Abstract
Cryo-electron microscopy is expanding its scope from macromolecules towards much larger and more complex cellular specimens such as organelles, cells and entire tissues. While isolated macromolecular specimens are typically composed of only very few different components that may be recognized by their shape, size or state of polymerization, cellular specimens combine large numbers of proteinaceous structures as well as nucleic acids and lipid arrays. Consequently, an unambiguous identification of these structures within the context of a whole cell may create a very difficult challenge. On plastic-embedded specimens, or Tokuyasu sections, epitopes that are exposed at the surface can be tagged by antibodies. However, vitrified sections have to be kept at strict cryo-conditions (below −140° C) and therefore do not allow any post-sectioning treatment of the specimens other than data acquisition in the microscope. Hence, the labels have to be placed into the specimen before freezing. Here we report on the application of a small metal-clustering protein, metallothionein (MTH), as a clonable label capable of clustering metal atoms into a high-density particle with high spatial resolution. We tested MTH as a label for kinesin-decorated microtubules (MTs) as well as the building blocks of desmin intermediate filaments (IFs).
Introduction
Recent years have seen a strong resurgence of interest in biological electron microscopy (EM, reviewed in Hoenger &McIntosh, 2009). In particular, cryo-electron microscopy (cryo-EM; Dubochet et al., 1988) based 3-D data analysis and cryo-electron tomography (cryo-ET: for examples see: Medalia et al., 2002; Nicastro et al., 2006; Cope et al., 2010) is increasingly popular for structural and functional investigations into macromolecular and cellular structures (reviewed in: Lucic et al., 2005). Technological advances in every aspect of the work from sample preparation and instrumentation to image acquisition and analysis have facilitated this renaissance. In addition, cryo-EM, which for a long time has been focusing predominantly on isolated macromolecular assemblies and viral particles now invades the field of cellular microscopy as well. This is achieved by preparing tissues, cells and cellular organelles by vitrified sectioning. Although this technique has been introduced quite some time ago by Christensen (1971), and a little later by McDowall et al. (1983) it only resurfaced again through the work of Al-Amoudi et al. (2004) and Hsieh et al. (2006; see also Bouchet-Marquis et al., 2006; Dubochet et al., 2007).
The desire to localize proteins in structures found in isolated complexes or in sections of cells is not new, and a variety of strategies have been developed previously. Most approaches are based on using antibodies (Kellenberger et al., 1987; reviewed in: Giddings et al., 2010), or chemical linkage of gold-maleimide clusters to exposed cysteines (e.g. see Hainfeld, et al., 1990; Milligan et al., 1990). However, with antibodies there are delivery issues and problems with compromising preservation to retain antigenicity. Delivery can be solved by utilizing specific antibody labels before embedding the samples in a polymerizable resin. Thereby the antibodies can get to the antigens easily but structural preservation is compromised (e.g. see Cooke et al., 1997). Macromolecular structures are better preserved in post-embedding labeling of the surface of sections but this limits delivery of antibodies to antigens located near the surface of the section (Kellenberger et al., 1987). Maleimide linkers require exposed cysteines often specifically placed in artificial Cys-light mutants (a construct that has been striped by all cysteines but a specifically placed one that is used for linking probes to it) that may compromise the structure of a protein domain. These strategies are definitely not optimal, particularly for tomography, which provides valuable information about biological structure by using comparatively thick sections.
Clonable tags that can be added to a gene of interest offer a solution to the delivery problem (see Wendt et al., 2002; Skiniotis et al., 2003), as so often demonstrated with fluorescent labels (e.g. GFP) in light microscopy. Excellent preservation techniques can be used with clonable tags, which are permanently attached to 100% of the target protein. While clonable tags solve the delivery issue, there is still the significant problem of creating sufficient electron density at the tag to allow its detection by EM. The density of protein labels alone does not stand out in a complex cellular environment. Previous approaches are based on photo-conversion of diaminobenzidine (DAB) mediated by GFP (Monosov et al., 1996; Grabenbauer et al., 2005) or by the biarsenical reagent ReAsH that binds a tetra-cysteine tag (Griffin et al., 1998). However, these approaches exhibit low spatial resolution, and none of them can be applied to vitrified specimens.
In this work we try to forgo photo-conversion and create high-density labels for imaging using metal clusters (here we used zinc and gold). An obvious candidate protein is metallothionein (MTH; Fig. 1; Sano et al., 1992; Sawyer et al., 1992; atomic NMR structure (4MT2): see Fig. 1 & Braun et al., 1992). These proteins are ubiquitous in eukaryotes where they bind a variety of metals often as a means of detoxification in peroxisomes and other compartments (Klaassen et al., 1999). It has been previously shown that MTHs are capable of binding gold atoms in high stoichiometry (Laib et al., 1985; Li et al., 1996). MTH genes are available and can be added to genes of interest to produce chimeras that may allow for direct gold labeling of the fusion protein via its MTH moiety. Mercogliano & DeRosier (2006) have demonstrated that purified MTH proteins can form gold clusters that can be imaged by EM. Here we demonstrated their visibility when cloned to components of macromolecular complexes (Fig. 5). Gold binding by MTH has been demonstrated by both ESI and MALDI mass spectrometry. These techniques revealed that recombinant maltose binding protein with two attached MTH domains next to each other (as we tested here as well) could cluster upwards of 40 gold atoms (Mercogliano & DeRosier; 2006). This would be nearly as much gold as is found in the EM reagent, Nanogold® (Nanoprobes Inc, Yaphank, NY).
Figure 1.
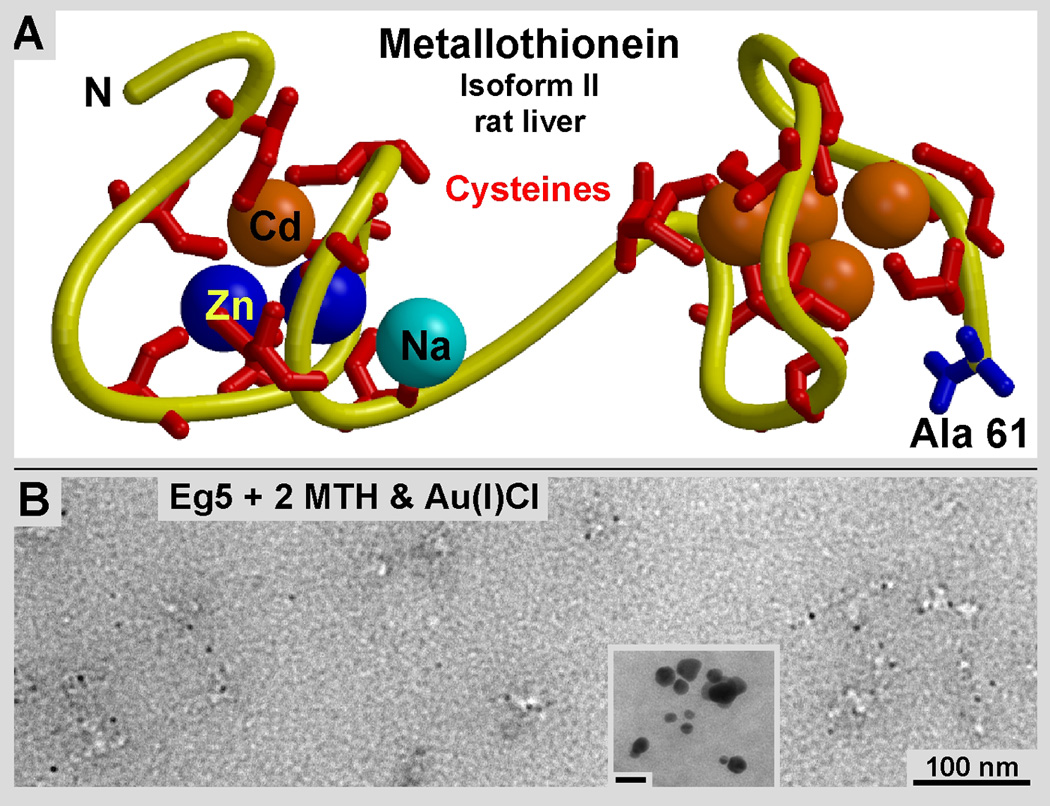
Structure and metal-clustering properties of MTH. A: Atomic structure of rat liver MTH (isoform II) at 2.0Å resolution (PDB: 4MT2; Braun et al., 1992). The structure was solved with the help of to five Cadmium ions (Cd: orange) clustered here within the molecule. MTH contains twenty cysteine residues that achieve the clustering of twenty or more gold atoms at the positions of the ions found here within the crystal structure. B: Purified Eg5 motor domains with two copies of MTH cloned to its C-terminal end (Eg5-2MTH). The sample was incubated with a 10 mM Au(1)Cl solution that was reduced by the MTH domains and formed small gold clusters in the order of 2–3 nm. Note that the clusters are all associated with protein mass. No self-reduction has occurred. However, when left alone without protein the gold solution tends to reduce itself into large clumps (inset in B), which fortunately are absent when MTH domains are present (B).
Figure 5.
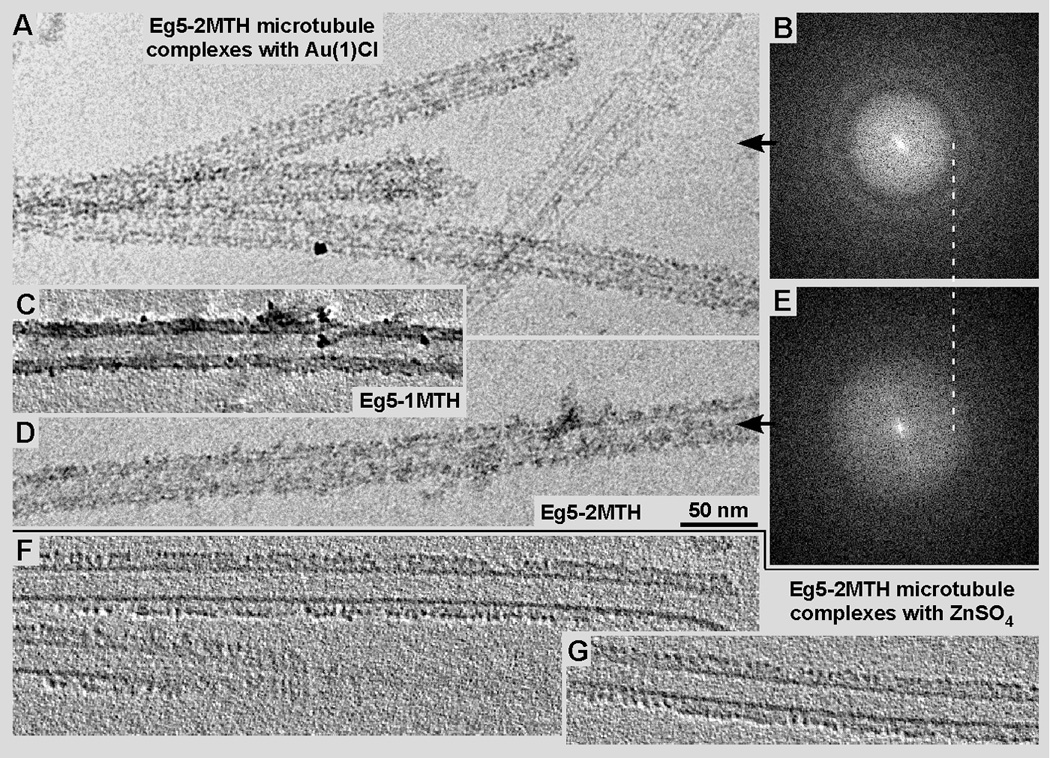
Cryo-EM (A & D) and cryo-ET (C, F & G) of microtubules decorated with Eg5-2MTH chimeras, after incubation with either a Au(I)Cl solution (A, C & D) or a ZnSO4 solution (F, G). Generally, the zinc solution (F, G) appears to better preserve the microtubule-motor complex structure than the gold solution (A, C & D). The visibility of the gold clusters varies slightly with the EM defocus chosen. Panel A was recorded at −4.5 µm defocus while panel D was taken at about −2.5 µm (see the corresponding diffraction patterns in B and E respectively). The MTH gold clusters form a stronger signal than the MTH-zinc particles, however, the intrinsic order of the motor-microtubule complex is changed visibly when incubated with gold (see tomographic ~9 nm slice in C) while the zinc clusters line up nicely (F, G).
Unfortunately our current success with MTH labeling is still limited to isolated macromolecular assemblies, while our attempts to express MTH in cells and form metal clusters in situ is still in its early development. Over-expression of MTH seems to produce stress to living cells. Hence recombinant expression of MTH in bacteria (e. g. E. coli) and other expression systems for protein purification or tagging structures in a cellular environment is still an emerging process. In both cases it seems that the presence of excess MTH alters the metal metabolism of the cells for the worse. If this problem can be overcome and normal cell-growth can be assured routinely, the next hurdle to overcome is the toxicity of gold and other metal solutions required for the formation of visible clusters in the tagged protein of interest inside living cells.
The issues discussed above may be the reason why there are only very few reports to date on MTH applications as molecular labels in cells (mostly E. coli) that have been studied in situ by EM (Nishino et al., 2007; Fukanaga et al., 2007; Diestra et al., 2009 a & b). So far we successfully cloned MTH to Eg5 kinesin motor domains (Fig. 2) and the intermediate filament (IF) protein desmin. The labeled Eg5 constructs were later complexed to microtubules for structural analysis by established methods (e.g. see: Krzysiak et al., 2006). Desmin-MTH molecules were purified for the polymerization into IFs (e.g. see: Kirmse et al., 2010). We were able to form visible gold clusters along the outer surface of desmin IFs as well as gold and zinc clusters on kinesin-MT complexes (see Fig. 3). Their density and precise location was analyzed by cryo-EM, cryo-ET, helical 3-D reconstruction (Figs. 3–5: DeRosier and Moore, 1970) and statistical difference mapping (Fig. 4: Milligan & Flicker, 1987).
Figure 2.
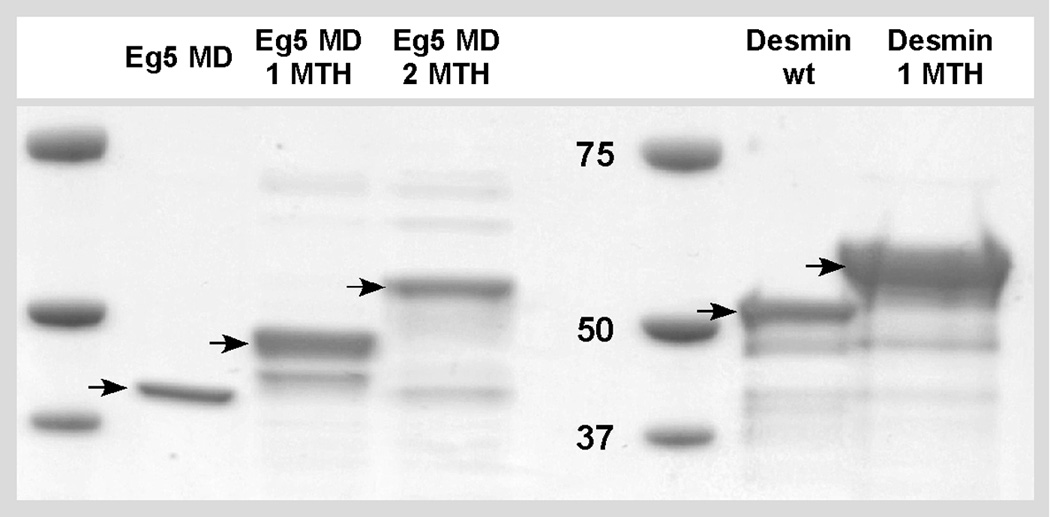
SDS-PAGE showing the Eg5 and IF constructs used: The Eg5–365 motor domain alone has a molecular weight of 42 kDa. The Eg5-1MTH chimera is 49 kDa, and Eg5-2MTH accounts for 56 kDa. Desmin wild-type is 53.5 kDa while desmin-1MTH forms a polypeptide of 60.5 kDa.
Figure 3.
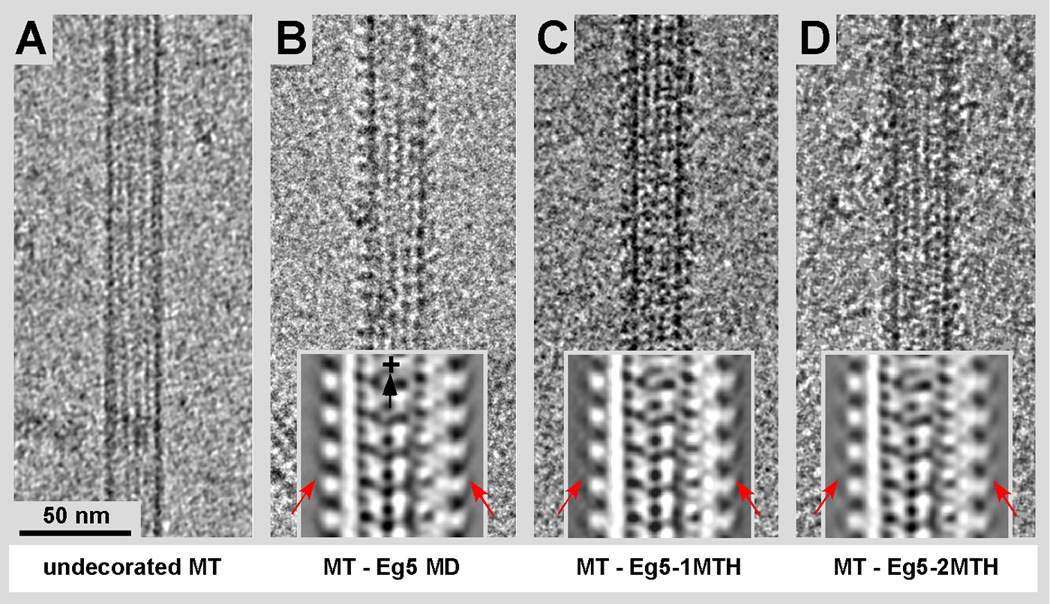
Cryo-EM of MTs complexed with Eg5 motor domains and Eg5-MTH chimeras. No gold solution or other metals have been added at this stage. The motor decoration of the MT binding sites was at or very near to stoichiometric ratios. A) Undecorated MT showing the moiré pattern of the 15-protofilament type. B) MT decorated with wild-type Eg5 motor domains (residues 1–365). C) MT decorated with the Eg5-1MTH motor domain chimera (MTH at the C-terminus) and D) two MTH domains in the same position (Eg5-2MTH). The insets show xz-projections of the corresponding 3D maps obtained from an average of 15-protofilament MTs. The MT-plus end is at the top. Arrows point at the regions where the additional mass from the MTH domains is visible.
Figure 4.
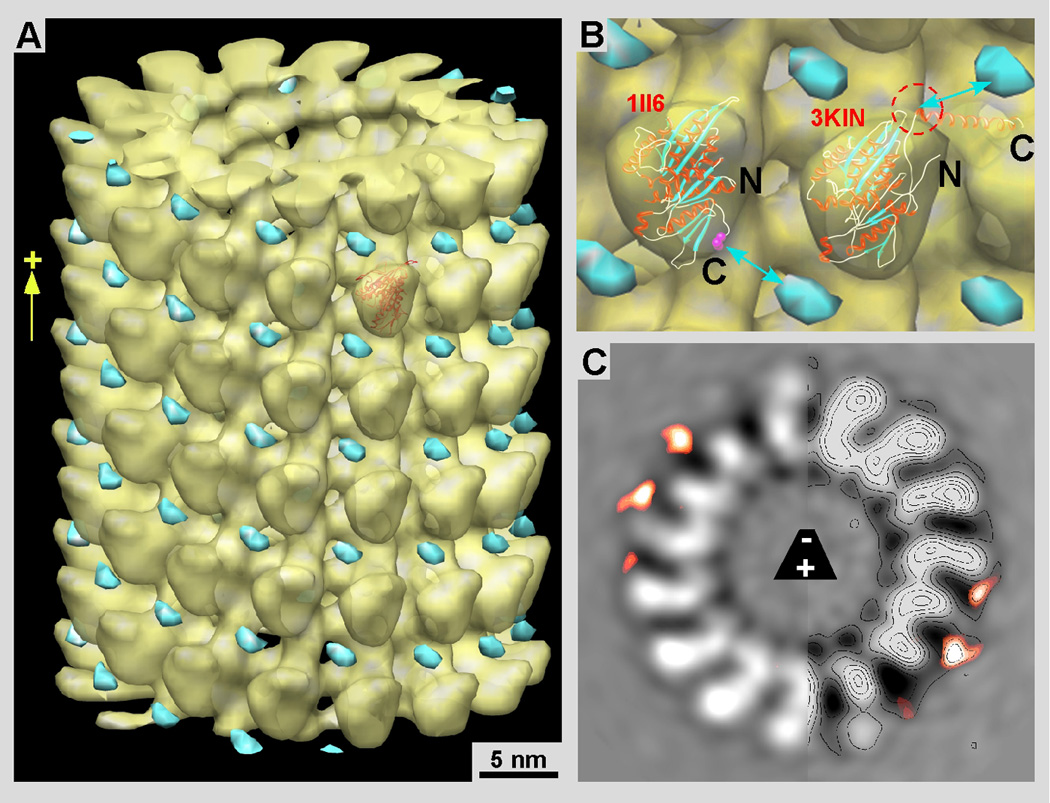
Difference mapping between wild-type and chimeric constructs reveals the location of the MTH domain. A) 3-D map of MT-Eg5 motor domains (yellow) overlaid with the difference density map (cyan) between MT-motor complexes of Eg5-MD and Eg5-2MTH. All the maps have been recorded from microtubule-motor complexes in the presence of 2mM AMP-PNP. B) Panel B shows the molecular docking of the kinesin motor-domain X-ray structures of Eg5 (left; 1II6: Turner et al., 2001) and rat kinesin-1 (right, 3KIN, chain A: Kozielski et al., 1997). We have added the rat kinesin structure for comparison to the Eg5 structure because the rat kinesin data shows the neck linker in the forward locked position. This configuration is expected for an ATP state and seems more likely than the backwards-pointing neck-linker in the Eg5 structure (the C-terminal Val-365 is marked in magenta). With the MT plus-end pointing up, the configuration of the kinesin neck-linker implies the connection between the head and the MTH domain (cyan) to go towards its upper right position. C) A 0.38 nm slice through the helical 3-D map of MT-Eg5 motor domains overlaid with the difference map as in A and B.
Results & Discussion
Expression and purification of Eg5 motor domains and Eg5-MTH constructs
Expression and purification of the chimeric constructs Eg5 motor domain (Eg5-MD) with a single MTH domain (Eg5-1MTH) as well as Eg5 motor domain with two MTH domains in tandem (Eg5-2MTH) proved to be rather problematic at the beginning. Eg5-2MTH was constructed with the intent to enlarge the metal cluster and thereby increase its visibility. Our initial experiments were carried out with the expression vector pRSETa (see M&M) that has been used for Eg5 motor domain expression elsewhere (e.g. see: Krzysiak et al., 2006). However, the expression of the Eg5-MTH constructs was low and the purification following the protocol for “Purification of Motor Proteins” (see M&M), using a S-Sepharose ion exchange column that was followed by a Ni-NTA column, yielded very low amounts of protein that were not sufficient for our binding experiments. Furthermore, cloning of the constructs into a pETa+ vector from Novagen was not achieving good results either. Although, with this vector expression of all of the three constructs was high, most of our protein was insoluble and ended in inclusion bodies from where it could not be re-folded properly. In addition, the cells often underwent a cycle of lysis before growing steadily, which renders the culture useless. One of the possible scenarios for that behavior might be that MTH scavenges too much metal ions from the cytosol and lowering their natural levels. This seems to be an intrinsic problem of metallothionein (MTH) as it is capable of adsorbing several different metals such as gold, zinc, sodium and cadmium (see Fig. 1; Braun et a., 1992).
The problems described above mostly disappeared after changing the vector for all three Eg5 constructs to the pQE80L expression system (see M&M). Now we were able to produce reasonable amounts of protein at good purity for all three constructs (Fig. 2). The expression levels of the proteins was high and the purification, desalting and concentration of the final proteins yielded high amounts of purified protein (1–1.5 mg/ml) that were sufficient for the metal clustering experiments shown below. Essentially, we followed again the protocol for “Purification of Motor Proteins”, using a S-Sepharose ion exchange column that was followed by a Ni-NTA column. Ideally the NTA column products should be further purified by size exclusion chromatography, which worked well for the Eg5 motor domain itself and the Eg5-1MTH chimera. However, size exclusion chromatography with Eg5+2MTH construct failed for yet unknown reasons. Nevertheless, despite a sub-optimal purification for this construct it still fully decorated MTs and allowed us to produce a conclusive 3-D map (Figs. 3 insets in B–D, and Fig. 4).
Microtubule decoration with Eg5-1MTH and Eg5-2MTH chimeras
Once the expression of the Eg5-MTH constructs was under control we were able to produce fully decorated MTs with all of the three constructs, Eg5-MD, Eg5-1MTH as well as Eg5-2MTH (Fig. 3B–D). Figure 3A shows an undecorated MT for comparison. All three constructs were further analyzed by cryo-EM and helical 3-D reconstruction or cryo-ET 3-D reconstruction. Thereby we could reproduce the typical regular binding pattern for all three constructs with the same binding configuration previously reported for Eg5 motor domains (e.g. see: Krzysiak et al., 2006) and other kinesin motor domains that all bind to the MT surface in a very similar conformation (reviewed in: Vale and Milligan, 2000).
3-D reconstruction and difference mapping reveals the location of the cloned MTH domains
We calculated 3-D maps by helical reconstruction from MTs decorated with Eg5 motor domain, and MTs decorated with Eg5-1MTH as well Eg5-2MTH. We determined statistically significant difference densities between the Eg5-MD map (Fig 4A & B, yellow; Fig. 4C grey) and Eg5-2MTH maps by a student T-test procedure optimized for EM 3-D data (Flicker & Milligan, 1987). The Eg5-MD map consisted of approximately 17,000 asymmetric units (36 individual datasets) while the Eg5-2MTH map was constructed from approximately 25,000 units. These quantities were sufficient to obtain a clearly identifiable difference density (Fig 4A & B, cyan; Fig. 4C, orange) that could be related to the MTH domains with a confidence level of 99.9%. The densities that come from the added MTH domains are also visible on the grey-level 3-D map representations within the insets of Figure 3 (compare insets Fig. 3B to C and D in particular (red arrows)). The difference mass locates almost exactly in-between adjacent axial rows of motor domains, right above the groove between protofilaments (Fig. 4). Hence, we docked the Eg5 and kinesin-1 motor domains into the motor head density according to the consensus orientation of a kinesin motor head as determined for many different kinesins (Vale and Milligan, 2000). For the molecular docking attempt in Figure 4B we used the 3KIN coordinates from Kozielski et al (1997: PDB 3KIN; chain A from the dimeric structure) next to the Eg5 monomer structure (such as PDB 1II6 that shows a monomeric Eg5 construct in the presence of MgADP; Turner et al., 2001). In the 3KIN structure the neck-linker (the polypeptide stretch between helix α6 and the neck coiled-coil) reflects more closely the configuration that is assumed for an ATP and AMP-PNP state, which is in a so-called “locked” position (Rice et al., 1999), despite having MgADP bound to the active site. Since our maps were obtained from MT-motor complexes in the presence of AMP-PNP, the neck-linker should be in this “locked” position. This constitutes a state where in the case of a dimeric motor complex both heads would be bound to the microtubule surface along a single protofilament as proposed for kinesin-1 in Hoenger et al. (1998) and confirmed for Eg5 dimeric head domains by surface shadowing (Krzysiak et al., 2006). A binding configuration with both heads touching the protofilament surface has the ATP head trailing with the neck-linker locked in a forward pointing position (towards the MT plus-end), and the nucleotide-free head leading towards the MT plus-end with its neck-linker pointing backwards to the trailing head (Hoenger et al., 1998; Skiniotis et al., 2003).
However, we cannot unambiguously determine the conformation of the neck-linker in our reconstructions, which are limited to a spatial resolution of about 2.5 nm. Therefore, if in our maps the neck-linker should be in a configuration similar to the 1II6 Eg5 map then it may well be that the connection is opposite to the suggestion we made in Figure 4B. This is not very likely, however, since the Eg5 1II6 structure has been determined in the presence of MgADP, and seems closer to a nucleotide-free state where the linker is dislocated (Rice et al., 1999) or, in the dimeric binding configuration connects backwards with the trailing head (Hoenger et al., 1998; Skiniotis et al., 2003).
Up to here the MTH domains in an empty configuration proved equally valuable as the SH3 domains we have used previously in combination with monomeric and dimeric rat kinesin (Skiniotis et al., 2003) and dimeric ncd (Wendt et al., 2002). The electron density of empty MTH domains is comparable to the surrounding protein structures and therefore barely visible in an in vitro approach as the one presented here, though averaging and difference mapping may determine their location unambiguously. However, within the dense and heterogeneous environment of an intact cell empty MTH domains would not be detectable.
Cryo-EM and Cryo-ET analysis of microtubule-Eg5-MTH complexes incubated with gold or zinc solutions
Obviously the main purpose of this project was to establish MTH with its metal-clustering capabilities as a high-electron density label with high spatial resolution for in vitro and in situ structural analyses of macromolecular complexes and cellular organelles. MTH has been shown to reduce several different metal ions into clusters and is therefore less selective than metal-clustering peptides (reviewed in: Heinz et al., 2009) which could provide a basis for clonable electron dense tags as well. Here we experimented with solutions of mono-valent gold in the form of Au(I)Cl and divalent zinc in the form of ZnSO4. We succeeded in placing metal-clustered labels to MT-motor complexes (Fig. 5A) and desmin intermediate filaments (Fig 6) incubated with Au(I)CI (Figs. 5A–E, & Fig. 6B), and in the case of the MT-motor complexes with ZnCl4 as well (Fig. 5F, G).
Figure 6.
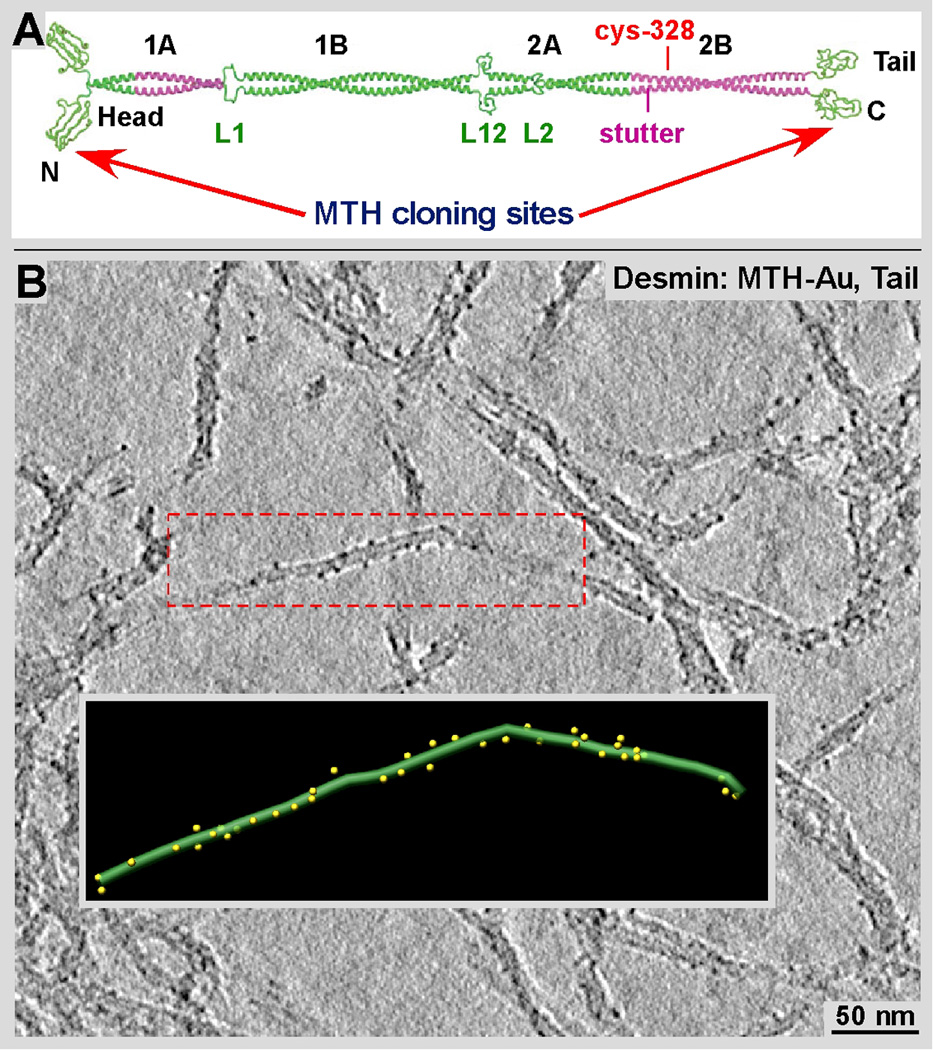
Gold clusters formed along desmin-MTH IFs. A) MTH domains were attached to monomeric desmin either at its N- or C-terminus. Desmin, like most other IF proteins form a strong coiled-coil dimer (adapted from Parry et al., 2007) Hence each desmin dimer carries two MTHs at ends of coiled-coil. B) Tomographic slice of desmin-MTH IFs after incubation at a final concentration of 200 µM of Au(I)Cl. The gold clusters are well visible along the outer surface of the filament. The inset shows a surface-rendered representation of the area marked by the red frame.
The advantage of gold is its high electron scattering potential that creates a well visible particle out of 40 or less atoms (Fig. 1B, comparable to a 1.4 nm Nanogold® particle (Nanoprobes Inc, Yaphank NY)). The disadvantage of Au(I)Cl is its solubility that has a realistically workable maximum at about 10 mM, which, in this form is not stable for longer than a few hours. Reduced gold-clusters may form easily by self-reduction as judged from the color change of the solution and with EM observations (see Fig. 1, inset). To our fortune, in the presence of Eg5-MTH constructs the gold-formed small, 2–3 nm clusters at the MTH domains and did not aggregate into large patches shown in Figure 1B inset. Furthermore, gold in excess is toxic to cells and may even disrupt large macromolecular assemblies, within cells but also in vitro (see Fig. 5A–E). We have incubated MTs decorated with Eg5-1MTH (Fig. 5C) and Eg5-2MTH (Fig. 5A & D; corresponding diffraction patterns: B & E). While the decoration with the MTH constructs worked flawlessly as demonstrated in Figures 3 and 4, the addition of a 10 mM Au(I)Cl solution produced high-density particles along the MT surface (Fig. 5A, C, D), but disrupted the underlying regularity of the MT-motor complex. Diffraction patterns at different defocus values (compare Fig. 5A–B with 5D–E) showed very little reflections that could be assigned to the axial 8 nm repeat of tubulin dimers decorated with motor domains. Cryo-ET of MTs complexed with Eg5-1MTH showed a similar picture. The edges of MTs in tomographic 9 nm slices showed high densities attributable to gold clusters but very little regularity that would reflect the axial tubulin dimer-motor repeat.
Zinc sulfate proved to be the much easier metal to work with. Not only is its solubility much higher, ZnSO4 remains stable in solution for a long time and does not reduce itself as quickly as Au(I)Cl. Also, the MT-motor complexes tolerated the zinc solution much better than the gold solution (compare Fig. 6F & G with A, C and D). Furthermore, by using ZnSO4 we were able to form MTH-metal clusters already during expression by adding 2 mM ZnSO4 (final concentration) to the medium. The E. coil cells that expressed the Eg5-MTH construct tolerated that salt much better than Au(I)Cl and we were able to form zinc clusters in situ. Zn-loaded Eg5-MTH protein could be purified and readily complexed to MTs. Cryo-ET revealed the zinc clusters as clear densities that follow nicely the 8 nm axial repeat along protofilaments (Fig. 5F&G). However, zinc clusters are much lower in density and therefore much harder to detect in a crowded environments like in the cytosol and cellular organelles. Zn ions (conc.: 0.25 mM) have been used to grow so-called Zn-sheets in vitro that exhibit an unnatural protofilament arrangement (Crepeau et al., 1977). Obviously, this may be a concern for using zinc in eukaryotes and therefore specimens should be monitored for potential effects of zinc on tubulin within the cytosol.
Tomographic 3-D analysis of desmin-MTH intermediate filaments incubated with Au(I)Cl
MTH should in the long run be useful as a general tag for EM. Thus, we did not confine our studies to one single macromolecular system and also tested the feasibility of MTH cloning and metal clustering to desmin intermediate filaments (IF). This system proved to be much easier to work with (see Fig. 6). The field of IFs has accumulated enormous amounts of physiological and clinical data over the last few decades, but it suffers from a lack of detailed structural information, both at the molecular and the atomic scales (reviewed in Herrmann et al., 2007). This is the motivation for our investigations into desmin IF structure by enhancing the visibility of the building block repeats utilizing electron-dense markers with a high spatial resolution (Fig. 5). Desmin is one of the earliest protein markers for muscle tissue in embryogenesis as it is detected in the somites of myoblasts (Bär et al., 2004). Vimentin and desmin appear to be inversely regulated during muscle cell differentiation. Both proteins are present early in the development of muscle cells but desmin is first expressed at very low levels and then increases as the cell completes differentiation. Once the muscle cell matures only desmin is present.
Our attempts to cluster gold onto the MTH domains cloned to either the N or the C-terminus was successful in both cases. Expression and purification of the desmin-MTH construct was much more straightforward than with the Eg5-MTH constructs (see M&M). Although we successfully expressed MTH on both ends (see Fig. 6A; N-terminal head group and C-terminal tail group) of the desmin polypeptide chain here we show the results for the C-terminal desmin-MTH clone only (Figure 6). Clearly visible gold-clusters line the outer surface of the desmin filaments (Fig. 6 B), although in a somewhat irregular order. Hence, we will improve the decoration efficiency of these constructs by further testing different labeling conditions and different metal solutions. In general the tagged IFs appear indistinguishable from wild-type filaments in shape and length, as well as with regard to the assembly properties and dynamics. Regardless the detailed impact of the additional MTH label on the IF structure has to be elucidated quantitatively. For example the growth rate between wild-type and tagged IFs has to be characterized.
Conclusions and Outlook
In our hands the use of MTH as a clonable high-density marker for cryo-EM and cryo-ET proved to be successful and promising for future attempts, but it remains a relatively tricky business with positive and negative aspects, in particular for intracellular applications. We are now able to successfully clone, express, and purify kinesin motor proteins as well as the building blocks for IFs. These components behave very similar to the wild-type constructs and can be incubated with gold or zinc to produce small clusters that are well visible by cryo-EM and may even be used as fiducial markers for the alignment of tiltseries during tomographic 3-D reconstruction. Hence, while the system seems to be promising for in vitro experiments the application of MTH in situ is still very challenging. Only very few reports exist from successful applications of MTH-mediated labeling directly within intact E. coli cells (by using Cadmium (CdCl2) on GroEL-MTH chimeras: Nishino et al., 2007; by using gold (Au(I)Cl) on Hfq-MTH chimeras: Diestra et al., 2009a & b), and in hippocampal neurons (by using Cadmium (CdCl2): Fukanaga et al., 2007). The difficulties of growing cells that express MTH at unnaturally large quantities may relate to the metal-scavenging effects of metallothionein itself when overexpressed in cells. The other problem may be toxic effects of providing a cell with enough metal solution (e.g. Ag(I)Cl, AuSO4, CdCl2, ZnSO4 etc.) that delivers sufficient ions to be reduced by MTH into visible clusters. We successfully cultured E. coli with ZnSO4, but these clusters are hard to see within a dense cellular environment. The future of clonable density markers may not only be MTH, but a variety of metal clustering peptides such as the A3 peptide (Slocic et al., 2005) and others (reviewed in Heinz et al., 2009) that exclusively cluster a particular type of metal. Clearly the demand for such labels is very present and will become even more sought after when vitrified sectioning leaves it’s stage of an emerging technology and develops into a routine application for ultrastructural investigations of cells, cellular organelles and large complex macromolecular assemblies.
Material & Methods
Cloning, expression and protein purification of Eg5-1MTH and Eg5-2MTH
The Kinesin motor domain of Eg5 (residues 1–365) was PCR amplified out of the pRSETa+2MTHs plasmid (generously provided by Alexander-Stemm-Wolf from Dr. Mark Winey lab at CU Boulder). 1MTH and 2MTHs were cloned at the C-terminus of the Eg5 motor domain with a short 6-residue linker sequence (RISRYT) between the C-terminus of Eg5 and the MTH sequence. SphI and HindIII restriction sites were introduced via PCR at the N and C termini of Eg5+2MTHs respectively. These enzymes were used for the cloning into the pQE80L vector, a 6×His tagged expression vector (Qiagen Inc., USA Valencia, CA). The amplified fragment was ligated with the digested vector (in a 5:1 molar ratio, fragment to vector) and was transformed into DH5a competent cells. The presence of the Eg5, Eg5+1MTH and/or Eg5+2MTHs was verified with SphI-HindII restriction digests and sequencing. Positive clones were transformed into the BL21-CodonPlus (DE3)-RIL cell line (Agilent Technologies, Inc. Santa Clara, CA) for protein expression experiments. A single colony was grown in 5 ml of LB (Invitrogen, USA) + kanamycin media over night (O/N) at 37° C and the next day a 1:50 500 ml culture was grown at 37° C until 0.35 OD (600 nm) and was induced with 0.4 mM IPTG. For the Zn binding experiment, 0.2 mM ZnSO4 was added during induction into the 500 ml O/N culture.
The purification protocol “Purification of Motor Proteins” was obtained from Kinesin Homepage methods page (protocol by Sharyn Endow: www.proweb.org/kinesin/Methods/bacterial_ex.html). Briefly, the cultures were grown O/N at 22° C and the protein was purified following the Qiagen protocol for “Purification under native conditions from E. Coli” using a Ni-column (Qiagen Inc., USA Valencia, CA). Immediately after purification, the fractions that contained sufficient amount of protein (Bradford reaction, Bio-Rad Laboratories, USA, Hercules, CA) were pooled and put through a PD10 desalting column to remove the Imidazole. The desalting buffer consisted of 20 mM Na2HPO4, 50 mM NaCl, 0.5 mM EDTA and 1 mM DTT at pH 8.0. The protein was concentrated using a 4 ml concentrator from Sartorius (Sartorius Stedim Biotech, Goettingen Germany). The concentrated solution was subsequently aliquoted and rapidly frozen in liquid N2 before being stored at −80° C. The small amount of the proteins (5–10 µl) that were used for each experiment was clarified from aggregates for 1 min in an airfuge at ~70,000g before every experiment. The final concentration of protein obtained for all three constructs was around 1.5 mg/ml.
Decoration of microtubules with Eg5, Eg5-1MTH and Eg5-2MTH for cryo EM, Au/Zn binding experiments, preparation of cryo-EM samples
Microtubules were polymerized for 30 min at 35° C in BRB80 (80 mM PIPES pH 6.8, 2 mM MgCl2) at a concentration of 5 mg/ml in the presence of 7.5% DMSO, 2 mM MgCl2, 1 mM GTP and 10 µM Taxol. The microtubules were left O/N room temperature to stabilize them. MTs at a concentration of 0.5 mg/ml were adsorbed to holey carbon grids previously grow discharged, for 1 min. After blotting the grid from the side, a mixture of Eg5, Eg5+1MTH or Eg5+2MTHs and 2.2 mM AMP-PNP was added on the grid for 1 min.
After blotting again the extra solution from the side, 5 µl of a 10 mM Au(I)Cl solution in plain water (pH 5) was added to the grid and the grid was incubated for 5 min in a humid chamber to prevent evaporation. The grids were plunge-frozen in liquid ethane using a homemade manual plunge freezer. To make sure that there was complete decoration, a 4:1 ration of motor to tubulin was used.
As a control, 5 µl of 10 mM Au(I)Cl (Au(I)Cl, 99% purity, VWR, USA) was added to the grid, blotted from the side washed with distilled H2O, stained with 5 µl of NanoVan® (Nanoprobes, Yaphank, NY, USA), and washed again with distilled H2O. (Fig. 1B, inset). In addition purified protein and Au(I)Cl,, was incubated at the same 4:1 ration (see above) for 2–5 min. in the absence of MTs and added to the grid (Fig. 1B).
In the case of using ZnSO4 instead of gold we were able to form the MTH-zinc clusters already in the cells before purification. To this end ZnSO4 was directly added to the expression media (described above) at a final concentration of 2mM. The purified Eg5-MTH constructs then had the clusters preformed and were directly added to the microtubules.
Electron microscopy and image processing
Cryo-EM and cryo-ET was performed on a TecnaiF20 (FEI-Company, Eindhoven, the Netherlands, and Hillsboro, OR). Samples were adsorbed onto holey carbon grids (4.0 µm holes, 2.0 µm spaces, C-Flat, Protochips Inc., Raleigh, NC, USA) with a GATAN-626 cryo-holder (GATAN Inc. Pleasanton, CA). Tomographic data recording was carried out with SerialEM (Mastronarde, 2005; available at http://bio3D.colorado.edu). Helical 3-D reconstruction was performed with the software suites Suprim (Schroeter & Bretaudiere, 1991) and Phoelix (Whittaker et al, 1995). Helical reconstruction was carried out on 15-protofilament microtubules as described in Beuron & Hoenger (2001). Tomographic 3-D reconstruction was carried out with IMOD (Kremer et al., 1996; available at http://bio3D.colorado.edu). Helical 3-D maps in figure 4 were supplemented with the X-ray structures 1II6 (Eg5 monomeric motor domain 1–365: Turner et al., 2001) and chain A from 3KIN (dimeric rat kinesin motor domain 1–372: Kozielski et al., 1997). The docking was carried out manually and the orientation was established according to Hoenger et al., 1998).
Purification, assembly, and gold labeling of Desmin Intermediate filaments
After cloning of the MTH tag to the tail domain of human desmin IF protein, desmin was produced and purified from E. coli strain BL21 CodonPlus -RIL competent cells (Agilent Technologies, former Stratagene, Santa Clara, CA, USA). Purification was done according to Herrmann et al. (1996). Desmin was stored in 8 M urea at −80° C after purification. Prior to the assembly of desmin into long filaments it was dialyzed in a stepwise fashion (8, 4, 2, and 1 M urea) into 2 mM sodium phosphate buffer at pH 7.5 (“Pi buffer”) as described previously (Mücke et al., 2004 & 2005). Assembly was initiated by adding KCl to a final concentration of 100 mM and the reaction was allowed to proceed at 37° C for 10–20 min.
Gold (I) Chloride (Au(I)Cl, 99% purity, VWR, USA) was dissolved in water at a final concentration of 10 mM and diluted into 100 mM KCl, 2 mM Pi buffer with a final concentration of 1 mM. For cryo-EM the gold was mixed at a 1:5 ratio with the assembled IFs in a volume of 10 µl and incubated for 2 min. at RT in a humidified chamber. 3–5 µl of the gold labeled filaments where then brought onto holey carbon grids (4.0 µm holes, 2.0 µm spaces, C-Flat, Protochips Inc., Raleigh, NC, USA) and plunge frozen in liquid ethane. Cryo-EM and cryo-ET was performed as described above.
Acknowledgments
We would like to thank Julia Cope and Miguel Gonzalez (both Univ. of Colorado, Boulder) for training CBM and MP on helical reconstruction methods. We are grateful to Susan P. Gilbert (Rensselaer Polytechnic Institute Troy NY) and Christopher Mercogliano (MRC-Cambridge, U.K.) for sharing the Eg5-MD and the MTH plasmids respectively with us. This project was supported by two NIH grants, 5R01-GM80993 (to A.H.) and 2P41-RR000592 (to A.H.) as well as by a grant from the German Research Foundation DFG KI 1464/2-1 to R.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, et al. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bär H, Strelkov S, Sjöberg G, Aebi U, Herrmann H. The biology of desmin filaments: how do mutations affect their structure, assembly, and organization? Journal of Structural Biology. 2004;148:137–152. doi: 10.1016/j.jsb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Beuron F, Hoenger A. Structural analysis of the microtubule-kinesin complex by cryo-electron microscopy. Methods Mol Biol. 2001;164:235–254. doi: 10.1385/1-59259-069-1:235. [DOI] [PubMed] [Google Scholar]
- Bouchet-Marquis C, Zuber B, Glynn AM, Eltsov M, Grabenbauer M, et al. Visualization of cell microtubules in their native state. Biol Cell. 2007;99:45–53. doi: 10.1042/BC20060081. [DOI] [PubMed] [Google Scholar]
- Braun W, Vasák M, Robbins AH, Stout CD, Wagner G, et al. Comparison of the NMR solution structure and the x-ray crystal structure of rat metallothionein-2. Proc Natl Acad Sci U S A. 1992;89:10124–10128. doi: 10.1073/pnas.89.21.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AK. Frozen thin sections of fresh tissue for electron microscopy, with a description of pancreas and liver. J. Cell Biol. 1971;51:772–804. doi: 10.1083/jcb.51.3.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- Crepeau RH, McEwen B, Dykes G, Edelstein SJ. Structural studies on porcine brain tubulin in extended sheets. J. Mol. Biol. 1977;116:301–315. doi: 10.1016/0022-2836(77)90218-2. [DOI] [PubMed] [Google Scholar]
- DeRosier DJ, Moore PB. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J Mol Biol. 1970;52:355–369. doi: 10.1016/0022-2836(70)90036-7. [DOI] [PubMed] [Google Scholar]
- Diestra E, Fontana J, Guichard P, Marco S, Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol. 2009a;165:157–168. doi: 10.1016/j.jsb.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Diestra E, Cayrol B, Arluison V, Risco C. Cellular electron microscopy imaging reveals the localization of the Hfq protein close to the bacterial membrane. PLoS One. 2009b;4(12):e8301. doi: 10.1371/journal.pone.0008301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Zuber B, Eltsov M, Bouchet-Marquis C, Al-Amoudi A, Livolant F. How to "read" a vitreous section. Methods Cell Biol. 2007;79:385–406. doi: 10.1016/S0091-679X(06)79015-X. [DOI] [PubMed] [Google Scholar]
- Fukunaga Y, Hirase A, Kim H, Wada N, Nishino Y, Miyazawa A. Electron microscopic analysis of a fusion protein of postsynaptic density-95 and metallothionein in cultured hippocampal neurons. J Electron Microsc (Tokyo) 2007;56:119–129. doi: 10.1093/jmicro/dfm027. [DOI] [PubMed] [Google Scholar]
- Giddings TH, Meehl JB, Pearson CG, Winey M. Electron tomography and immuno labeling of Tetrahymena thermophila basal bodies. " Methods in Cell Biol. 2010;96:117–141. doi: 10.1016/S0091-679X(10)96006-8. [DOI] [PubMed] [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nature Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Hainfeld JF, Foley CJ, Maelia LE, Lipka JJ. Eleven tungsten atom cluster labels: High-resolution, site-specific probes for electron microscopy. J. Histochem. Cytochem. 1990;38:1787–1793. doi: 10.1177/38.12.1701458. [DOI] [PubMed] [Google Scholar]
- Heinz H, Farmer BL, Pandey RB, Slocik JM, Patnaik SS, et al. Nature of molecular interactions of peptides with gold, palladium, and Pd-Au bimetal surfaces in aqueous solution. J Am Chem Soc. 2009;131:9704–9714. doi: 10.1021/ja900531f. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Häner M, Brettel M, Müller SA, Goldie KN, et al. Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Bär H, Kreplak L, Strelkov S, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- Hoenger A, Sack S, Thormählen M, Marx A, Müller J, et al. Image reconstructions of microtubules decorated with monomeric and dimeric kinesins: comparison with x-ray structure and implications for motility. J Cell Biol. 1998;141:419–430. doi: 10.1083/jcb.141.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, McIntosh JR. Probing the macromolecular organization of cells by electron tomography. Mol Biol Cell. 2009;20:963–972. doi: 10.1016/j.ceb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CE, Leith A, Mannella CA, Frank J, Marko M. Towards high-resolution three-dimensional imaging of native mammalian tissue: electron tomography of frozen-hydrated rat liver sections. J Struct Biol. 2006;153:1–13. doi: 10.1016/j.jsb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Dürrenberger M, Villiger W, Carlemalm E, Wurtz M. The efficiency of immunolabel on Lowicryl sections compared to theoretical predictions. J. Histochem. Cytochem. 1987;35:959–969. doi: 10.1177/35.9.3302020. [DOI] [PubMed] [Google Scholar]
- Kirmse R, Bouchet-Marquis C, Page C, Hoenger A. Three-dimensional cryo-electron microscopy on intermediate filaments. Methods Cell Biol. 2010;96:565–589. doi: 10.1016/S0091-679X(10)96023-8. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, J. Liu J, S. Choudhuri S. Metallothionein: An intracellular protein to protect against cadmium toxicity. Annu. Rev. Pharmacol. Toxicol. 1999;39:267–294. doi: 10.1146/annurev.pharmtox.39.1.267. [DOI] [PubMed] [Google Scholar]
- Kozielski F, Sack S, Marx A, Thormählen M, Schönbrunn E, et al. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Krzysiak TC, Wendt T, Sproul LR, Tittmann P, Gross H, et al. A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 2006;25:2263–2273. doi: 10.1038/sj.emboj.7601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laib JE, Shaw CF, 3rd, Petering DH, Eidsness MK, Elder RC, Garvey JS. Formation and characterization of aurothioneins: Au,Zn,Cd-thionein, Au,Cd-thionein, and (thiomalato-Au)chi-thionein. Biochemistry (Mosc) 1985;24:1977–1986. doi: 10.1021/bi00329a027. [DOI] [PubMed] [Google Scholar]
- Li H, Otvos JD. HPLC characterization of Ag+ and Cu+ metal exchange reactions with Zn- and Cd-metallothioneins. Biochemistry (Mosc) 1996;35:13937–13945. doi: 10.1021/bi961402f. [DOI] [PubMed] [Google Scholar]
- Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McDowall AW, Chang JJ, Freeman R, Lepault J, Walter CA, Dubochet J. Electron microscopy of frozen hydrated sections of vitreous ice and vitrified biological samples. J Microsc. 1983;131:1–9. doi: 10.1111/j.1365-2818.1983.tb04225.x. [DOI] [PubMed] [Google Scholar]
- McIntosh JR. Electron microscopy of cells: a new beginning for a new century. J. Cell Biol. 2001;153:F25–F32. doi: 10.1083/jcb.153.6.f25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercogliano CP, DeRosier DJ. Gold nanocluster formation using metallothionein: mass spectrometry and electron microscopy. J. Mol. Biol. 2006;355:211–223. doi: 10.1016/j.jmb.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Milligan RA, Flicker PF. Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol. 1987;105:29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan RA, Whittaker M, Safer D. Molecular structure of F-actin and location of surface binding sites. Nature. 1990;348:217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- Monosov EZ, Wenzel TJ, Luers GH, Heyman JA, Subramani S. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J. Histochem. Cytochem. 1996;44:581–589. doi: 10.1177/44.6.8666743. [DOI] [PubMed] [Google Scholar]
- Moor H. Theory and practice of high-pressure freezing. In: Steinbrecht RA, Zierhold K, editors. Cryotechniques in Biological Electron Microscopy. Berlin: Springer-Verlag; 1987. pp. 175–191. [Google Scholar]
- Mücke N, Kreplak L, Kirmse R, Wedig T, Herrmann H, et al. Assessing the flexibility of intermediate filaments by atomic force microscopy. J. Mol. Biol. 2004;335:1241–1250. doi: 10.1016/j.jmb.2003.11.038. [DOI] [PubMed] [Google Scholar]
- Mücke N, Kirmse R, Wedig T, Leterrier JF, Kreplak L. Investigation of the morphology of intermediate filaments adsorbed to different solid supports. J. Struct. Biol. 2005;150:268–276. doi: 10.1016/j.jsb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Nishino Y, Yasunaga T, Miyazawa A. A genetically encoded metallothionein tag enabling efficient protein detection by electron microscopy. J Electron Microsc (Tokyo) 2007;56:93–101. doi: 10.1093/jmicro/dfm008. [DOI] [PubMed] [Google Scholar]
- Parry DA, Strelkov SV, Burkhard P, Aebi U, Herrmann H. Towards a molecular description of intermediate filament structure and assembly. Exp Cell Res. 2007;313:2204–2216. doi: 10.1016/j.yexcr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Sano T, Glazer AN, Cantor CR. A streptavidin-metallothionein chimera that allows specific labeling of biological materials with many different heavy metal ions. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1534–1538. doi: 10.1073/pnas.89.5.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JR, Tucker PW, Blattner FR. Metal-binding chimeric antibodies expressed in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9754–9758. doi: 10.1073/pnas.89.20.9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter JP, Bretaudiere JP. SUPRIM: easily modified image processing software. J Struct Biol. 1996;116:131–137. doi: 10.1006/jsbi.1996.0021. [DOI] [PubMed] [Google Scholar]
- Skiniotis G, Surrey T, Altmann S, Gross H, Song Y-H, et al. Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J. 2003;22:1518–1528. doi: 10.1093/emboj/cdg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocik JM, Stone MO, Naik RR. Synthesis of gold nanoparticles using multifunctional peptides. Small. 2005;1:1048–1052. doi: 10.1002/smll.200500172. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT. Immunochemistry on ultrathin frozen sections. Histochem J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- Turner J, Anderson R, Guo J, Beraud C, Fletterick R, Sakowicz R. Crystal structure of the mitotic spindle kinesin eg5 reveals a novel conformation of the neck-linker. J.Biol. Chem. 2001;276:25496–25502. doi: 10.1074/jbc.M100395200. [DOI] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Whittaker M, Carragher BO, Milligan RA. PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy. 1995;58:245–259. doi: 10.1016/0304-3991(95)00057-8. [DOI] [PubMed] [Google Scholar]


