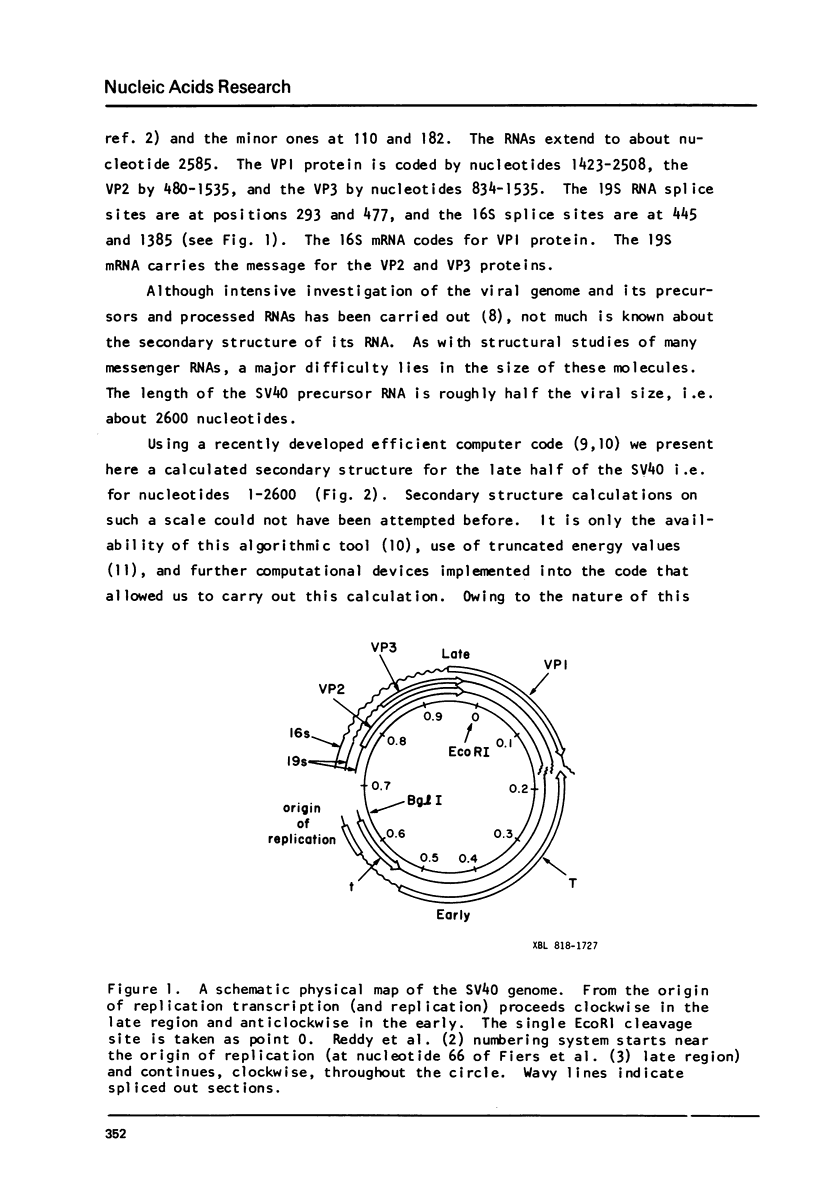
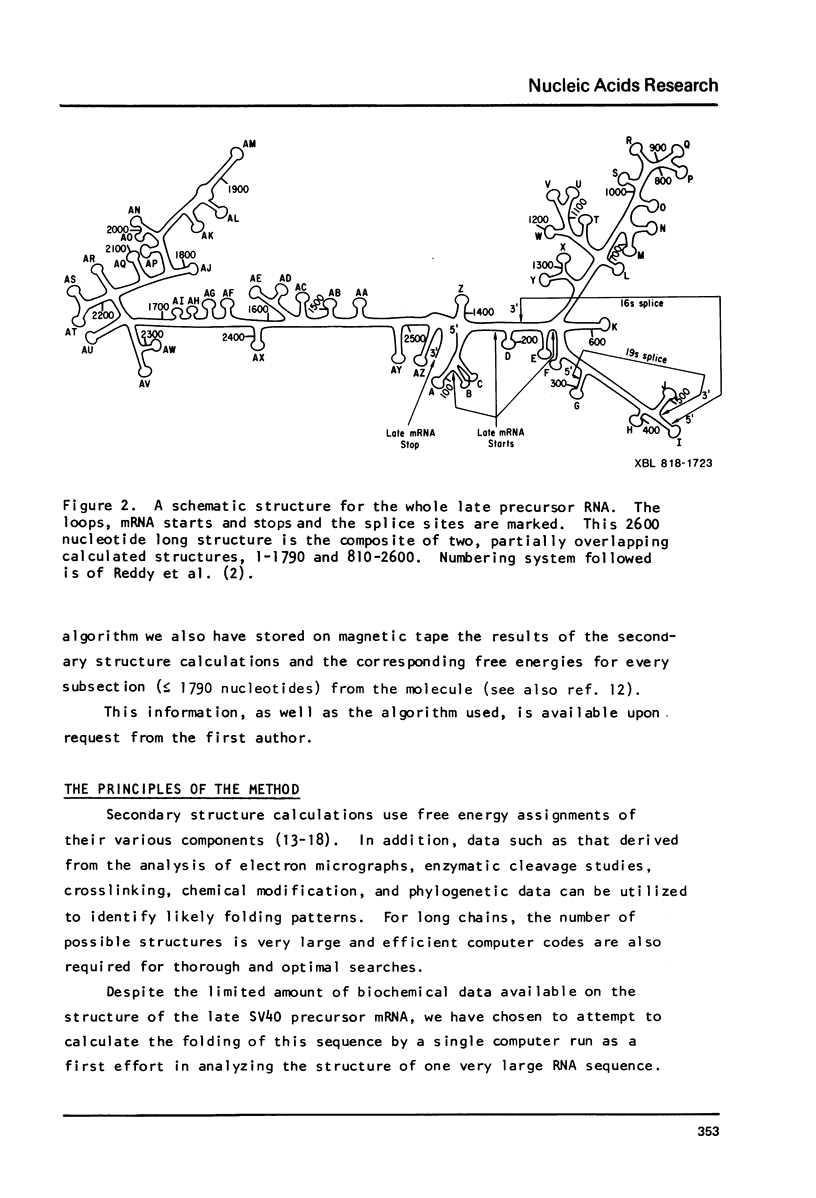
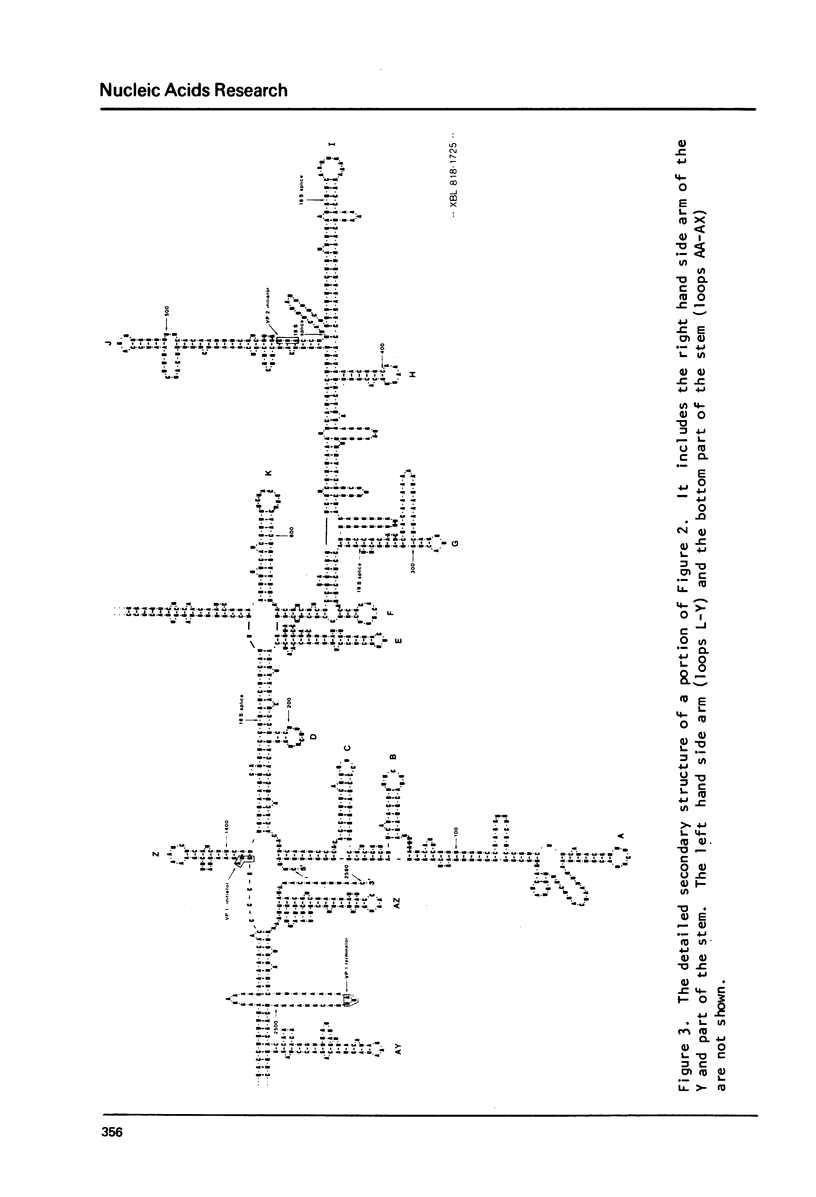
Abstract
Structures for all sequences containing less than 1790 nucleotides in the 2600 nucleotide late region of the SV40 virus have been computed and saved on magnetic tape. Previously the longest sequence whose secondary structure was calculated in a single computer run contained 950 nucleotides. In the past, analysis of long molecules required numerous repeated, partially overlapping computations on much shorter segments. The structure obtained for the late half of the SV40 is Y-shaped with two unequal arms. It has 52 short hairpins. Two long range interactions between nucleotides near 650 and 1350 and between 1450 and 2450 appear to play an important role. The first is within the 16S intron; the second is in the 3' exon. The 5' and 3' ends of the molecule are close to each other and are found in the major elongated stem in the vicinity of the fork.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bina-Stein M., Thoren M., Salzman N., Thomspon J. A. Rapid sequence determination of late simian virus 40 16S mRNA leader by using inhibitors of reverse transcriptase. Proc Natl Acad Sci U S A. 1979 Feb;76(2):731–735. doi: 10.1073/pnas.76.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Delisi C., Crothers D. M. Prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 1971 Nov;68(11):2682–2685. doi: 10.1073/pnas.68.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Glotz C., Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980 Jun 11;8(11):2377–2395. doi: 10.1093/nar/8.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Hamer D. H., Leder P. Splicing and the formation of stable RNA. Cell. 1979 Dec;18(4):1299–1302. doi: 10.1016/0092-8674(79)90240-x. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Jelinek W. R. Mapping of inverted repeated DNA sequences within the genome of simian virus 40. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1631–1634. doi: 10.1073/pnas.74.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Dhar R., Khoury G. Mapping the spliced and unspliced late lytic SV40 RNAs. Cell. 1978 Aug;14(4):971–982. doi: 10.1016/0092-8674(78)90351-3. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Uhlenbeck O. C., Doty P. Self-complementary oligoribonucleotides: adenylic acid-uridylic acid block copolymers. J Mol Biol. 1971 Apr 28;57(2):201–215. doi: 10.1016/0022-2836(71)90341-x. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Jacobson A. B. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. Three dimensional structure and sequence homology determine splicing sites in eukaryotic precursor RNA. J Theor Biol. 1980 Apr 21;83(4):647–662. doi: 10.1016/0022-5193(80)90193-9. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Shen C. K., Hearst J. E. A technique for relating long-range base pairing on single-stranded DNA and eukaryotic RNA processing. Anal Biochem. 1979 May;95(1):108–116. doi: 10.1016/0003-2697(79)90192-1. [DOI] [PubMed] [Google Scholar]
- Shen C. K., Hearst J. E. Mapping of sequences with 2-fold symmetry on the simian virus 40 genome: a photochemical crosslinking approach. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1363–1367. doi: 10.1073/pnas.74.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Zuker M., Ebel J. P., Ehresmann C. Structural organization of the 16S ribosomal RNA from E. coli. Topography and secondary structure. Nucleic Acids Res. 1981 May 11;9(9):2153–2172. doi: 10.1093/nar/9.9.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Villarreal L. P., White R. T., Berg P. Mutational alterations within the simian virus 40 leader segment generate altered 16S and 19S mRNA's. J Virol. 1979 Jan;29(1):209–219. doi: 10.1128/jvi.29.1.209-219.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



