrane proteins of the nuclear envelope with potential disease links using subtractive proteomics. Novartis Found Symp. 2005;264:63–76. discussion 76-80, 227-30. [PubMed] [Google Scholar]
- Segura-Totten M, Wilson KL. BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 2004;14:261–266. doi: 10.1016/j.tcb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Solimando L, Sengupta K, Shimi T, Adam SA, et al. The highly conserved nuclear lamin Ig-fold binds to PCNA: its role in DNA replication. J Cell Biol. 2008;181:269–280. doi: 10.1083/jcb.200708155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Zastrow MS, Wilson KL. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus. 2010;1:264–272. doi: 10.4161/nucl.1.3.11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann TP, Goldman AE, Wang C, Huang S, Goldman RD. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J Cell Biol. 2002;156:603–608. doi: 10.1083/jcb.200112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CL, Kozlov S, Fong LG, Young SG. Mouse models of the laminopathies. Exp Cell Res. 2007;313:2144–2156. doi: 10.1016/j.yexcr.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkov SV, Schumacher J, Burkhard P, Aebi U, Herrmann H. Crystal structure of the human lamin A coil 2B dimer: implications for the head-to-tail association of nuclear lamins. J Mol Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Meister P, Gasser SM. The nuclear envelope--a scaffold for silencing? Curr Opin Genet Dev. 2009;19:180–186. doi: 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci U S A. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorburger K, Lehner CF, Kitten GT, Eppenberger HM, Nigg EA. A second higher vertebrate B-type lamin. cDNA sequence determination and in vitro processing of chicken lamin B2. J Mol Biol. 1989;208:405–415. doi: 10.1016/0022-2836(89)90505-6. [DOI] [PubMed] [Google Scholar]
- Wheeler MA, Ellis JA. Molecular signatures of Emery-Dreifuss muscular dystrophy. Biochem Soc Trans. 2008;36:1354–1358. doi: 10.1042/BST0361354. [DOI] [PubMed] [Google Scholar]
- Wiesel N, Mattout A, Melcer S, Melamed-Book N, Herrmann H, et al. Laminopathic mutations interfere with the assembly, localization, and dynamics of nuclear lamins. Proc Natl Acad Sci U S A. 2008;105:180–185. doi: 10.1073/pnas.0708974105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. "Laminopathies": a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest. 2009;119:1825–1836. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lian Q, Zhu G, Zhou F, Sui L, et al. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerger M, Kolb T, Richter K, Karakesisoglou I, Herrmann H. Induction of a massive endoplasmic reticulum and perinuclear space expansion by expression of lamin B receptor mutants and the related sterol reductases TM7SF2 and DHCR7. Mol Biol Cell. 2010;21:354–368. doi: 10.1091/mbc.E09-08-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]


