Abstract
The draft genome of Dietzia cinnamea strain P4 was determined using pyrosequencing. In total, 428 supercontigs were obtained and analyzed. We here describe and interpret the main features of the draft genome. The genome contained a total of 3,555,295 bp, arranged in a single replicon with an average G+C percentage of 70.9%. It revealed the presence of complete pathways for basically all central metabolic routes. Also present were complete sets of genes for the glyoxalate and reductive carboxylate cycles. Autotrophic growth was suggested to occur by the presence of genes for aerobic CO oxidation, formate/formaldehyde oxidation, the reverse tricarboxylic acid cycle and the 3-hydropropionate cycle for CO2 fixation. Secondary metabolism was evidenced by the presence of genes for the biosynthesis of terpene compounds, frenolicin, nanaomycin and avilamycin A antibiotics. Furthermore, a probable role in azinomycin B synthesis, an important product with antitumor activity, was indicated. The complete alk operon for the degradation of n-alkanes was found to be present, as were clusters of genes for biphenyl ring dihydroxylation. This study brings new insights in the genetics and physiology of D. cinnamea P4, which is useful in biotechnology and bioremediation.
Electronic supplementary material
The online version of this article (doi:10.1007/s10482-011-9633-7) contains supplementary material, which is available to authorized users.
Keywords: Dietzia cinnamea P4, Genome sequence, Biotechnology, Bioremediation
Introduction
Dietzia species are Gram-positive bacteria with high G+C content, which belong to the order actinomycetales. They are widely distributed in nature, as they have been isolated from soil, deep sea sediment, soda lakes, reed rhizomes, skin and the intestinal tracts of marine fish (Yassin et al. 2006). Dietzia cinnamea strain P4 was isolated in a study on the diversity of hydrocarbon degraders in a tropical rainforest soil in Brazil, with no history of oil contamination (Evans et al. 2004). In subsequent work, von der Weid et al. (2007) demonstrated that D. cinnamea strain P4 is able to degrade a wide range of n-alkanes, pristane as well as phytane, whereas it grows in mineral liquid media amended with aromatic hydrocarbons as sole carbon sources. It thus may have applications in the bioremediation of contaminated soils.
The ability to degrade a wide range of organic compounds probably derives from the lifestyle of D. cinnamea like bacteria, which may assimilate a variety of different carbon sources and may also respond rapidly to environmental stressors, thus coming up with an excellent candidate in bioremediation processes in environments that have suffered oil spills. Recently, different strains belonging to Dietzia sp. were also described as petroleum hydrocarbon degraders, confirming their potential for the recovery of areas where oil leakage occurred (Bihari et al. 2011; Wang et al. 2011).
To the best of our knowledge, so far not a single Dietzia genome has been published. In an effort to better understand the relationship between Dietzia-like soil actinomycetes and the tropical soil habitat, including the response to oil contamination, we pyrosequenced and annotated the genome of D. cinnamea P4. Our results not only confirm the potential for bioremediation of hydrocarbon contamination, but also the ability to degrade other xenobiotic compounds and biotechnology potential, e.g. biosynthesis of compounds of industrial interest, such as frenolicin, nanaomycin and avilamycin antibiotics, the antitumor compound azinomycin, and rhamnolipids. Furthermore, a complex metabolic network tuned to energy acquisition and biosynthesis of products of commercial interest was revealed.
Materials and methods
Strain and growth
D. cinnamea strain P4 was previously isolated from samples of an acidic sandy loam (Cambisol) soil collected from an environmentally protected area of tropical Atlantic forest (Biological Reserve of Poço das Antas) located in Rio de Janeiro state, Brazil. It was cultivated and maintained in Luria–Bertani (LB; tryptone 1%, NaCl 0.5%, yeast extract 0.5%) medium at 30°C under shaking up to exponential phase. Genomic DNA was obtained from strain P4 as described by Seldin and Dubnau (1985).
Escherichia coli K12A15 wild-type is proficient in all DNA repair/damage tolerance systems. This strain was used as a positive control in tests for cellular resistance to stressors (Nghiem et al. 1988). In contrast, E. coli BH980 (mutY−) mutant strain is deficient in base excision repair mediated by the MutY protein. It was used as the positive control in tests of spontaneous mutagenesis (Nghiem et al. 1988).
Hydrocarbon degradation and growth experiments with D. cinnamea P4 were performed in minimal Bushnell Haas Broth (BHB), supplemented with the appropriate carbon source. Strain P4 was initially grown in LB broth until mid-exponential phase, then cells were harvested and cell pellets washed 3× in 1× phosphate-buffered saline (PBS). About 104 cells were used as an inoculum into 30 ml of BHB in 250-ml bottles. n-Hexadecane and benzene were added at final concentrations of 1 and 0.5%, respectively, and cultures were incubated at 28°C with shaking at 200 rpm. Samples from each bottle were taken on a daily basis to assess growth by CFU counting. All experiments were conducted in triplicate and at least two independent growth experiments were conducted for each carbon source. Growth experiments were also performed in 2-l bioreactors in aerobic conditions using 1 l of BHB supplemented with n-hexadecane at 1%. Here, we analyzed growth at pH 5–10, and temperatures from 25 to 45°C. At least two independent growth experiments were conducted for each condition.
Survival experiments
Cell cultures in exponential growth phase were exposed to a series of physical and chemical agents. Surviving cell fractions were determined by the ratio between the number of remaining cells after and the number of viable cells before each treatment. Ultraviolet A (UV-A), ultraviolet B (UV-B) and ultraviolet C (UV-C) were used as the stress factors, as follows. Cultures in the exponential growth phase (~2e8 CFU/ml in 10 ml LB) were centrifuged at 6,000×g at 4°C, washed twice with phosphate buffer (PB), as described above. For UV-C, cell suspensions were irradiated with a General Electric 15-W germicidal lamp (G15T8, 254 nm) with increasing doses. After each treatment, aliquots were taken and plated on solid LB. Plates were incubated for 24–48 h and scored for remaining CFUs. UV strength was determined by means of a Latarjet dosimeter. For UV-B, a VL215 lamp (312 nm) was used. The strength was also determined by using a VLX-3W dosimeter with a UV-B photocell. For UV-A, irradiation was performed with a VL212 lamp (365 nm), while fluence was determined with a Black Ray Dosimeter with a long wave photocell.
Chemical agents
Hydrogen peroxide (H2O2) and cumene hydroperoxide (CH)—Resistance to the oxidative agents H2O2 (inorganic) and CH (organic) was determined at three different concentrations: 0.1, 1 and 10 M. Cells were grown in LB medium as described. After this, 50 μl were taken and added to 2.5 ml of molten top agar. The mixture was poured onto LB plates and left at room temperature for 5 min. Whatman filters (#1) were placed on the plates (four per plate) and soaked with 10 μl of each H2O2 or CH solution. Plates were kept in the dark at 4°C for 1 h, then incubated for 24 h at 30°C and finally observed to determine growth inhibition halos. A relative scale was used, ranging from one to four crosses (+), respectively 2 up to 12 mm halos.
Mutagenesis
Both spontaneous and induced mutagenesis were evaluated using the rifampicin (Rp) resistance system. Cells were challenged with UV-A, UV-B and UV-C to evaluate induced mutagenesis and then monitored for the appearance of Rp-resistant mutants (Rp) over the spontaneous levels. Cells in the exponential growth phase were diluted and plated on solid LB, while undiluted aliquots were plated on LB containing 100 μg/ml Rp. After incubation at 30°C for 24–48 h, colonies were scored and mutation frequency was determined. Mutagenesis was expressed as the frequency of Rp mutants per 108 cells (Rp/108 cells).
Statistical analyses of survival and mutagenesis experiments
The results are presented as means of at least three experiments with respective standard errors. Data were analyzed by ANOVA followed by the Student–Newman–Keuls multiple comparison test using the statistical program InStat version 3.01 (GraphPad Software, San Diego, CA, USA). A 5% level of significance was adopted to evaluate the data.
Pyrosequencing, genome analysis and annotation
Pyrosequencing was performed (one run) on a FLX Roche apparatus at the University of Copenhagen, Copenhagen, Denmark. Over 500,000 reads of good quality were obtained. Following contig formation by automated software, the genome was automatically annotated, after which all annotation was manually controlled and analyzed. The total of sequenced bases was 3,555,295 which correspond to about 99% of the D. cinnamea P4 genome. Predicted protein-encoding genes were identified using GLIMMER v3.02 (Delcher et al. 2007) and EMBOSS for prokaryotes, followed by automatic annotation using search results against Pfam (Finn et al. 2010), Smart (Letunic et al. 2009), COG (Tatusov et al. 2003) and conserved Domains Database (CDD, Marchler-Bauer et al. 2010) databases. Metabolic pathways found were categorized using the KEGG database. Transmembrane domains were identified using TransportDB (Ren and Paulsen 2007) or THMMH (Krogh et al. 2001) and SignalP was used to predict signal peptide regions (Emanuelsson et al. 2007). All annotation was done manually by using NCBI BLASTP against the non-redundant database. Orthologous genes were identified by cut-off scores of e < 10−5 and 30% identity. In addition, we only considered the putative genes of D. cinnamea P4 when they showed coverage of at least 30% with orthologous genes. Insertion sequences were found by using the IS finder database (Siguier et al. 2006). Ribosomal RNA (rRNA) genes were detected by searching the whole genome sequence against rRNA databases with NCBI BLASTN and transfer RNAs by using tRNAscan-SE (Schattner et al. 2005). The G+C content was determined by using the software EMBOSS (see website). The total nucleotide sequence and annotation of D. cinnamea P4 is deposited in GenBank under accession no. AEKG01000000.
Phylogenetic analysis
Closely related sequences of Cox form I and Cox form II were recovered from the GenBank database and aligned to ZP_08023611 sequence of strain D. cinnamea P4 using the Clustal W software (Larkin et al. 2007). The phylogenetic tree was constructed using the neighbor-joining method. The MEGA 4.0 software was used to calculate pairwise P-distance value (Tamura et al. 2007). Bootstrap analysis was performed with 1,000 repetitions and scale bar indicates the distance in substitutions per amino acids.
Results and discussion
Ecology and physiology of D. cinnamea P4
The habitat from which D. cinnamea strain P4 was isolated, a tropical rainforest soil (sandy loam Cambisol; pH 5.5), revealed the presence of high levels of humic acids and heavy metals (Evans et al. 2004). The forest soil further showed oxygen characteristics that rapidly fluctuate between anaerobic/microaerobic and aerobic conditions, as a result of rapid fluctuations in moisture content. Hence, cells were likely exposed to periodic fluctuations in water and osmotic potential, impacting their physiology. Strain P4 was non-spore forming, non-motile and strictly aerobic. It was also able to grow at pH values up to 10 but not at pH below 6.0 and tempetures of 25–40°C. Furthermore, it showed growth at NaCl concentrations in the range 0–10%. Assays of degradation of different petroleum hydrocarbons (e.g. hexadecane and benzene) revealed the ability of strain P4 to use compounds from oil and diesel, in particular benzene and alkane compounds, as sole carbon sources (von der Weid et al. 2007).
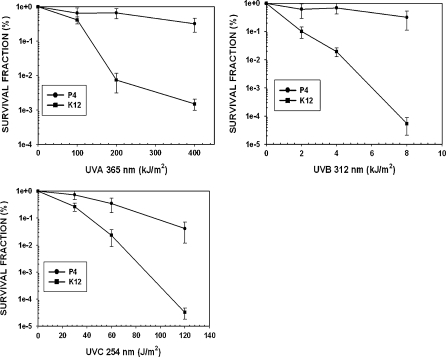
In order to evaluate the biological responses of strain P4 to environmental stresses conferred by physical and oxidative chemical mutagens, exponential phase cultures (next to those of E. coli wild type strains K12A15 and BH980) were exposed to UV-A, UV-B and UV-C irradiations, as well as oxidative stress (using H2O2 and CH). Surviving fractions in the treated populations were determined by using the ratio between the number of viable cells before and after each treatment (see Supporting information). D. cinnamea P4 revealed to be quite resistant to the stressors. In fact, it performed as well as, or better than, the E. coli wild type strains with all physical agents tested (P < 0.05), indicating that it is endowed with efficient tolerance mechanisms. In particular, its resistance to UV-A was remarkable (Fig. 1). Regarding the mutability of strain P4, spontaneous mutation to Rp resistance occurred at the same level as that of E. coli K12A15. The latter strain is not considered to be a mutator phenotype, especially when compared to strain BH980 (data not shown). Interestingly, strain P4 was extremely refractory to UV-A-induced mutagenesis, which was significantly different from that of wild-type strain K12A15 (P < 0.05). Whereas strain K12A15 revealed a 10.8-fold increase of mutation to Rp after UV-A treatment, strain P4 displayed no increase in the frequency of Rp mutants. Under UV-B, P4 was quite susceptible to mutagenesis, but to a lesser extent than K12A15, both in terms of absolute numbers of Rp mutants as in fold increase (P < 0.05). A similar response was observed between strains P4 and K12A15 after challenge with UV-C, and these two strains did not reveal the Rp mutant levels of the BH980 mutator strain. Overall, these data indicate that Dietzia cinnemea P4 is not a spontaneous mutator. It was particularly resistant to UV-induced mutagenesis, especially to the UV-A part of the solar UV spectrum.
Fig. 1.
Survival of D. cinnamea P4 and E. coli AB1157 after UV-A, UV-B and UV-C treatments. Cultures in exponential growth phase were treated with increasing doses of UV radiation (365, 312, 254 nm, respectively) as described in “Materials and methods” section
General genome features
Using high-coverage pyrosequencing on a FLX machine, we assembled the genome (coverage 30×). The sequencing yielded 565,157 shotgun reads for a total of 3,555,295 nucleotides (Table 1). The resulting assembly finally consisted of 428 supercontigs, with a length range of >1 to 61 kb. Given the difficulty to completely close the genome (due to repeats and extremely-high G+C% regions), we focused on the roughly 3.5 Mb in 1 repllicon evidenced by pulse-field gel electrophoresis, of sequence for annotation. The average G+C content of the genome was 70.9%, similar to that of the genomes of related actinomycetes, such as Rhodococcus jostii RHA1 and Nocardia farcinica IFM 10152 (Table 2).
Table 1.
Summary of genomic content of D. cinnamea strain P4
| Features | Value |
|---|---|
| Total size (bp) | 3,555,295 |
| G+C% | 70.96 |
| CDSsa | 3,328 |
| Predicted proteins | 2,665 |
| Conserved proteins | 389 |
| Orphan genes | 484 |
| rRNA operons | 2 |
| tRNAs | 49 |
aCDS: protein-encoding gene
Table 2.
Comparison between the D. cinnamea strain P4 genome and those other hydrocarbon degrader microorganisms
| Organism | Size (Mb)a | %G+Ca | Regulatorsa | Oxygenasesa | Transport proteinsa |
|---|---|---|---|---|---|
| Dietzia cinnamea P4 | 3.5b | 70.9 | 88 | 67 | 311 |
| Rhodococcus jostii RHA1 | 9.7 | 67.0 | 705 | 203 | 890 |
| Nocardia farcinica IFM 10152 | 6.3 | 70.8 | 448 | 96 | 838 |
| Bulkholderia xenovorans LB400 | 9.7 | 62.6 | 745 | 134 | 1,139 |
| Corynebacterium glutamicum ATCC 13032 | 3.3 | 65.6 | 148 | 27 | 402 |
| Acinetobacter sp. ADP1 | 3.6 | 40.4 | 188 | 52 | 368 |
| Streptomyces coelicolor A3(2) | 8.7 | 72.1 | 671 | 70 | 838 |
| Polaromonas sp. JS666 | 5.2 | 62.5 | 405 | 63 | 801 |
aData collected from www.cbs.dtu.dk/services/
bPredicted data
Similarity and domain searches showed that the D. cinnamea strain P4 genome contains 3,328 predicted protein-encoding genes (CDS). The putative CDSs encode 2,665 proteins of known function, next to 389 conserved hypothetical proteins without function (source: NCBI, NR database and CDD). A total of 484 proteins were unique to D. cinnamea strain P4, with no significant similarity to sequences that represent known functions in the database (Table 1). Two rRNA operons were found, next to 49 tRNA genes for all 20 natural amino acids. The number of rRNA operons is right within the range of such numbers across the actinobacteria, whereas the number of tRNA sites is slightly below that of actinobacteria (estimated average around 70).
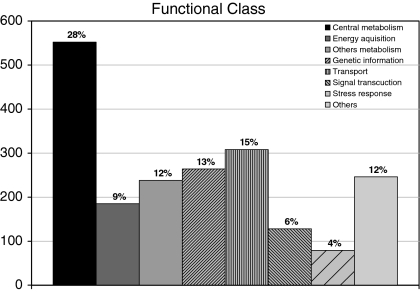
A major part of the strain P4 genome revealed suites of genes encoding putative proteins devoted to energy acquisition and compound transport (Fig. 2). We did not find any gene encoding flagellum synthesis and motility, which is consistent with the lack of motility as described by von der Weid et al. (2007). The high number of genes devoted to different carbon acquisition pathways, coupled to the versatile pathways for degradation of hydrocarbons and plant compounds, and elaborate stress response systems, indicated the presence of potential strategies that allow the survival and competitiveness of strain P4 in soil settings. The elevated number of uncharacterized genes encoding proteins of unknown function (14%) and hypothetical proteins (10%) is in accordance with findings for other soil actinomycetes (McLeod et al. 2006; Mongodin et al. 2006; Takarada et al. 2008).
Fig. 2.
Functional distribution of protein classes over the 428 contigs based on KEGG categorization and COG classifications. The “Others” column includes oxygenases, cell division, multidrug resistance and cell wall biogenesis proteins
Considering the cut-off scores described in “Materials and methods”, among the 3,328 CDSs of the D. cinnamea P4 genome, about 42% of them showed highest similarity to genes of Rhodococcus sp., 16% to those of Mycobacterium sp. and 13% to those of Corynebacterium sp., while 14% were similar to genes of Gordonia bronchiales DSM43247, N. farcinica IFM10152 and Tsukamurella paurometabola DSM20162. The high degree of similarity to genes of Rhodococcus is understandable given its taxonomic proximity. In fact, D. cinnamea strain P4 was initially classified as a member of the genus Rhodococcus (Evans et al. 2004).
Alternative energy generation pathways
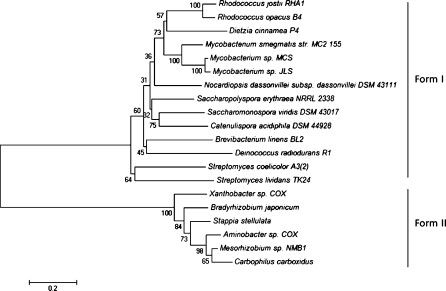
In spite of the fact that gene annotations do not necessarily imply functionality, genome sequences of other bacteria have provided insights into key potential functions. Thus, aerobic CO oxidizers have been uncovered, including several actinomycetes from soils (King and Weber 2007). We here indicate the presence of a CO oxidation pathway in the strain P4 genome by the identification of genes that likely encode the aerobic oxidation of CO to CO2, which is catalyzed by the so-called carbon monoxide dehydrogenase complex (CODH). The genome of strain P4 further revealed the presence of all genes for the large, medium and small subunits of this complex, i.e. the coxSLM genes (ZP_08023608, ZP_08023611–12) (Table S1). The distribution of these genes may differ between species (Santiago et al. 1999; Fuhrmann et al. 2003; Pelzmann et al. 2009), however the typically clustered coxSLM organization was indeed found in the strain P4 genome. Two forms of CODH, i.e. form I (which oxidizes CO) and form II (function unknown) have been described (King and Weber 2007). Our analysis also revealed that the large subunit of the Cox complex of strain P4 matched form I (Fig. 1S; Fig. 3). One putative accessory gene coxE (ZP_08022289) as well as a single xanthine and CODH maturation factor (xdhC/coxF—ZP_08023609) were also identified. The presence of accessory genes in addition to the CODH complex supports the proposition of Santiago et al. (1999), that the presence of cox accessory genes is essential, with a CuMoO2 metal centre, composed by the Cox complex, for complete CO oxidation.
Fig. 3.
Phylogenetic relationship between amino acid sequence ZP_08023611 of D. cinnamea P4 and the sequences of form I and II large CODH proteins (CoxL) of related organisms
The genome of D. cinnamea strain P4 also showed the possible use of nitrogenous organic compounds as terminal electron acceptors in oxygen-limiting conditions by dissimilatory nitrate reduction. The capacity to reduce NO3 − to NO2 − by strain P4 was suggested by the finding of large and small subunit membrane-bound NarGH complexes and the accessory soluble proteins NarIJ clustered in the narGHIJ operon (ZP_08022539–42). In addition, we found a single gene moaA (ZP_08022534), one moaE (ZP_08022522), one moeA (ZP_08022532), one moeA1 (ZP_08024513), and a single copy of moaC/mogC (ZP_08022523). These genes clustered together with a gene for multicopper oxidase, CopA (ZP_08022537), which is involved in the synthesis of molybdenum cofactor, and is essential for nitrate reductase activities (Moreno-Vivián et al. 1999). The supply of nitrate for use by the NarGH omplex in the reduction reaction is probably performed by the ABC-type transporter NarK (ZP_08022543, ZP_08024598), which clustered along the narGHIJ operon, and a second copy which was localized in another site in the strain P4 chromosome.
Autotrophic carbon fixation
The energy generated and conserved via aerobic CO oxidation can be directed to fix CO2 into biomolecules. In strain P4, this may proceed by two different mechanisms, i.e. via the reductive TCA cycle (rTCA) and/or the 3-hydroxypropionate cycle. Here, we found evidence for the ability of strain P4 to employ both metabolic pathways by the presence of the main genes that are required for them (Table S1).
Although these cycles are prevalent among anaerobic bacteria, e.g. inhabitants of hydrothermal vents, in actinomycete species it was shown that the key enzymes for autotrophic CO2 fixation can operate under microaerophilic conditions, sometimes using sulfide or hydrogen as the preferred electron donors (Rothschild 2008). Our genome analysis revealed the presence of the multisubunit pyruvate:ferredoxin oxidoreductase (ZP_08023401), phosphoenolpyruvate synthase (ZP_08024792) and phosphoenolpyruvate carboxylase (ZP_08025255), which are indicative for the presence of this branch of the TCA cycle, in addition to both 2-oxoglutarate:ferredoxin oxidoreductase subunits (ZP_08023094–5), isocitrate lyase (ZP_08022794) and citrate lyase (ZP_08024811). The latter are both constituents of the normal TCA cycle, but operating in reverse mode. Autotrophic CO2 fixation by the 3-hydroxypropionate cycle, which was first described for Chloroflexus aurantiacus (Eisenreich et al. 1993; Herter et al. 2001), is possible by a reaction in which two molecules of CO2 are converted into glyoxylate by action of the large biotin-dependent acetyl/propionyl-CoA carboxylase enzyme and malonyl-CoA reductase. In the strain P4 genome, we found one cluster of genes containing malonyl-CoA reductase next to genes for both subunits of acetyl/propionyl-CoA carboxylase (ZP_08022855–57), and the complete pathway for the biosynthesis of biotin compounds (see below). Besides these results, we assessed the utilization of CO2 at different concentrations as the sole carbon source to spur the growth of strain P4 (see supporting information).
The ability of D. cinnamea strain P4 to use C1 compounds as energy and carbon sources is suggested by our finding of genes involved in formate oxidation and formaldehyde transfer (CH2O). In formate oxidation, there are two possible mechanisms: formate can be taken up from the environment by the ABC transport system and subsequently become oxidized to CO2 by the formate dehydrogenase complex, which is encoded by the fdhABD genes (ZP_08023469, ZP_08023479–80) (Lenger et al. 1997). The second mechanism utilizes formaldehyde as the substrate, which can be converted to formate by the bifunctional enzyme 5,10-methylene-tetrahydrofolate dehydrogenase/methenyl tetrahydrofolate cyclohydrolase (ZP_08022869). The subsequent reaction catalyzes the oxidation of formate to CO2 by the formate dehydrogenase complex (Eisenreich et al. 1993). The formaldehyde transfer mechanism involves the participation of three different metabolic cycles: serine, glyoxylate regeneration and poly-β-hydroxy isobutyryl-CoA (PHB) cycles, with genes overlapping between distinct pathways. This complex mechanism found in the strain P4 genome is similar to the one described in Methylobacterium extorquens strain AM1 (Chistoserdova et al. 2003). This strain utilizes tetrahydrofolate (H4F) as a cofactor and 5,10-methylene-tetrahydrofolate dehydrogenase/methenyl tetrahydrofolate cyclohydrolase catalyzes the formation of methylene-H4F from formaldehyde and H4F. Moreover, it is directed to C1 assimilation cycles by the serine, glyoxylate regeneration and PHB cycles (Table S1).
Hydrocarbon metabolism
Alkane compounds are the most abundant components of crude oil, making up over 50% of total oil mass. Alkane molecules are chemically rather inert, but in the presence of O2 they can be activated by oxygenases and directed to complete oxidation (van Beilen et al. 2001). In a previous study, strain P4 showed to be capable of degrading different compounds present in crude oil, such as differently-sized alkanes (C-11 up to C-36) and aromatic hydrocarbons (von der Weid et al. 2007). In the current genome analysis, we identified one gene cluster containing genes for enzymes that are likely involved in n-alkane degradation. These encompassed one TetR family transcriptional regulator (ZP_08022270) and one alkane 1-monooxygenase-rubredoxin (Rub) fusion protein (ZP_08022271). We also noted the presence of the gene for a lipid transporter (ZP_08022272), which clusters with the previous genes. In agreement with the phylogenetic relatedness of the putative transcriptional regulator TetR with AlkS transcriptional regulators of several actinomycetes, this suggested a likely regulatory function of ZP_08022270 in the alkane biodegradation pathway (Canosa et al. 2000). The putative monooxygenase region revealed 96% amino acid identity with the alkane hydroxylase-Rub fusion protein of Dietzia sp. E1. A comparison with the THMMH database indicated that the protein encoded by this gene contains six transmembrane loops, which is similar to other AlkB proteins. The Rub region protein, which is involved in electron transfer, showed similarity with the RubA protein of other actinomycetes.
Analysis of the P4 genome further confirmed the presence of genes encoding enzymes involved in the biphenyl and benzoate degradation pathways. One chromosomal region was identified, which was found to contain the 2,3-dihydrobiphenyl 1,2-dioxygenase, bphB (ZP_08022749), 2,3-dihydroxybiphenyl 1,2-dioxygenase, bphC (ZP_08022738, ZP_08024903) and 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, bphD (ZP_08024298) genes. These three genes make part of the upper biphenyl pathway. In addition, we found a LysR transcriptional regulator (ZP_08022766) clustering with these genes, indicating involvement in the regulation of expression of the bphBCD operon. The metabolites resulting from the upper pathway are 2-hydroxy-penta-2,4-dienoate and benzoate. The first compound can be converted to TCA intermediates by the lower biphenyl pathway, which is encoded by bphEFG genes that were also found (ZP_08022744–45, ZP_08022748). Moreover, benzoate is converted to catechol by the benzoate dioxygenase complex, benABC (ZP_08023230–32), which is directed to the catechol branches of the β-ketoadipate central pathway (Table S1) (Gonçalves et al. 2006; McLeod et al. 2006). In addition, two genes probably encoding MFS family benzoate transporters were found (ZP_08022153), which supports the importance of this pathway for strain P4.
Other oxygenases
Oxygenases are enzymes that catalyze the insertion of oxygen atoms from O2 into an organic substrate. They consist of two types: enzymes that catalyze the insertion of a single oxygen atom, monooxygenases, and enzymes that catalyze the insertion of two oxygen atoms, dioxygenases. Reactions in which organic compounds are oxygenated have great value for organic synthesis, thus there is great interest of biotechnology industries. Genomic analysis of P4 revealed the presence of 45 monooxygenases and 22 dioxygenases (Table S2). The number of oxygenases present in the D. cinnamea strain P4 genome is relatively high, as judged by comparing the number of oxygenases per Mb across the genomes of different species (Table S2). R. josti RHA1 and Burkholderia xenovorans LB400, strains known to have large numbers of oxygenases in their genomes, have 20.9 and 13.8 oxygenases per Mb respectively, while strain P4 has 19.1. The number of cytochrome P450 genes in the strain P4 genome was nine, which is in line with that in other actinomycetes (Mongodin et al. 2006). Among the cytochrome P450 of strain P4, genes encoding linalool monooxygenase and cypX stood out. Both are involved in secondary metabolism (Table S2).
We identified coding genes for the enzyme 3-keto 9-β-hydroxylase, which is responsible for opening the ring of cholesterol compounds. In addition, we noted two coding genes for cholesterol oxidase. There are two different classes of cholesterol oxidase, each with two subclasses. Class I contains the FAD cofactor non-covalently bound to the enzyme; this class occurs mainly in actinomycetes. Class II contains the cofactor covalently linked to the enzyme, with Gram-negative bacteria as main representatives. The strain P4 genome was found to contain two different cholesterol oxidases of class I (ZP_08024621, ZP_08025358). Phylogenetic and similarity analyses suggested that CDSs ZP_08025358 and ZP_08024621 were distant from each other, being the first similar to class I-1, and the second to class I-2. The latter lacks a signal sequence, suggesting cytoplasmic localization (Navas et al. 2001). In addition, strain P4 also possessed genes described for catabolic pathways: specifically, genes encoding lactate monooxygenase, dimethylalanine monooxygenase, glycolate oxidase and lysine/ornithine monooxygenase (Table S2).
Secondary metabolism
The genome sequence of strain P4 revealed genes coding for the biosynthesis of terpene compounds, non-ribosomal peptide and polyketide synthases (PKS), which were distributed non-uniformly over the strain P4 genome. We found genes of the so-called DOXP/MEP pathways of terpenoid backbone biosynthesis, responsible for the synthesis of geranyl biphosphate and undecaprenyl diphosphate compounds (ZP_08022053, ZP_08022609, ZP_08022222, ZP_08022224, ZP_08022981–82, ZP_08022587, ZP_08023982, ZP_08024149, ZP_08025190). Subsequently, the geranyl can be converted into (S)-carvone by limonene 6-monooxygenase and carveol dehydrogenase genes (ZP_08023075, ZP_08024532). The full pathway for l-rhamnose biosynthesis was also detected in the P4 genome (ZP_08021925, ZP_08021927, ZP_08023460–62, ZP_08024663, ZP_08024641). l-rhamnose is a component of cell walls of pathogens which spurs virulence, and so this characteristic might confer virulence characteristics on this bacterium. We also found the complete pathway for the secondary carotenoid metabolite that is probably connected to UV-resistance (see below).
We identified 10 putative PKSs of PKS type II, and a single regulator of PKS expression (ZP_08022881). PKSs catalyze a decarboxylative condensation of simple metabolites like acetate, malonate, propionate, butyrate and their derivatives into growing polyketide chains (van der Ploeg et al. 2001). Microbial PKSs are divided into three major types. The PKS type I system consists of one or more multifuntional proteins that contain a different active site for each modification. PKS type II comprises individual proteins for the synthesis of aromatic products. PKS type III is composed of homodimers grouped in the synthase superfamily. In addition, there is an atypical PKS type, which catalyzes iterative condensation of extenders in a polyketide chain, called iterative type I PKS. We deduced the presence of two coding genes that likely catalyze the synthesis of new antibiotics: a gene homologous to aviGT4, the product of which is responsible for the synthesis of avilamycin A (ZP_08024273), and a gene homologous to ncnH, possibly involved in biosynthesis of the aromatic antibiotic naphthocyclinone (ZP_08022739). The latter antibiotic exhibits activity against Gram-positive bacteria (Sthapit et al. 2004). The genome annotation further identified a single copy of a gene for naphthoate synthase (ZP_08024639), a component of the interactive type I PKS family that participates in the biosynthetic pathway of Azinomycin B, a complex natural polyketide with strong antitumor activity (Brünke et al. 2001).
Stress responses and DNA repair mechanisms
The strain P4 genome showed elaborate systems that allow tolerance to environmental stresses. Also, multiple DNA repair mechanisms were found. Strain P4 may resist reactive oxygen radicals [produced by redox reactions in aerobic metabolism] using two superoxide dismutases, namely one SodA (ZP_08023526) and a single Cu–Zn SodC (ZP_08021853). Furthermore, three heme-catalases (ZP_08022106, ZP_08022939, ZP_08025226), one Mn-catalase (ZP_08022130) and two peroxidases (ZP_08023084, ZP_08021854) may be involved. We also noted the transcriptional regulator OxyR (ZP_08023122), which—at high levels of hydrogen peroxide (H2O2)—activates the detoxifying enzyme alkyl peroxidase (ZP_08023120).
The strain P4 genome was further found to contain genes involved in thiol redox control, e.g. thioredoxin (ZP_08022440, ZP_08022475, ZP_08022586, ZP_08023318, ZP_08023810, ZP_08024053, ZP_08024653) and thioredoxin reductase (ZP_08023294, ZP_08023504, ZP_08024475), glutathione peroxidase (ZP_08023200, ZP_08024106), glutathione reductase (ZP_08022216) and glutathione S-transferase (ZP_08024195). Also, we found the organic hydroperoxide resistance gene (ZP_08023119–20), which is involved in detoxifying organic oxidative agents and methionine sulfoxide reductase (ZP_08023523). In addition, we detected a gene cluster composed of genes homologous to sufBCDSE (ZP_08022081–85, ZP_08022076, ZP_08023534, ZP_08025308). This cluster is likely expressed under conditions of iron starvation and oxidative stress, working in protection or repair of [Fe–S] proteins (Zhao et al. 2008). Another important system involved in the acquisition of iron present in the genome of D. cinnamea strain P4 is the ABC siderophore transport system (Table S1).
Strain P4 probably repairs damage to its DNA through nucleotide excision, as evidenced by the finding of uvrA, uvrBC and uvrD-like helicases, genes for DNA alkylation repair and DNA glycosylases. Furthermore, pyrimidine dimer resolving enzymes, e.g. photo lyase, and recombination repair proteins—ruvAB and recAB—were found. Also, other components of the recombination apparatus, recNOQ, and other enzymes of Holliday junction resolution were found. Four copies of the Nudix family of genes (which has as a role in cleaning the cell of toxic nucleotide metabolites), were found. In addition, we noted the presence of phytoene synthetase (ZP_08023983), phytoene dehydrogenase (ZP_08022383, ZP_08023928), lycopene cyclase (ZP_08022078) and lycopene elongase (ZP_08024381) genes. These genes compose the β-carotene biosynthesis pathway, which is suggested to confer radiation resistance by scavenging electrons from reactive oxygen species (Bessman et al. 1996).
The ability of D. cinnamea P4 to protect itself from toxic effects of heavy metals was evidenced by the 32 putative coding genes identified that are potentially involved in conferring resistance against these compounds (Table S1). Genes for arsenic resistance (through extrusion) and inactivation by reduction were localized at two different sites in the genome. The first was a region composed of genes for an arsenite export protein (ZP_08022909) and two arsenate reductases (ZP_08022908, ZP_08023703), the second contained two transport systems in tandem, probably involved as an efflux pump of arsenic compounds (ZP_08023621). Also, genes for mercury resistance were detected: one putative mercury reductase protein (ZP_08021869) and one transporter system involved in heavy metal translocation (ZP_08023622). Interestingly, two transcriptional regulators were found to be localized at different sites in the genome (ZP_08021962, ZP_08024385), beside other transport systems involved in heavy metal translocation, such as copper, zinc and cadmium. Although we found three copies of terC genes (ZP_08025049, ZP_08025084, ZP_08025329), responsible for resistance to tellurium ions, other genes involved in this, e.g. terBDE, were missing. Resistance to copper was associated with the presence of the sensor genes copRS (ZP_08023436–7) and cusS (ZP_08022249), two copies of copper-translocating P-type ATPase copA (ZP_08024603, ZP_08024655) and one of copC (ZP_08021960). This suggested one mechanism of copper trafficking, triggered by redox-dependent coordination properties of the metal (Phillips et al. 2002). In spite of some metals being necessary for cell homeostasis, a single gene coding for a protein involved in the efflux of iron was detected, which probably has a role in the resistance to iron at high concentrations. Another seven ATPase efflux transporter systems for cadmium, cobalt and zinc were found in the P4 genome, showing the importance of metal resistance for strain P4 ecology (Table S1).
The genome information further showed that strain P4 may sense osmotic stress by virtue of an osmolarity sensor protein (ZP_08022040), after which a response is triggered. The internal osmotic equilibrium is probably maintained by accumulation of organic osmolytes (compatible solute strategy), which are synthesized by the enzymes choline dehydrogenase (ZP_08024621) and betaine aldehyde dehydrogenase (ZP_08023435). We also noted the presence of genes encoding transport mechanisms for these compounds (ZP_08024080–82, ZP_08024126, ZP_08024243, ZP_08024347, ZP_08024500). Also, we identified the so-called ehuA gene, which is involved in the uptake of extracellular ectoine compounds (ZP_08022970). Intracellular accumulation of trehalose and glycogen has been reported under water stress conditions. The genes in the strain P4 genome responsible for trehalose biosynthesis from glycogen were found to be clustered in the treYXZ operon (ZP_08022640, ZP_08022646–47). TreX is a glycogen-debranching enzyme, yielding 1,4-α-glucan compounds, TreY reduces the end of 1,4-α-glucan to form trehalose residues and TreZ liberates trehalose from the product of the TreY-catalyzed reaction (Arnesano et al. 2003). In addition, the presence of an aquaporine Z gene (ZP_08024724) suggests the role of control of water movement in efforts to osmotic stress tolerance.
One of the desirable characteristics in a strain with potential for bioremediation/biocatalysis is tolerance to solvents. Strain P4 is able to grow in oil, diesel, benzene and alkanes (von der Weid et al. 2007). This tolerance is probably due to the presence of an ABC transporter system encoded by the ttgABC operon (ZP_08024403–11). These genes may be constitutively expressed, as their control is normally accomplished by the transcriptional repressor ttgR, which we could not find in the genome annotation. The genome of strain P4 further possesses universal stress-related proteins (USPs) (ZP_08022937, ZP_08023009, ZP_0802386, ZP_08025031), which are involved in a wide variety of responses to environmental stress, such as carbon, nitrogen and phosphate starvation, exposure to heat shock and UV radiation. Also, we found the gene for the transcriptional regulator of heat shock, HcrA (ZP_08022061), dnaJ-dnaK (ZP_08022060, ZP_08023538, ZP_08023540), clpBCX (encoding chaperonins) (ZP_08022156, ZP_08022993, ZP_08023310), groEL (ZP_08022808), hsp90 (ZP_08022887), and cold shock (ZP_08022237, ZP_08023098, ZP_08023522).
Among the genes that are potentially involved in survival and adaptation of strain P4 in soil, a wide repertoire of antibiotic resistance genes was found. In total, 12 genes encoding B and C classes of beta-lactamases and penicillin biosynthesis were found, next to at least 12 putative ATP-ABC and 10 MFS transport systems (Table S1). The latter are likely involved in the efflux of drugs like enterobactin (ZP_08023706) and daunorubicin (ZP_08024809). We found four copies of genes coding for aminoglycoside 3`-phosphotransferases (ZP_08022108, ZP_08022904, ZP_08022978, ZP_08023139), which are involved in the modification/efflux of neomycin and kanamycin, next to two genes involved in export of polyketide antibiotics (ZP_08023291, ZP_08025214). One gene coding for the radical SAM 23-S rRNA methyltransferase of the Cfr family (ZP_08022226) was found. This is thought to perform post-transcriptional modification of rRNA, thus conferring resistance to ribosome-targeting antibiotics (Kaminska et al. 2010). Also, we identified a single copy of a gene for erythromycin esterase (ZP_08022936), which presents a likely role against the action of this antibiotic.
Transport of molecules
Analysis of the strain P4 genome using the THMMH, NR, CDD and TransportDB databases revealed the presence of about 311 CDSs with putative transport roles (Table S1). This value represents about 8.8% of the whole strain P4 genome, which is consistent with values from other sequenced actinomycete genomes. The best-represented transporter class was the ABC family (49%), followed by the MFS family (13%). In both families, the presence of genes involved in drug export, solvent and metal efflux stood out. This may reflect a requirement for these systems in an environment like soil, where the presence of antibiotic producers is likely. Hence, the ability to compete in the niche is enhanced by the use of efficient antibiotic efflux systems next to the synthesis of own antimicrobials. In addition to the uptake transporters described above, the strain P4 genome contains several genes for permeases for diverse sugar classes, glycerol, nucleosides, amino acids, acetate, urea, vitamin B12 and C4-dicarboxylate. The secADEFY genes (Table S1), which encode the Type II secretion system, were also detected in strain P4 genome. The tatAC translocase genes (Table S1) were also found. They encode a component of the Sec-independent protein translocation system, which is involved in translocation of folded proteins across the periplasmic membrane.
Concluding remarks
The tropical soil from which D. cinnamea P4 was isolated is characterized as a complex environment with frequent chemical and physical inputs. The resulting energy flux is mainly due to the primary production, which is reflected in root exudation as well as in the decomposition of plant parts. The current annotation of the strain P4 genome assists us in gaining new insights in the mechanisms employed by D. cinnamea P4 in its adaptation to soil. The presumably saprophytic lifestyle of D. cinnamea P4 is reflected in the complex network of gene products involved in the acquisition of carbon, as well as in the seemingly efficient processes of protection against environmental stress. The diverse array of ABC- and MFS-like transporters, which are predicted to be involved in the acquisition of different materials from soil, next to the complex catabolic mechanisms for the (simultaneous) use of plant compounds like limone, pinene, alkane/alkanesulfonate, cyclic alkanes, nitrogen compounds and complex carbohydrates, highlights the strategies for growth and survival in a complex habitat. The genome sequence also confirmed the existence of pathways for the transformation of aromatic and phenolic compounds which are generated following decomposition of plant materials. Here, the assimilation of aromatic compounds like biphenyl, benzoate and phenol converge to the common β-ketoadipate pathway. The ability to use aerobic CO oxidization and a possible anaerobic dissimilatory nitrate reduction pathway as energy generation mechanisms, and the fixation of CO2, support the capacity to grow autotrophically. However, an aerobic heterotrophic lifestyle may predominate in the organism. The harsh climate can be inhospitable to microorganisms dwelling in tropical soils, as alternating periods of drought and high humidity, high sun incidence raising the temperature, and the presence of chemicals constitute typical stressors. Another factor of importance for the fitness of strain P4 is the high number of genes devoted to a rapid response to stress signals. Strain P4 was shown to possess elaborate systems for osmotic stress control, also revealing the complete pathway for synthesis of carotenoid compounds that confer UV and oxidative stress resistance. Moreover, the multiplicity of genes devoted to antibiotic resistance in the P4 genome reveals the importance of drug tolerance in its habitat.
Finally, the D. cinnamea P4 genome was found to encode a high number of enzymes with potential application in biocatalysis. Oxygenases involved in secondary metabolite biosynthesis, e.g. of carveol compounds and genes for the biosynthesis of aromatic antibiotics exemplify some products of commercial interest. Finally, the unexpected metabolic capabilities of D. cinnamea P4, combined with the tolerance of this organism to environmental stresses and its features of easy cultivation in bioreactors, provides a stimulus to future applications in biocatalysis and bioremediation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1. (a) Alignment between putative coxL form I genes of D. cinnamea strain P4 and related organisms; (b) between putative coxL form II genes among related organisms. (DOC 316 kb)
Acknowledgments
Thanks are due to CNPq and Coimbra Institute for providing a fellowship to L. Procópio to come over to the Groningen lab, where the genome annotation and physiological experiments were performed. This study was carried out as part of a research project supported by PETROBRAS.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Arnesano F, Banci L, Bertini I, Mangani S, Thompsett AR. A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc Natl Acad Sci USA. 2003;100(7):3814–3819. doi: 10.1073/pnas.0636904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O’Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271(41):25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- Bihari Z, Szabó Z, Szvetnik A, Balázs M, Bartos P, Tolmacsov P, Zombori Z, Kiss I. Characterization of a novel long-chain n-alkane-degrading strain, Dietzia sp. E1. Z Naturforsch C. 2011;65(11–12):693–700. doi: 10.1515/znc-2010-11-1210. [DOI] [PubMed] [Google Scholar]
- Brünke P, Sterner O, Bailey JE, Minas W. Heterologous expression of the naphthocyclinone hydroxylase gene from Streptomyces arenae for production of novel hybrid polyketides. Antonie van Leeuwenhoek. 2001;79(3–4):235–245. doi: 10.1023/A:1012037329949. [DOI] [PubMed] [Google Scholar]
- Canosa I, Sánchez-Romero JM, Yuste L, Rojo F. A positive feedback mechanism controls expression of AlkS, the transcriptional regulator of the Pseudomonas oleovorans alkane degradation pathway. Mol Microbiol. 2000;35(4):791–799. doi: 10.1046/j.1365-2958.2000.01751.x. [DOI] [PubMed] [Google Scholar]
- Chistoserdova L, Laukel M, Portais JC, Vorholt JA, Lidstrom ME. Multiple formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J Bacteriol. 2003;186(1):222–822. doi: 10.1128/JB.186.1.22-28.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W, Strauss G, Werz U, Fuchs G, Bacher A. Retrobiosynthetic analysis of carbon fixation in the phototrophic eubacterium Chloroflexus aurantiacus. Eur J Biochem. 1993;215(3):619–632. doi: 10.1111/j.1432-1033.1993.tb18073.x. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Evans FF, Rosado AS, Sebastián GV, Casella R, Machado PL, Holmström C, Kjelleberg S, Elsas JD, Seldin L. Impact of oil contamination and biostimulation on the diversity of indigenous bacterial communities in soil microcosms. FEMS Microbiol Ecol. 2004;49:295–305. doi: 10.1016/j.femsec.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:276–280. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann S, Ferner M, Jeffke T, Henne A, Gottschalk G, Meyer O. Complete nucleotide sequence of the circular megaplasmid pHCG3 of Oligotropha carboxidovorans: function in the chemolithoautotrophic utilization of CO, H(2) and cO(2) Gene. 2003;322:67–75. doi: 10.1016/j.gene.2003.08.027. [DOI] [PubMed] [Google Scholar]
- Gonçalves ER, Hara H, Miyazawa D, Davies JE, Eltis LD, Mohn WW. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl Environ Microbiol. 2006;72(9):6183–6193. doi: 10.1128/AEM.00947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herter S, Farfsing J, Gad’On N, Rieder C, Eisenreich W, Bacher A, Fuchs G. Autotrophic CO(2) fixation by Chloroflexus aurantiacus: study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J Bacteriol. 2001;183(14):4305–4316. doi: 10.1128/JB.183.14.4305-4316.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska KH, Purta E, Hansen LH, Bujnicki JM, Vester B, Long KS. Insights into the structure, function and evolution of the radical-SAM 23S rRNA methyltransferase Cfr that confers antibiotic resistance in bacteria. Nucleic Acids Res. 2010;38(5):1652–1663. doi: 10.1093/nar/gkp1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GM, Weber CF. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol. 2007;5(2):107–118. doi: 10.1038/nrmicro1595. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lenger R, Herrmann U, Gross R, Simon J, Kröger A. Structure and function of a second gene cluster encoding the formate dehydrogenase of Wolinella succinogenes. Eur J Biochem. 1997;246(3):646–651. doi: 10.1111/j.1432-1033.1997.t01-2-00646.x. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:229–232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010;39:225. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod MP, Warren RL, Hsiao WW, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJ, Holt R, Brinkman FS, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci USA. 2006;103(42):15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongodin EF, Shapir N, Daugherty SC, DeBoy RT, Emerson JB, Shvartzbeyn A, Radune D, Vamathevan J, Riggs F, Grinberg V, Khouri H, Wackett LP, Nelson KE, Sadowsky MJ. Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. PLoS Genet. 2006;2(12):e214. doi: 10.1371/journal.pgen.0020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Vivián C, Cabello P, Martínez-Luque M, Blasco R, Castillo F. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol. 1999;181(21):6573–6584. doi: 10.1128/jb.181.21.6573-6584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas J, González-Zorn B, Ladrón N, Garrido P, Vázquez-Boland JA. Identification and mutagenesis by allelic exchange of choE, encoding a cholesterol oxidase from the intracellular pathogen Rhodococcus equi. J Bacteriol. 2001;183(16):4796–4805. doi: 10.1128/JB.183.16.4796-4805.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem Y, Cabrera M, Cupples CG, Miller JH. The mutY gene: a mutator locus in Escherichia coli that generates GC—TA transversions. Proc Natl Acad Sci USA. 1988;85(8):2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzmann A, Ferner M, Gnida M, Meyer-Klaucke W, Maisel T, Meyer O. The CoxD protein of Oligotropha carboxidovorans is a predicted AAA+ ATPase chaperone involved in the biogenesis of the CO dehydrogenase [CuSMoO2] cluster. J Biol Chem. 2009;284(14):9578–9586. doi: 10.1074/jbc.M805354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RW, Wiegel J, Berry CJ, Fliermans C, Peacock AD, White DC, Shimkets LJ. Kineococcus radiotolerans sp. nov, a radiation-resistant, gram-positive bacterium. Int J Syst Evol Microbiol. 2002;52(Pt 3):933–938. doi: 10.1099/ijs.0.02029-0. [DOI] [PubMed] [Google Scholar]
- Ren Q, Paulsen IT. TransportDB: large-scale comparative genomic analyses of cytoplasmic membrane transport systems in prokaryotes. J Mol Microbiol Biotechnol. 2007;12(3–4):165–179. doi: 10.1159/000099639. [DOI] [PubMed] [Google Scholar]
- Rothschild LJ. The evolution of photosynthesis…again? Philos Trans R Soc Lond B Biol Sci. 2008;363(1504):2787–2801. doi: 10.1098/rstb.2008.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago B, Schuebel U, Egelseer C, Meyer O. Sequence analysis, characterization and cO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene. 1999;236:115–124. doi: 10.1016/S0378-1119(99)00245-0. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33(Web Server issue):W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin L, Dubnau L. DNA homology among Bacillus polymyxa, Bacillus macerans, Bacillus azotofixans and other nitrogen fixing Bacillus strains. Int J Syst Bacteriol. 1985;35:151–154. doi: 10.1099/00207713-35-2-151. [DOI] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:32–36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sthapit B, Oh TJ, Lamichhane R, Liou K, Lee HC, Kim CG, Sohng JK. Neocarzinostatin naphthoate synthase: an unique iterative type I PKS from neocarzinostatin producer Streptomyces carzinostaticus. FEBS Lett. 2004;566(1–3):201–206. doi: 10.1016/j.febslet.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Takarada H, Sekine M, Kosugi H, Matsuo Y, Fujisawa T, Omata S, Kishi E, Shimizu A, Tsukatani N, Tanikawa S, Fujita N, Harayama S. Complete genome sequence of the soil actinomycete Kocuria rhizophila. J Bacteriol. 2008;190(12):4139–4146. doi: 10.1128/JB.01853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;11:4–41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beilen JB, Panke S, Lucchini S, Franchini AG, Röthlisberger M, Witholt B. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology. 2001;147(Pt 6):1621–1630. doi: 10.1099/00221287-147-6-1621. [DOI] [PubMed] [Google Scholar]
- van der Ploeg JR, Barone M, Leisinger T. Functional analysis of the Bacillus subtiliscysK and cysJI genes. FEMS Microbiol Lett. 2001;201(1):29–35. doi: 10.1111/j.1574-6968.2001.tb10728.x. [DOI] [PubMed] [Google Scholar]
- von der Weid I, Marques JM, Cunha CD, Lippi RK, Dos Santos SC, Rosado AS, Lins U, Seldin L. Identification and biodegradation potential of a novel strain of Dietzia cinnamea isolated from a petroleum-contaminated tropical soil. Syst Appl Microbiol. 2007;30(4):331–339. doi: 10.1016/j.syapm.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Wang XB, Chi CQ, Nie Y, Tang YQ, Tan Y, Wu G, Wu XL. Degradation of petroleum hydrocarbons (C6–C40) and crude oil by a novel Dietzia strain. Bioresour Technol. 2011;102(17):7755–7761. doi: 10.1016/j.biortech.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Yassin AF, Hupfer H, Schaal KP. Dietzia cinnamea sp. nov, a novel species isolated from a perennial swab of a patient with a bone marrow transplant. Int J Syst Evol Microbiol. 2006;56(3):641–645. doi: 10.1099/ijs.0.63863-0. [DOI] [PubMed] [Google Scholar]
- Zhao Q, He Q, Ding W, Tang M, Kang Q, Yu Y, Deng W, Zhang Q, Fang J, Tang G, Liu W. Characterization of the azinomycin B biosynthetic gene cluster revealing a different iterative type I polyketide synthase for naphthoate biosynthesis. Chem Biol. 2008;15(7):693–705. doi: 10.1016/j.chembiol.2008.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (a) Alignment between putative coxL form I genes of D. cinnamea strain P4 and related organisms; (b) between putative coxL form II genes among related organisms. (DOC 316 kb)