Abstract
The peroxisome-proliferator activated receptor alpha (PPARα) is a member of the nuclear receptor family with many important physiologic roles related to metabolism and inflammation. Previous research in pediatric patients with septic shock revealed that genes corresponding to the PPARα signaling pathway are significantly downregulated in a subgroup of children with more severe disease. In this study, PPARα expression analysis using whole blood derived RNA revealed that PPARα expression was decreased in patients with septic shock and that the magnitude of that decrement correlated with the severity of disease. In a mouse model of sepsis, induced by cecal ligation and puncture (CLP), knockout mice lacking PPARα had decreased survival compared to wild type animals. Plasma cytokine analysis demonstrated decreased levels of IL-1β, IL-6, IL-17, KC, MCP-1, MIP-2, and TNFα at 24 hours in PPARα knockout animals. Cell surface markers of activation on splenic dendritic cells, macrophages, and CD8 T-cells were reduced in PPARα null animals and the bacterial load in lung and splenic tissues was increased. These data indicate that reduced or absent PPARα expression confers a survival disadvantage in sepsis and that PPARα plays a role in maintaining appropriate immune functions during the sepsis response.
Keywords: pediatric sepsis, septic shock, PPARα, nuclear hormone receptors, bacterial load
Introduction
The peroxisome-proliferator activated receptor alpha (PPARα) belongs to a family of nuclear receptors identified initially by their ability to induce peroxisome proliferation in rodents. This family, in turn, comprises part of the greater superfamily of ligand activated nuclear receptor transcription factors which also includes the receptors for steroid and thyroid hormones. There are three known PPAR receptors: α, β/δ, and γ, which are all broadly involved with the regulation of various metabolic processes, especially those related to lipid and glucose homeostasis (1–4). In recent years PPAR signaling has also been shown to modulate cell proliferation and differentiation, apoptosis, and importantly, the regulation of inflammation (5–9).
Sepsis is an unfortunately common, life threatening syndrome that afflicts thousands of adults and children each year. Despite decades of research, little headway has been made in understanding the mechanisms that underlie the complex host:pathogen interactions that result in such morbidity and mortality, much less provide effective therapy thereto. Recent discovery-oriented clinical investigations using genome-wide expression profiling in children with septic shock have revealed a subset of patients with increased severity of disease, worse multisystem organ failure, and decreased survival (10, 11). One notable characteristic of this subset of septic children was profound downregulation of multiple genes involved with cell signaling of the immune system. The PPARα/RXR (retinoic acid X receptor) pathway was among those most affected by this suppression (11).
With this information, we hypothesized that PPARα plays an important role in the host response to a septic challenge and that its downregulation or absence impairs survival. The investigations described herein give insight into possible mechanisms of that role and provide new targets for future inquiry.
Materials and Methods
PPARα expression analysis
With Institutional Review Board approval as described previously, (10–13) children less than 10 years of age who were admitted to a pediatric intensive care unit and met established criteria for the systemic inflammatory response syndrome (SIRS), sepsis, or septic shock (14) were enrolled in the study between March 2003 and June 2010. Age matched controls were recruited from the ambulatory departments of participating institutions using previously published inclusion and exclusion criteria (10). Additional information regarding the patient cohort is included in the Table. Each child had blood samples drawn for gene expression analysis within 24 hours of presentation. Total RNA was isolated from whole blood samples using the PaxGene Blood RNA System (PreAnalytiX, Qiagen/Becton Dickson, Valencia, CA) according the manufacturer’s specifications and microarray hybridization was performed by the Affymetrix Gene Chip Core facility at Cincinnati Children’s Hospital Research Foundation as previously described using the Human Genome U133 Plus 2.0 Gene-Chip (Affymetrix, Santa Clara, CA) (10, 15). For leukocyte subset PPARα expression, specific populations were isolated from blood drawn from 13 additional children with septic shock and 5 controls using the Miltenyi autoMACS cell separation platform and MACS Microbeads targeted at specific isolation of neutrophils, monocytes, and lymphocytes according to the manufacturer’s specifications (Miltenyi Biotec Inc., Auburn, CA). After leukocyte subset isolation, leukocyte subset total RNA was isolated using the RNeasy RNA Isolation Kit (Qiagen, Valencia, CA) and expression analyzed on the Affymetric Gene Chip as noted above. Analyses were performed using one patient sample per chip with GeneSpring GX 7.3 software (Agilent Technologies, Palo Alto, CA).
Table.
Demographics of study cohort
| Controls | SIRS | Sepsis | Septic Shock | |
|---|---|---|---|---|
| # of subjects | 52 | 21 | 36 | 179 |
| Median age (years)a | 2.2 (0.7 – 4.7) | 4.4 (2.1 – 8.4) | 2.6 (1.2 – 5.9) | 2.4 (0.9 – 6.2) |
| # of males/females | 29/23 | 11/10 | 28/8 | 108/71 |
| Median PRISM score | n/a | 10 (4 – 13) | 10 (5 – 14) | 15 (10 – 21)b |
| # of deaths (%) | n/a | 1 (5) | 1 (3) | 29 (16)c |
| # with gram negative organism (%) | n/a | n/a | 14 (39) | 36 (20)d |
| # with gram positive organism (%) | n/a | n/a | 12 (33) | 52 (29) |
| # with negative cultures (%) | n/a | n/a | 10 (28) | 91 (51) |
All medians reported with interquartile ranges in parentheses.
p < 0.05 vs SIRS and Sepsis (ANOVA on Ranks).
p < 0.05 vs SIRS and Sepsis (Chi-square with 2 degrees of freedom).
p < 0.05 vs Sepsis (Chi-square with 2 degrees of freedom).
Mouse Cecal Ligation and Puncture (CLP)
Mouse experiments were conducted in accordance with appropriate guidelines under approval of the Institutional Animal Care and Use Committee of the Cincinnati Children’s Hospital Research Foundation. PPARα knockout mouse breeding pairs (B6.129S4-Pparatm1Gonz/J) were obtained from Jackson Laboratories and 6–12 week old male offspring were used for all experiments. Age matched male C57BL/6J mice, which are of identical genetic background to the knockout mice, were obtained from Harlan Laboratories and housed in the same room for at least a week prior to experimentation. Polymicrobial sepsis was induced via CLP (16). Briefly, mice were anesthetized with 1.5% halothane in 50% oxygen via nose cone throughout the surgical procedure. A left paramidline laparotomy was performed and the cecum exteriorized and ligated with 3.0 silk suture immediately distal to the ileocecal junction. Care was taken not to obstruct bowel continuity. Two punctures were made with a 21-gauge needle along the antimesenteric aspect of the cecum and light pressure applied to express a small amount of fecal material. The cecum was replaced in the abdomen and the abdominal wall closed in two layers with suture and then topical tissue adhesive. Sham operated animals had the same procedure performed except that after exteriorization the cecum was only gently squeezed. All animals were resuscitated with 20 ml/kg of normal saline solution (roughly 0.5 ml per mouse) delivered subcutaneously in the nape of the neck. Each further received subcutaneous antibiotic injections (imipenem/cilastatin 25 mg/kg, Merk) in the nape of the neck one hour after CLP and then every 12 hours until sacrifice or until four days had passed. The purpose of the antibiotics was to moderate the severity of the model and mimic the clinical treatment of sepsis in humans. Mice were allowed free access to food and water after the procedure.
Survival Studies
After CLP, mice were observed and mortality documented every 12 hours for the first 4 days and then every 24 hours until 7 days had passed since CLP. Surviving mice were then euthanized.
Cytokine and Chemokine Measurement
Mice were euthanized with CO2 asphyxiation at 3, 6, and 24 hours after CLP. Whole blood was harvested via sterile, percutaneous cardiac puncture, and placed in lithium heparin plasma separator tubes (BD Vacutainer). Tubes were centrifuged at 2,500 rpm for 10 minutes. The plasma was collected into Eppendorf tubes and frozen at −80° C until batch analysis could be performed at a later date. Aliquots were subsequently analyzed using a magnetic bead immunoassay kit (Millipore, Billerica, MA) according to manufacturer instructions on a Luminex multiplex instrument (Luminex, Austin, TX). The cytokines and chemokines analyzed included interferon gamma (IFN-γ), IL-1β, IL-6, IL-10, IL-17, KC, macrophage chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1α (MIP-1α), MIP-2, and TNFα.
Flow Cytometry
Analyses of cell surface antigen expression were performed as previously described (17) on spleen homogenate samples. Flow cytometry data acquisition and analysis were performed on an LSR II using FACS Diva software (BD Biosciences, Mountain View, Calif) using the following antibodies: CD11b (clone M1/70), CD11c (clone Hl3), MHCII (clone AF6-120.1), TCR (clone H57-597), CD8a (clone 53-6.7), CD4 (clone RM4-5), CD69v (clone H1.2F3). All antibodies were obtained from BD Biosciences.
Bacterial Load Determination
Blood was harvested aseptically as described above. Peritoneal lavage fluid was collected by injecting 2 mls of sterile PBS into the peritoneum and then aspirating at least 1 ml back. Lungs and spleens were collected and homogenized in sterile PBS. All samples were serially diluted in sterile PBS and cultured on 5% sheep blood agar plates. Plates were incubated for 24 hours in 37° C and colony enumeration was performed the next day.
Statistical Analysis
The student’s t-test and Mann-Whitney Rank Sum analyses were used to describe normally distributed and non-parametric data respectively. A Kaplan-Meier curve was generated to assess survival between study groups. Calculations were performed using SigmaStat software (Systat Software Inc., San Jose, CA). A P value less than 0.05 was considered statistically significant.
Results
PPARα expression
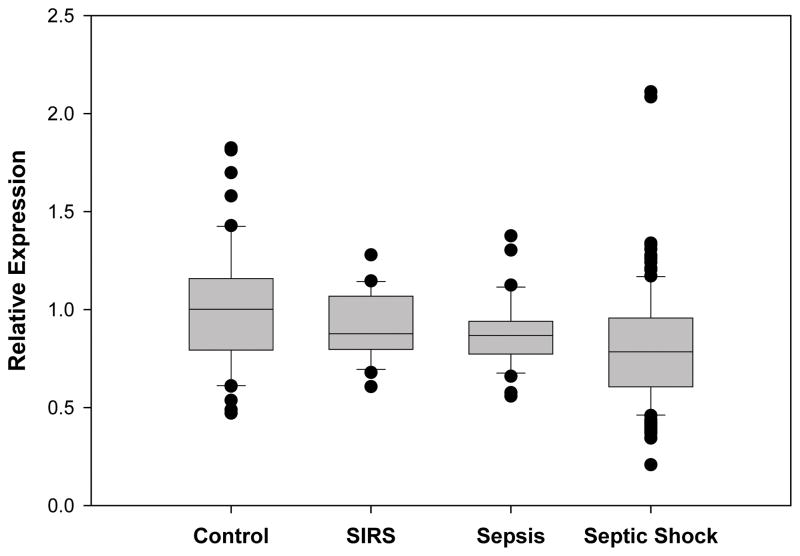
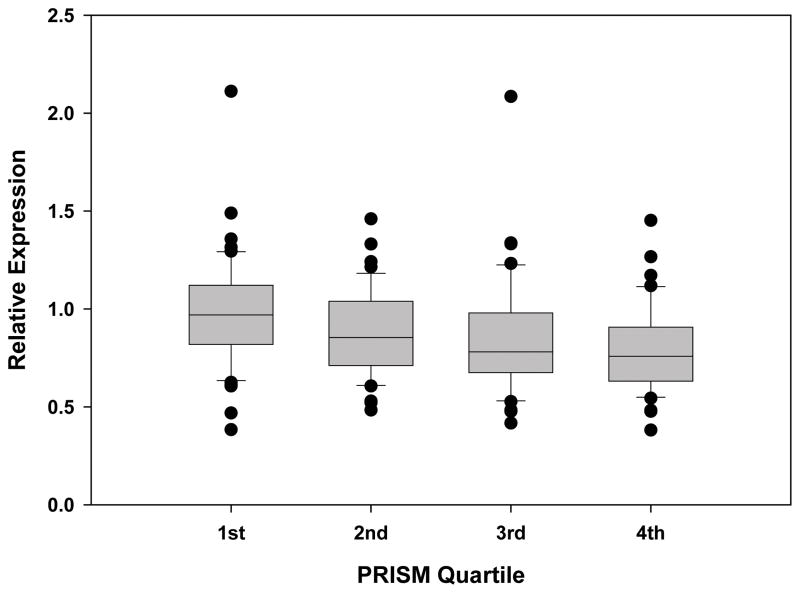
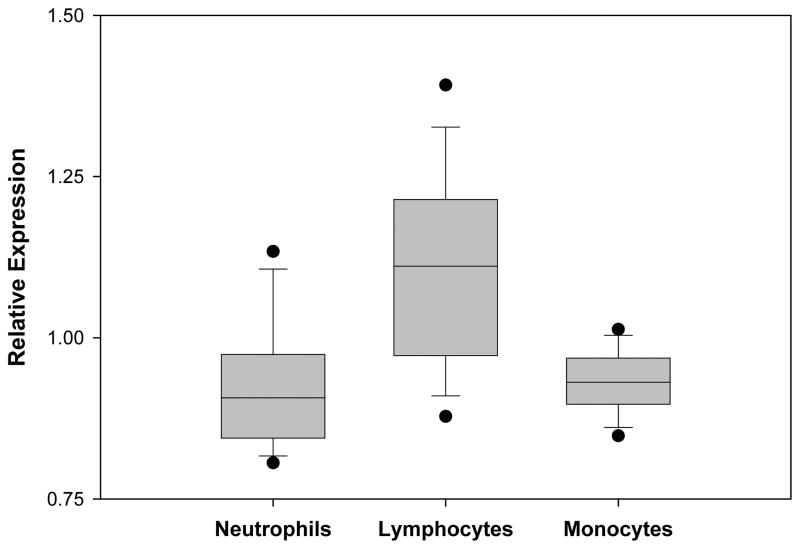
The relative expression of PPARα from whole blood derived mRNA in 21 children with SIRS, 36 children with sepsis, and 180 children with septic shock was compared with PPARα expression from 52 healthy controls. PPARα expression decreased as the severity of the sepsis syndrome increased and was significantly depressed in the children with septic shock (Figure 1. This is a box and whisker plot. The boxes span the 25th and 75th percentile values with the median indicated by the horizontal line within the box. The bars represent the 10th and 90th percentile values. The filled circles represent outliers.). Patients with septic shock were then categorized by severity of disease according to PRISM score quartiles. Those in the 3rd and 4th PRISM quartiles (i.e. higher illness severity) had significantly lower PPARα expression compared to children in the 1st PRISM quartile (Figure 2). Leukocyte subpopulation specific expression data normalized to the median expression of control patients reveal that PPARα repression is greatest in neutrophils and macrophages (Figure 3). These clinical data indicate that PPARα expression is decreased in children with septic shock and that the degree of PPARα repression correlates with illness severity.
Figure 1.
Relative expression of PPARαmRNA in controls (n = 52), patients with SIRS (n = 21), patients with sepsis (n = 36), and patients with septic shock (n = 179). Data are derived from whole blood RNA and represent the first 24 hours of admission to the PICU. P < 0.001 for all pairwise comparisons (ANOVA on Ranks). P < 0.05 for septic shock vs. controls (post hoc Dunn’s test).
Figure 2.
Relative expression of PPARα mRNA in 180 patients with septic shock grouped according to PRISM quartiles. Data are derived from whole blood RNA and represent the first 24 hours of admission to the PICU with septic shock. P < 0.001 for all group comparisons (Kruskal-Wallis ANOVA on Ranks with 3 degrees of freedom). P < 0.05 3rd quartile vs. 1st quartile, and 4th quartile vs. 1st quartile.
Figure 3.
Relative expression of PPARαmRNA in leukocyte subsets of 13 patients with septic shock. Data are derived from the indicated leukocyte subset RNAs and represent the first 24 hours of admission to the PICU. P < 0.001 for all pairwise comparisons (ANOVA on Ranks). P < 0.05 for neutrophils vs. lymphocytes; and for monocytes vs. lymphocytes (post hoc Dunn’s test).
CLP Survival
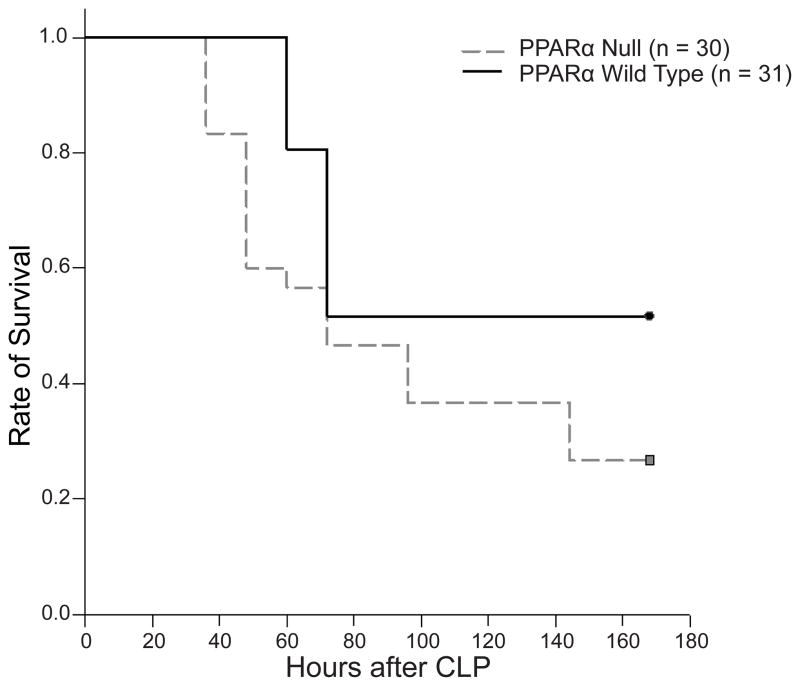
With this clinical data in hand, we sought to further evaluate the role PPARα plays in sepsis by studying mice lacking the PPARα gene. Groups of PPARα null mice and PPARα wild type mice of identical genetic background were subjected to cecal ligation and puncture, which induces a polymicrobial sepsis within hours of surgery. Imipenem 25 mg/kg was administered subcutaneously one hour after CLP and then every twelve hours for four days. Mice were observed for a total of seven days. PPARα null mice demonstrated decreased survival compared to wild-type mice and onset of significant mortality occurred earlier for the PPARα null group (Figure 4). This same pattern of mortality was observed across two other models of sepsis including cecal slurry (intraperitoneal injection of a suspension of donor mouse cecal contents) without antibiotics and CLP without antibiotics (data not shown).
Figure 4.
PPARα null mice exhibit decreased survival (27%) in CLP induced sepsis compared to wild type mice (52%, p = 0.021).
Cytokine and chemokine levels
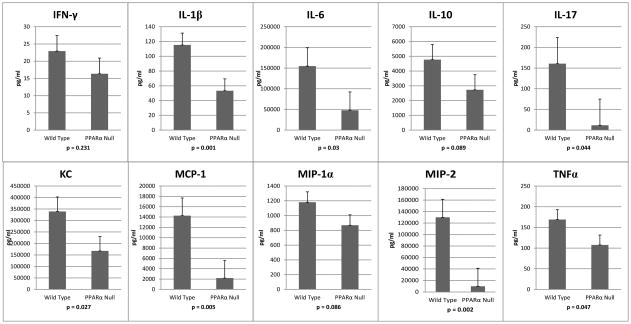
Hypothesizing that the increased mortality observed in the survival studies of PPARα knockout mice was due to differences in the inflammatory/immune response during the first 24 hours (knockout mice started dying at around 36 hours) we evaluated multiple inflammatory cytokines and chemokines (IFN-γ, IL-1β, IL-6, IL-10, IL-17, KC, MCP-1, MIP-1α, and TNFα) at 3, 6, and 24 hours using a multiplex bead array. There were no statistical differences between the groups at any time point for IFN-γ IL-10 or MIP-1α. At 6 hours, however, IL-1β and IL-17 were both reduced in the knockout mice. At 24 hours the knockout mice demonstrated an almost global reduction in the cytokines assayed when compared to wild type mice with IL-1β, IL-6, IL-17, KC, MCP-1, and TNFα all lower in the PPARα null group (Figure 5). The cytokine levels in sham operated mice were dramatically lower than in the CLP mice (data not shown). There were no statistically significant differences between the sham operated PPARα knockout and wildtype mice except for IL-6 and MCP-1 at 6 hours only. These cytokines were both decreased in the PPARα knockout animals (135 vs. 518 pg/ml, p = 0.030 for IL-6; 83 vs. 189 pg/ml, p = 0.009 for MCP-1).
Figure 5.
PPARα null mice compared to wild type mice demonstrate significantly decreased levels in the majority of inflammatory cytokines assayed at 24 hours after CLP.
Flow cytometry
We next sought to characterize the cellular immunologic response using flow cytometric analysis of splenocytes and peritoneal lavage. Upon laparotomy after euthanasia it was immediately apparent by visual inspection that the size of the spleen in the knockout animals was markedly increased. This discrepancy was observed at all time points and in sham operated animals as well as those who received CLP. At 24 hours the average weight of the spleens from the knockout animals was more than twice that of the wild types (109 mg vs. 52 mg, p = 0.004). Total splenic leukocyte counts were dramatically elevated in the knockout mice as well (173 vs. 54, p = 0.0005). Even when the leukocyte count was normalized to splenic tissue weight, the PPARα null mice showed statistically more leukocytes per mg of tissue.
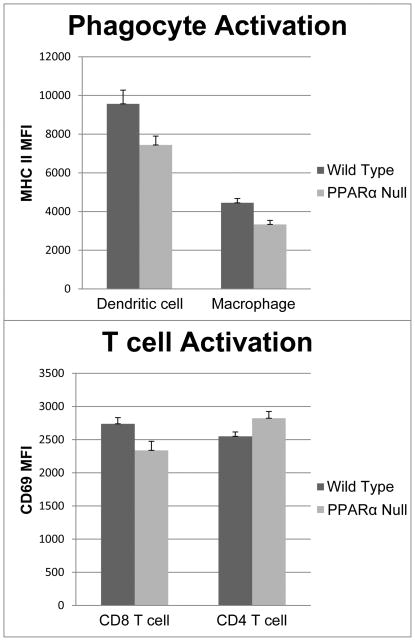
At 24 hours splenic dendritic cells and macrophages demonstrated reduced activation compared to their wild-type counterparts (MHC II mean fluorescence intensity 7,443 vs. 9,567; p = 0.036 for DCs and 3,332 vs. 44,446; p = 0.006 for macrophages respectively). CD8 T cells also showed evidence of decreased activation in the PPARα knockout mice (CD69 MFI was 2,339 and 2,739 respectively; p = 0.041). CD4 cells had no difference in their activation between groups as measured by CD69 intensity (Figure 6). There were no differences in any of these parameters between PPARα knockout and wildtype sham operated mice (data not shown).
Figure 6.
MHC II surface expression is significantly decreased on splenic dendritic cells and macrophages in PPARα null mice compared to controls at 24 hours. CD69 is also significantly decreased in CD8 T cells, but not CD4 T-cells.
Bacterial Load
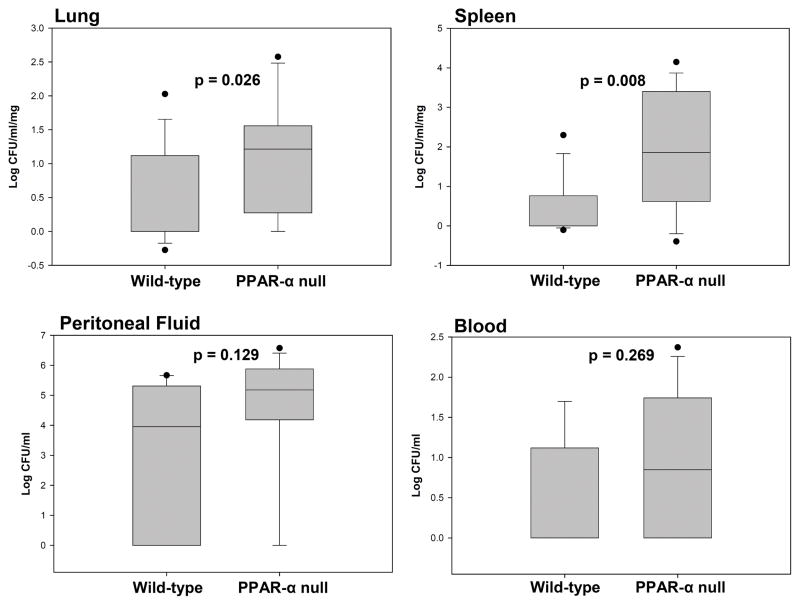
Bacterial load was increased in the PPARα knockout mouse tissues. At 24 hours after CLP blood, peritoneal fluid, lung, and spleen were collected, the organs homogenized, and dilutions plated on agar. Bacterial colonies were counted the next day. No significant difference in colony counts was detected between groups in the blood and peritoneal fluid. In the lung and splenic tissues, however, it was apparent that PPARα null mice had increased bacterial loads compared to wild type mice. The lungs of PPARα knockout mice had over twice the numbers of colony counts on average than wild type mice (p = 0.026) and in the spleen there was over 4 times as much bacteria (p = 0.008, Figure 7).
Figure 7.
Tissue bacterial load is increased in PPARα null animals compared to wild types. No statistical difference was noted in bacterial load between groups in the peritoneal fluid and blood compartments. Y-axis is log scale.
No differences between PPARα null mice and wild types were detected in whole lung MPO levels, macrophage phagocytic capability, or in phagocyte oxidative burst (data not shown).
Discussion
With this research we demonstrate that children with septic shock have reduced PPARα expression in whole blood derived RNA. The magnitude of that reduction is commensurate with disease severity as measured by PRISM scores; the sicker the child the lower the PPARα expression. To our knowledge, this is the first reported association between PPARα and severity of disease in sepsis.
These findings are significant because they indicate that dysregulation or loss of PPARα activity may play a central role in the immunopathogenesis of sepsis beyond its known function as a metabolic regulator. Indeed, these two activities of potential immune and metabolic coordination may be intimately associated. Recently, the intersection between metabolic disease and inflammation has been an active field of research and a high degree of crosstalk is observed between specific cells and tissues of both systems (9, 18–21). PPARα activity may be necessary for the initiation, maintenance, or resolution of an appropriate immune response and the absence of PPARα from the regulatory milieu can contribute substantially to life threatening disease processes. Evidence in support of this hypothesis was provided in our animal studies.
Mice that lack a functional PPARα gene are more susceptible to death using a model of surgically induced polymicrobial sepsis treated with antibiotics. Antibiotics were used specifically to mimic the clinical arena where physicians encounter and treat sepsis syndromes. PPAR alpha null mice experienced earlier onset of mortality than wild type mice and had overall increased mortality, indicating that PPARα is involved early in the development of sepsis. This pattern of mortality was conserved across two other models of sepsis, which strengthens the conclusion that PPARα null mice have a more susceptible phenotype.
We have also shown that PPARα null mice, compared to wild type mice, have reduced levels of many inflammatory cytokines at 24 hours after the initiation of sepsis. There were no statistically significant differences in cytokine levels between the two groups at 3 or 6 hours after CLP. This was a surprising observation due to the significant body of research that ascribes an anti-inflammatory role to PPARα in other models of disease and inflammation (22–29). Thus, its absence in sepsis should have permitted the evolution of a hyperinflammatory state after CLP. We initially anticipated that an early hypercytokinemia would have contributed substantially to the observed increase in mortality. The results obtained, however, indicated that a hypoinflammatory state is developing even at this early time point. This conclusion is corroborated by evidence of decreased splenocyte activation and increased tissue bacterial load.
Splenic dendritic cells and macrophages from PPARα null animals, when compared to those from wild type mice, exhibited decreased surface expression of MHCII, an antigen presentation protein upregulated in activated antigen presenting cells. CD69, a marker of T cell activation, was lower as well on PPARα null CD8 T cells. It is not known whether decreased activation results in decreased cytokine levels or vice versa.
The finding of increased tissue bacterial load is important. PPARα knock out animals had more bacteria in their lung and splenic tissues at 24 hours than wild type counterparts. No statistical difference in bacterial load was seen between study groups in the blood and peritoneal fluid. This is likely due to the administration of antibiotics which preferentially cleared those body fluids, but probably had less tissue penetration. Even if overwhelming bacteremia was not a factor in sepsis mortality in our antibiotic treated model, tissue microbial persistence points toward a functional deficit in host defense mechanisms in the PPARα knockout mice. This may be attributable to increased bacterial tissue penetration and colonization, permissive bacterial replication, or decreased immunologic bacterial clearance compared to wildtype mice. Reduced bacterial killing could be due to the decreased level of circulating chemokines which possibly impaired phagocyte tissue extravasation and chemotaxis. It does not appear to be directly related to macrophage phagocytosis or oxidative burst.
Despite the association observed between PPARα expression levels and disease severity in septic children, it is yet unclear whether the relationship is causal or coincidental. PPARα expression in the blood could possibly be reduced by any number of variables that are also known to be associated independently with severe sepsis and increased mortality including lymphocyte apoptosis, mitochondrial bioenergetic failure, and metabolic derangements induced by progressive organ failure. Furthermore, whole blood derived mRNA represents only the expression patterns of circulating leukocytes and may not mirror PPARα expression or activity in other tissues less accessible to sampling. Although PPARα in those tissues is likely relevant to organ function and survival in sepsis we feel that our initial approach of evaluating whole blood derived mRNA is viable as a hypothesis-generating tool because of its relative ease of sampling and because it allows a biologically relevant assessment of the immunologic response in sepsis.
However, our experimental data from mice indicate that individuals without PPARα are less able to survive in the face of a septic challenge. As noted above, we did not observe the anticipated hyperinflammatory storm, but rather a relative immunosuppressed state in the PPARα null animals. This finding, though unexpected, fits well with much current thinking that a significant portion of actual human mortality from sepsis results from a state of relative immunoparalysis (30–32). A possible pro-inflammatory role for PPARα, however, does stand at variance with what is currently understood about its related family members PPAR. A substantial body of research has ascribed to PPAR an anti-inflammatory influence on multiple cellular processes (6, 33–36). It is not currently known how PPARα and PPARγ interact with each other or with the third member of the family PPARβ/δ. It may be that while PPAR has potent anti-inflammatory properties, those of PPARα are less significant, but that PPARα’s activation of important bioenergetic pathways in leukocytes is essential for their normal activation and function.
In conclusion, our data indicate that decreased or absent PPARα expression confers a survival disadvantage in sepsis and potentiates a functionally immunosuppressed state. Further research is needed to evaluate the effect of PPARα activation and to elucidate the specific mechanisms by which this nuclear receptor impinges upon the immunologic response in sepsis.
Acknowledgments
This work was supported by: RO1 GM067202 (B.Z.) and R01 GM064619 (H.R.W.)
References
- 1.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405(6785):421–4. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 2.Lee C-H, Olson P, Evans RM. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144(6):2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- 3.Ferré P. The biology of peroxisome proliferator-activated receptors: relationship with lipid metabolism and insulin sensitivity. Diabetes. 2004;53 (Suppl 1):S43–50. doi: 10.2337/diabetes.53.2007.s43. [DOI] [PubMed] [Google Scholar]
- 4.Brown JD, Plutzky J. Peroxisome proliferator-activated receptors as transcriptional nodal points and therapeutic targets. Circulation. 2007;115(4):518–33. doi: 10.1161/CIRCULATIONAHA.104.475673. [DOI] [PubMed] [Google Scholar]
- 5.Kota BP, Huang TH-W, Roufogalis BD. An overview on biological mechanisms of PPARs. Pharmacol Res. 2005;51(2):85–94. doi: 10.1016/j.phrs.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Bailey ST, Ghosh S. “PPAR”ting ways with inflammation. Nat Immunol. 2005;6(10):966–967. doi: 10.1038/ni1005-966. [DOI] [PubMed] [Google Scholar]
- 7.Glass CK, Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol. 2006;6(1):44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- 8.Michalik L, Wahli W. Involvement of PPAR nuclear receptors in tissue injury and wound repair. J Clin Invest. 2006;116(3):598–606. doi: 10.1172/JCI27958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bensinger SJ, Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454(7203):470–7. doi: 10.1038/nature07202. [DOI] [PubMed] [Google Scholar]
- 10.Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Tagavilla MA, Odoms K, Dunsmore K, Barnes M, Aronow BJ. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30(2):146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, Freishtat RJ, Anas N, Meyer K, Checchia PA, Monaco M, Odom K, Shanley TP. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong HR, Cvijanovich N, Allen GL, Lin R, Anas N, Meyer K, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Shanley TP. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med. 2009;37(5):1558–1566. doi: 10.1097/CCM.0b013e31819fcc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong HR, Freishtat RJ, Monaco M, Odoms K, Shanley TP. Leukocyte subset-derived genomewide expression profiles in pediatric septic shock. Pediatr Crit Care Med. 2010;11(3):349–355. doi: 10.1097/PCC.0b013e3181c519b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein B, Giroir B, Randolph A. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6(1):2–8. doi: 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- 15.Shanley TP, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Odoms K, Barnes M, Sakthivel B, Aronow BJ, Wong HR. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med. 2007;13(9–10):495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard WJ, Choudhry M, Schwacha MG, Kerby JD, Rue LW, 3rd, Bland KI, Chaudry IH. Cecal ligation and puncture. Shock. 2005;24 (Suppl 1):52–57. doi: 10.1097/01.shk.0000191414.94461.7e. [DOI] [PubMed] [Google Scholar]
- 17.Caldwell CC, Kojima H, Lukashev D, Armstrong J, Farber M, Apasov SG, Sitkovsky MV. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. J Immunol. 2001;167(11):6140–6149. doi: 10.4049/jimmunol.167.11.6140. [DOI] [PubMed] [Google Scholar]
- 18.Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44(6):968–75. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Odegaard JI, Chawla A. Mechanisms of macrophage activation in obesity-induced insulin resistance. Nat Clin Pract Endocrinol Metab. 2008;4(11):619–626. doi: 10.1038/ncpendmet0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 21.Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest. 2011;121(6):2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunard R, DiCampli D, Archer DC, Stevenson JL, Ricote M, Glass CK, Kelly CJ. WY14,643, a PPAR alpha ligand, has profound effects on immune responses in vivo. J Immunol. 2002;169(12):6806–6812. doi: 10.4049/jimmunol.169.12.6806. [DOI] [PubMed] [Google Scholar]
- 23.Di Paola R, Esposito E, Mazzon E, Genovese T, Muià C, Crisafulli C, Malleo G, Sessa E, Meli R, Cuzzocrea S. Absence of peroxisome proliferators-activated receptors (PPAR)alpha enhanced the multiple organ failure induced by zymosan. Shock. 2006;26(5):477–484. doi: 10.1097/01.shk.0000230299.78515.2c. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Gocke AR, Lovett-Racke A, Drew PD, Racke MK. PPAR Alpha Regulation of the Immune Response and Autoimmune Encephalomyelitis. PPAR Res. 2008;2008:546753. doi: 10.1155/2008/546753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smeets PJH, Teunissen BEJ, Planavila A, de Vogel-van den Bosch H, Willemsen PHM, van der Vusse GJ, van Bilsen M. Inflammatory pathways are activated during cardiomyocyte hypertrophy and attenuated by peroxisome proliferator-activated receptors PPARalpha and PPARdelta. J Biol Chem. 2008;283(43):29109–29118. doi: 10.1074/jbc.M802143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaefer MB, Pose A, Ott J, Hecker M, Behnk A, Schulz R, Weissmann N, Günther A, Seeger W, Mayer K. Peroxisome proliferator-activated receptor-alpha reduces inflammation and vascular leakage in a murine model of acute lung injury. Eur Respir J. 2008;32(5):1344–1353. doi: 10.1183/09031936.00035808. [DOI] [PubMed] [Google Scholar]
- 27.Patel NSA, di Paola R, Mazzon E, Britti D, Thiemermann C, Cuzzocrea S. Peroxisome proliferator-activated receptor-alpha contributes to the resolution of inflammation after renal ischemia/reperfusion injury. J Pharmacol Exp Ther. 2009;328(2):635–643. doi: 10.1124/jpet.108.146191. [DOI] [PubMed] [Google Scholar]
- 28.Riccardi L, Mazzon E, Bruscoli S, Esposito E, Crisafulli C, Di Paola R, Caminiti R, Riccardi C, Cuzzocrea S. Peroxisome proliferator-activated receptor-alpha modulates the anti-inflammatory effect of glucocorticoids in a model of inflammatory bowel disease in mice. Shock. 2009;31(3):308–316. doi: 10.1097/SHK.0b013e31818339e7. [DOI] [PubMed] [Google Scholar]
- 29.Gocke AR, Hussain RZ, Yang Y, Peng H, Weiner J, Ben L-H, Drew PD, Stuve O, Lovett-Racke AE, Racke MK. Transcriptional modulation of the immune response by peroxisome proliferator-activated receptor-{alpha} agonists in autoimmune disease. J Immunol. 2009;182(7):4479–4487. doi: 10.4049/jimmunol.0713927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348(2):138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 31.Ward NS, Casserly B, Ayala A. The compensatory anti-inflammatory response syndrome (CARS) in critically ill patients. Clin Chest Med. 2008;29(4):617–625. viii. doi: 10.1016/j.ccm.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hotchkiss RS, Coopersmith CM, McDunn JE, Ferguson TA. The sepsis seesaw: tilting toward immunosuppression. Nat Med. 2009;15(5):496–497. doi: 10.1038/nm0509-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zingarelli B, Sheehan M, Hake PW, O’Connor M, Denenberg A, Cook JA. Peroxisome proliferator activator receptor-gamma ligands, 15-deoxy-Delta(12,14)-prostaglandin J2 and ciglitazone, reduce systemic inflammation in polymicrobial sepsis by modulation of signal transduction pathways. J Immunol. 2003;171(12):6827–37. doi: 10.4049/jimmunol.171.12.6827. [DOI] [PubMed] [Google Scholar]
- 34.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature. 1998;391(6662):79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 35.Zingarelli B, Cook JA. Peroxisome proliferator-activated receptor-gamma is a new therapeutic target in sepsis and inflammation. Shock. 2005;23(5):393–9. doi: 10.1097/01.shk.0000160521.91363.88. [DOI] [PubMed] [Google Scholar]
- 36.Straus DS, Glass CK. Anti-inflammatory actions of PPAR ligands: new insights on cellular and molecular mechanisms. Trends Immunol. 2007;28(12):551–558. doi: 10.1016/j.it.2007.09.003. [DOI] [PubMed] [Google Scholar]