Abstract
Sulfide-oxidizing bacteria of the genus Beggiatoa are known to accumulate phosphate intracellularly as polyphosphate but little is known about the structure and properties of these inclusions. Application of different staining techniques revealed the presence of unusually large polyphosphate inclusions in the marine Beggiatoa strain 35Flor. The inclusions showed a co-occurrence of polyphosphate, calcium and magnesium when analyzed by scanning electron microscopy and energy dispersive X-ray analysis. Similar to polyphosphate-enriched acidocalcisomes of prokaryotes and eukaryotes, the polyphosphate inclusions in Beggiatoa strain 35Flor are enclosed by a lipid layer and store cations. However, they are not notably acidic. 16S rRNA gene sequence-based phylogenetic reconstruction showed an affiliation of Beggiatoa strain 35Flor to a monophyletic branch, comprising other narrow vacuolated and non-vacuolated Beggiatoa species. The polyphosphate inclusions represent a new type of membrane surrounded storage compartment within the genus Beggiatoa, distinct from the mostly nitrate-storing vacuoles known from other marine sulfide-oxidizing bacteria of the family Beggiatoaceae.
Keywords: Beggiatoa, Polyphosphate, Acidocalcisome, Confocal laser scanning microscopy
Introduction
Filamentous members of the family Beggiatoaceae are ubiquitous in freshwater (Kojima et al. 2003; Strohl and Larkin 1978), marine (Jørgensen 1977; Mußmann et al. 2003; Rosenberg and Diaz 1993) and hypersaline sediments (Hinck et al. 2007). Depending on the sulfide level in the environment they occur as single filaments in the interstitial space of sediments (Mußmann et al. 2003) or as dense mats at hydrothermal vents (Kalanetra et al. 2004; Nelson et al. 1989) and along coasts with local upwelling (Schmaljohann et al. 2001; Schulz et al. 2000). Filamentous Beggiatoaceae consist of up to several hundreds of cylindrical cells and can reach a length of more than 1 cm. The filament widths range in marine strains from 1 to 200 μm, whereas freshwater strains mostly exhibit widths less than 5 μm (Strohl 2005).
The filamentous Beggiatoaceae are typically sulfide-oxidizing autotrophic bacteria but most isolates so far are organoheterotrophic. Some of the larger lithotrophic species may use both CO2 and organic compounds as carbon sources (Mußmann et al. 2007; Teske and Nelson 2006). Sulfide can be oxidized with oxygen (Nelson et al. 1986) or alternatively nitrate as electron acceptor (Kamp et al. 2006; Sweerts et al. 1990). In the larger marine representatives, nitrate was shown to be stored in a vacuole, which makes up most of the cell volume (Fossing et al. 1995; McHatton et al. 1996; Schulz et al. 1999). Elemental sulfur is stored as an intermediate compound within the periplasm in the form of spherical inclusions, which are enclosed by invaginations of the cytoplasmic membrane (Strohl et al. 1981). Short-chain fatty acids are stored as polyhydroxyalkanoates (PHA) (Strohl and Larkin 1978). Other genera of sulfide-oxidizing bacteria of the family Beggiatoaceae show similar characteristics (Salman et al. 2011). However, Thiomargarita namibiensis stores glycogen instead of PHA (Schulz and Schulz 2005) and species of the genus “Candidatus Marithrix” (originally described as vacuolate, attached filaments) do not store nitrate in their vacuole (Kalanetra et al. 2004; Kalanetra and Nelson 2010; Salman et al. 2011).
Some Beggiatoa strains accumulate phosphate as polyphosphate. This was shown by staining with methylene blue (Strohl and Larkin 1978) and 4′,6-diamidino-2-phenylindole dihydrochlorid (DAPI) (Brock and Schulz-Vogt 2011), by transmission electron microscopy (TEM) of thin sections (Maier and Murray 1965) and by TEM combined with energy-dispersive X-ray microanalysis (EDXA) of whole filaments (de Albuquerque et al. 2010). Polyphosphate is a polymer of many tens or hundreds of orthophosphate residues linked by high-energy phosphoanhydride bonds (Kornberg 1995). It is assumed to be a molecule of many functions such as an ATP substitute, phosphate reservoir, chelator of metals and may play an important role in the survival and fitness of bacterial cells in general (Ault-Riche et al. 1998; Kornberg et al. 1999; Seufferheld et al. 2008). In a recent study, we showed that the decomposition of internally accumulated polyphosphate within the marine Beggiatoa strain 35Flor and the subsequent release of phosphate are mediated by a change from oxic to anoxic cultivation conditions at high sulfide concentrations (Brock and Schulz-Vogt 2011).
In the present study, we investigated the polyphosphate inclusions of the same Beggiatoa strain in more detail. We stained filaments with DAPI for the detection of polyphosphate inclusions simultaneously with dyes specific for lipid layers and acidic cell compartments. By means of scanning electron microscopy (SEM) in combination with EDXA we studied the elemental composition of these inclusions. Further, we investigated the phylogenetic affiliation of Beggiatoa strain 35Flor based on its 16S rRNA gene sequence, enabling the comparison of intracellular structures with closely related strains of the same genus.
Materials and methods
Cultivation of Beggiatoa strain 35Flor
We used the marine Beggiatoa strain 35Flor, which has been maintained in modified gradient medium according to Nelson and Jannasch (1983) for 9 years. Originally, it was collected from a microbial consortium of a black band disease at scleractinian corals from Florida Keys. Beside the Beggiatoa strain, the culture contains a Pseudovibrio sp., which seems to be required for the growth of the Beggiatoa strain (A. Schwedt, personal communication). To simulate natural growth conditions, the Beggiatoa strain was cultivated in sterile aged natural seawater obtained from the North Sea near the island of Helgoland, Germany. A sulfide/oxygen gradient (Nelson and Jannasch 1983) was generated as follows: 4 ml of a solid bottom agar (1.5% Bacto Agar, BD, Franklin Lakes, USA), containing 2 mmol l−1 Na2S, 0.5 mmol l−1 (NH4)2SO4 and 40 μmol l−1 K2HPO4, were overlaid with 8 ml of a semi solid top agar (0.75% Bacto Agar), containing vitamins, 0.5 mmol l−1 (NH4)2SO4, 2 mmol l−1 NaHCO3 and 40 μmol l−1 K2HPO4. The pH of both agars was adjusted to around 8 using HCl. To allow gas exchange with ambient air, the caps of the cultivation tubes were loosely closed. The sulfide gradient in the culture tubes was allowed to establish for one day before inoculation with Beggiatoa filaments.
Staining of polyphosphate
Granules of polyphosphate were detected by staining with toluidine blue and DAPI. Toluidine blue reveals red inclusions due to a metachromatic effect when binding to polyphosphate (Kulaev et al. 2004). One part of the Beggiatoa culture was mixed with two parts of 96% ethanol to dissolve stored sulfur globules. After 2 days at room temperature, the filaments were heat fixed on a glass slide, washed with water, stained for 30 s with toluidine blue (0.3% in 0.5% acetic acid), washed with 0.5% acetic acid and finally with water. The filaments were observed by bright field microscopy using an Axioplan universal microscope (Zeiss, Oberkochen, Germany) equipped with a 100-fold Zeiss NA 1.3 oil immersion objective lens.
DAPI is a common stain for DNA and shows a shift from a blue DNA-specific signal with an emission maximum around 460 nm to a yellow signal with an emission maximum around 525 nm when binding to polyphosphate (Tijssen et al. 1982). A stock solution containing 2.8 mmol l−1 DAPI was added to a sample of the Beggiatoa culture to a final concentration of 140 μmol l−1. After incubation for 6 h at room temperature in the dark the filaments were inspected for polyphosphate inclusions using a confocal laser scanning microscope (CLSM) LSM 510 (Zeiss) equipped with a 100-fold Zeiss Apochromat NA 1.4 oil immersion objective lens. Excitation wavelengths of 351 and 364 nm were used and emission was detected by a Zeiss BP filter between 505 and 550 nm. In parallel, a transmission image was collected on the in-build photodiode at a wavelength of 630 nm to check for cell compartments such as vacuoles and sulfur globules. Due to light refraction, sulfur globules appear as dark spots or spheres. Contrast of images was adjusted and an overlay of polyphosphate-DAPI fluorescence and transmission image was created in Canvas X (ACD Systems, Seattle, USA).
Staining of cell membranes and acidic cell compartments
To study the structure of the polyphosphate inclusions, dual staining with DAPI for polyphosphate and several fluorochromes, specific for different cell constituents, was carried out. Imaging was performed with the LSM 510 CLSM using the multi track mode. Lipids were stained with the lipophilic dye Nile Red (final concentration 8 μmol l−1 in 10% DMSO, Sigma Aldrich, St Louis, USA; CLSM excitation: 488 and 543 nm, CLSM emission: Zeiss LP 585 nm), which is suitable for lipid layer detection at an emission wavelength above 590 nm (Greenspan and Fowler 1985). Furthermore, the yeast vacuole membrane marker MDY-64 (final concentration 100 μmol l−1 in 1% DMSO, Invitrogen, Carlsbad, USA; CLSM excitation: 458 nm, CLSM emission: Zeiss BP 475–525 nm) was used for staining membranes (Hinck et al. 2007). Acidic cell compartments were visualized by two pH-sensitive dyes. Acridine Orange (final concentration 6 μmol l−1, Sigma Aldrich; CLSM excitation: 488 nm, CLSM emission: Zeiss LP 505 nm) accumulates in acidic cell compartments dependent on the pH difference. At higher concentrations Acridine Orange shows a shift in emission from green to red because of aggregate formation (Han and Burgess 2010). As it also binds to DNA and RNA the interpretation of images can be difficult. Therefore, LysoSensor Green DND-189 (final concentration 2 μmol l−1 in 0.2% DMSO, Invitrogen; CLSM excitation: 488 nm, CLSM emission: Zeiss BP 505–530 nm), which has an emission maximum around 510 nm at pH values of 4−5 (Lin et al. 2001), was applied. Each of the dyes was added to a DAPI treated Beggiatoa sample as described above and incubated for at least 30 min at room temperature. Contrast of images was adjusted and overlays of fluorescent images of polyphosphate-DAPI and particular fluorochromes were created in Canvas X (ACD Systems).
Scanning electron microscopy (SEM) and energy dispersive X-ray analysis (EDXA)
To detect the main elements of the polyphosphate inclusions, Beggiatoa filaments were observed by SEM in combination with EDXA. After 7 days of growth, filaments were fixed by addition of glutaraldehyde to the growth medium to a final concentration of 2.5% for at least 3 h. Fixed and unfixed filaments were picked out of their growth medium by means of self-made glass needles, washed in water, placed onto 10 × 10 mm silicon wafer supports (Plano, Wetzlar, Germany) that were attached to aluminum stubs and subsequently dried in a desiccator. SEM was performed in a Hitachi S3200N scanning electron microscope (Hitachi, Tokyo, Japan) operating at 20 kV. The uncoated specimens were investigated either immediately in the low vacuum mode at 5 Pa or after desiccation in the high vacuum mode using a Robinson BSE detector. Digitized pictures were recorded using either the DISS and DIPS software packages (point electronic GmbH, Halle, Germany) or the INCA software package (Oxford Instruments, Wiesbaden, Germany). EDXA measurements of whole filaments or selected spots and areas were performed using an Oxford INCA system (PentaFET Precision INCA X-act detector, INCA software package).
16S rRNA gene sequence amplification and phylogenetic analysis
Twenty μl of the mat were removed from a Beggiatoa culture with a pipette and collected in 10 ml sterile artificial seawater. With a self-made glass needle, a single filament was removed and washed by dragging it through fresh artificial seawater. Subsequently, the filament was directly used for polymerase chain reaction (PCR). The reaction mix in a final volume of 25 μl contained 0.2 mmol l−1 dNTPs, 1 μmol l−1 each of forward and reverse primer GM3F and GM4R (Muyzer et al. 1995), 1 × high fidelity PCR reaction buffer with 1.5 mmol l−1 MgCl2 and 1.25 U High Fidelity enzyme mix (Fermentas, St. Leon-Rot, Germany). The filament was disrupted inside the reaction vial using a sterile needle before applying the PCR program with the following steps: initial denaturation (94°C, 2 min), followed by 25 cycles of 94°C for 30 s, 48°C for 30 s and 72°C for 2 min. The program was terminated with a final elongation step (7 min, 72°C). The PCR product was separated on a 1% agarose gel, cut out and extracted with a kit (Quiagen, Hilden, Germany). The amplicon was directly sequenced using the Big Dye Cycle Sequencing Kit (Applied Biosystems, Carlsbad, USA) and further analyzed on an ABI Genetic Analyzer 3130× (Applied Biosystems). The sequence was deposited in the GenBank/EMBL/DDBJ databases under accession number FR717278.
The phylogenetic affiliation of the sequence was inferred with the ARB software package (Ludwig et al. 2004) based on release 102 of the SILVA SSURef database (Pruesse et al. 2007). For tree reconstruction, 229 nearly full-length 16S rRNA gene sequences and nucleotide positions between 252 and 1436 (according to Escherichia coli numbering) were included. Calculation was based on neighbor joining, maximum parsimony and maximum likelihood methods applying 10, 30 and 50% positional conservatory filters. After comparison of the retrieved trees, a multifurcation tree based on the 10% neighbor joining tree was manually constructed according to the Standard Operating Procedure for Phylogenetic Inference (SOPPI) (Peplies et al. 2008). A distance matrix with similarity correction of the obtained sequence and its closest relatives was calculated with neighbor joining criteria in the ARB program.
Results
Polyphosphate inclusions
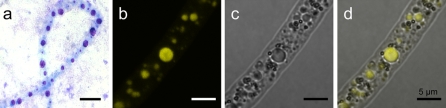
Staining of Beggiatoa filaments with toluidine blue and DAPI revealed the existence of several polyphosphate inclusions within the cells (Fig. 1). The width of the filaments was around 6 μm. The size of the polyphosphate bodies was highly variable and ranged from barely visible in the CLSM to inclusions of more than 3 μm (Fig. 1b). They were irregularly distributed in the cells. Within a single cell, large inclusions were predominantly found in the center whereas smaller inclusions were found throughout the cell volume. Some cells either lacked polyphosphate inclusions completely or possessed only few small ones. In the transmission mode of the CLSM, different cell compartments such as sulfur globules, which were clearly visible as dark spots or small circles, were detected. Large spherical areas in the cell showed congruence with large polyphosphate inclusions stained with DAPI (Fig. 1b–d). Some cell compartments could neither be related to sulfur globules nor to polyphosphate inclusions.
Fig. 1.
Polyphosphate inclusions within the marine Beggiatoa strain 35Flor after 7 days of growth. a Bright field image of a toluidine blue-stained Beggiatoa filament. Due to metachromasy polyphosphate inclusions of different sizes appear violet. b–d CLSM images of a DAPI-stained Beggiatoa filament. b Staining with DAPI reveals polyphosphate inclusions of different sizes by a yellow fluorescence signal. c In the transmission mode of the CLSM, many spherical cell compartments of different sizes are visible. d The overlay of (b) and (c) reveals congruence of polyphosphate inclusions with the large and with some of the smaller cell compartments. Small dark spots probably represent sulfur granules. Scale bars represent 5 μm
Membrane and pH staining
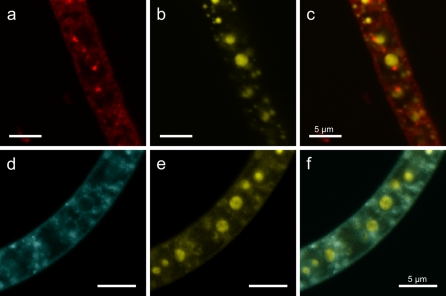
Both the lipid-staining dyes Nile Red and MDY-64 revealed several lipid layer enclosed cell areas and some small completely stained inclusions (Fig. 2). The areas enclosed by a lipid layer were of different sizes and mostly of spherical shape. Dual staining with DAPI and Nile Red or MDY-64 revealed that polyphosphate inclusions were enclosed by a lipid layer, probably a membrane. This was less obvious for smaller polyphosphate inclusions as their size was close to the resolution limit of the CLSM.
Fig. 2.
Fluorescence of Beggiatoa strain 35Flor filaments obtained by dual staining with DAPI for polyphosphate and Nile Red or MDY-64 for lipid detection. a Lipid layers of spherical structure and different sizes are visible by staining with Nile Red. b In the same filament, stained with DAPI, polyphosphate inclusions of different sizes are visible by a yellow fluorescence signal. c An overlay of the Nile Red and DAPI fluorescence shows the existence of lipid layers for most of the large and some of the small polyphosphate inclusions. d Staining with MDY-64 reveals the same pattern of lipid layers as for Nile Red staining. e Polyphosphate inclusions of different sizes. f The overlay of MDY-64 and DAPI fluorescence reveals that most polyphosphate inclusions are enclosed by a lipid layer indicating a membrane. Note The detected internal lipid layers do not exclusively surround polyphosphate inclusions. Scale bars represent 5 μm
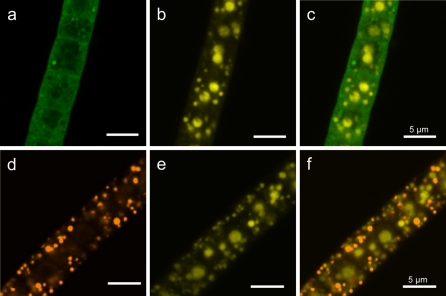
By staining with LysoSensor Green DND-189, some intense fluorescing spots were visible (Fig. 3a). Furthermore, spherical areas, which were mostly distributed within the cytoplasmic cell periphery, were detected. The cytoplasm itself showed a weak fluorescent signal. No enhanced fluorescence signal for LysoSensor Green DND-189, indicating a notably acidic milieu, was revealed for cell areas with polyphosphate inclusions, which were detected in the same filament by DAPI staining (Fig. 3a–c). A similar result was obtained with the pH sensitive dye Acridine Orange, as this fluorochrome did not accumulate within the large central area of the cell (Fig. 3d–f), which consisted of polyphosphate inclusions, as shown by DAPI staining. In contrast, it highlighted numerous inclusions, which were neither polyphosphate nor sulfur globules. These inclusions showed an intensive fluorescence and burst within seconds under laser illumination.
Fig. 3.
Fluorescence of Beggiatoa strain 35Flor filaments obtained by dual staining with DAPI for polyphosphate and the pH-sensitive dyes LysoSensor Green DND-189 or Acridine Orange. a LysoSensor Green DND-189 shows fluorescence mostly in the cell periphery, indicating a low pH. b DAPI reveals polyphosphate inclusions of different sizes in the same filament. c The overlay of (a) and (b) shows no fluorescence for LysoSensor Green DND-189 within polyphosphate inclusions, indicating that the pH is not notably acidic. d Acridine Orange reveals several spheres of acidic pH in the cell periphery. e DAPI reveals polyphosphate inclusions of different sizes in the same filament. f The overlay of (d) and (e) reveals no fluorescence for Acridine Orange in polyphosphate inclusions, indicating that the pH in the polyphosphate inclusions is not notably acidic. Scale bars represent 5 μm
SEM and EDXA
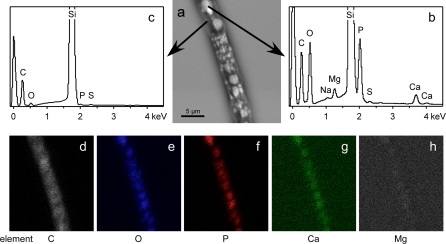
The electron micrograph obtained by back-scattered electrons showed a heterogeneous distribution of elements in the Beggiatoa cells (Fig. 4a). Areas with higher concentrations of heavier elements are indicated by a lighter grey value (Fig. 4a). These areas were either detected as a single large spot in the center of a cell or as many small spots distributed in the entire cell. EDXA revealed significantly higher peaks of phosphorus and oxygen compared to carbon in these areas (Fig. 4b). An association with calcium and magnesium was also shown by peaks of these elements at the respective sites (Fig. 4b). The cytoplasm surrounding these inclusions, as indicated by darker grey values in Fig. 4a, showed much smaller peaks for phosphorus and oxygen than for carbon and no peaks for calcium and magnesium were detected (Fig. 4c). In accordance to the EDXA spectra, element mapping revealed higher amounts of phosphorus, oxygen and calcium for the lighter areas of the electron micrograph as compared to the ambient cytoplasm (Fig. 4d–h). Magnesium, which showed a distinct peak by EDXA revealed a very weak signal by element mapping. The pattern of the cell areas with higher concentrations of heavier elements and the distribution of phosphorus, oxygen and calcium was similar to the distribution of polyphosphate inclusions observed in DAPI stained cells.
Fig. 4.
Back-scattered electron micrograph, EDXA spectra and elemental mapping of a Beggiatoa strain 35Flor filament. a A micrograph of an intact, fixed and air-dried Beggiatoa filament obtained by back-scattered electrons reveals areas of lighter grey value which indicate high concentrations of elements with higher atomic weight. b EDXA spectra of these areas reveal large peaks for phosphorus and oxygen and small peaks for calcium and magnesium. c In adjacent darker areas of the filament neither calcium nor magnesium peaks are detectable. Very large peaks of silica originate from the silicon wafer support. d–h A clear spatial correlation of oxygen, phosphorus and calcium is obtained by elemental mapping of the filament. h The signal for magnesium is much weaker than observed for the other elements
16S rRNA-based phylogeny
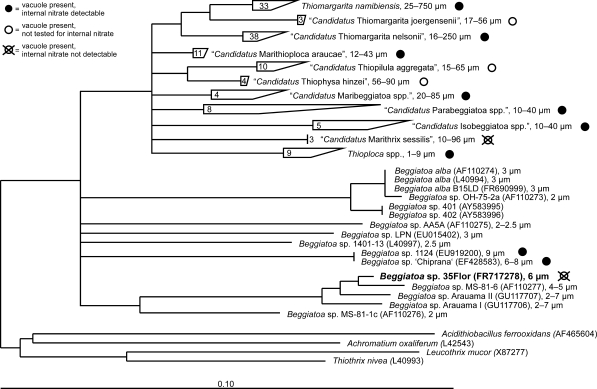
Phylogenetic analysis based on the nearly full-length 16S rRNA gene sequence obtained from Beggiatoa strain 35Flor confirmed its close affiliation to a monophyletic cluster including other narrow vacuolated and non-vacuolated Beggiatoa spp. within the family Beggiatoaceae (Fig. 5). Within this cluster, the obtained sequence was affiliated to a distinct sub-cluster and was most closely related to the sequence of the marine strain Beggiatoa sp. MS-81-6 (Ahmad et al. 2006), sharing 98.1% sequence identity. The latter strain was isolated from a salt March (Nelson et al. 1982) and exhibited a similar diameter (4–5 μm) to that of the strain investigated in this study. The sub-cluster contained sequences of other narrow Beggiatoa spp. originating from marine and hypersaline habitats. Interestingly, Beggiatoa strain 35Flor was the only one within this monophyletic sub-cluster featuring intracellular vacuole-like structures. Within the other sub-cluster of the genus Beggiatoa, internal vacuoles were reported for only for two other strains, Beggiatoa sp. 1124 and Beggiatoa sp. “Chiprana” (6–8 μm diameter). However, in these bacteria the vacuoles were proposed to contain nitrate as high internal nitrate concentrations were detected (Hinck et al. 2007).
Fig. 5.
Phylogenetic tree based on 16S rRNA gene sequences showing the affiliation of Beggiatoa strain 35Flor to other Beggiatoa spp. sequences. The closest relative is the Beggiatoa strain MS-81-6. Beggiatoa strain 35Flor is the only known species within the monophyletic sub-cluster which contains intracellular vacuoles. Vacuolation is a common characteristic within the family of Beggiatoaceae. Only within the genera Beggiatoa and Thioploca narrow non-vacuolated representatives have been found so far. For most vacuolated representatives, storage of nitrate within the vacuole is proposed (filled circles), whereas the strain described here is the only one so far having central vacuoles filled with polyphosphate. Nodes displayed in this multifurcation tree correspond to bootstrap values equal or higher than 75% confidence using maximum likelihood bootstrap calculation. The nomenclature of sulfide-oxidizing bacteria presented here follows a recently proposed reclassification of the family Beggiatoaceae by Salman et al. (2011)
Discussion
Staining of filaments of the marine Beggiatoa strain 35Flor with DAPI revealed polyphosphate inclusions of up to 3 μm in diameter (Fig. 1). Compared to previous studies on polyphosphate inclusions in bacteria and acidocalcisomes in eukaryotic protists, polyphosphate inclusions of such large size are very unusual. In Agrobacterium tumefaciens, polyphosphate inclusions have been reported with diameters around 210 nm (Seufferheld et al. 2003). Larger polyphosphate bodies of 400–500 nm maximum size were found in the cyanobacterium Synechococcus sp. (Lawry and Jensen 1979). The largest polyphosphate inclusions of more than 2 μm diameter have so far been observed in the non-filamentous, giant sulfide-oxidizing bacterium Thiomargarita namibiensis (Schulz and Schulz 2005), a close relative of Beggiatoa strain 35Flor (Fig. 5). In narrow freshwater Beggiatoa spp. polyphosphate inclusions of less than 1 μm have been reported by Maier and Murray (1965). In some narrow, hypersaline Beggiatoa spp. comparable inclusions of similar size were recently reported by de Albuquerque et al. (2010). However, both size and number of polyphosphate inclusions greatly depend on the environmental conditions the bacteria are exposed to, such as the sulfide and oxygen regime (Brock and Schulz-Vogt 2011). Therefore, it is difficult to compare the storage capabilities for polyphosphate between organisms which have experienced different environmental conditions. In conclusion, we propose that the polyphosphate inclusions detected in the marine Beggiatoa strain 35Flor are the largest described so far in the family Beggiatoaceae and probably in bacteria in general.
The polyphosphate inclusions analyzed in this study are similar to acidocalcisomes of eukaryotes as they are also enclosed by a lipid layer, likely a membrane (Fig. 2), and store cations such as calcium and magnesium within the inclusion (Fig. 4). Bacterial organelles similar to acidocalcisomes have already been found in other bacteria such as A. tumefaciens (Seufferheld et al. 2003). Acidocalcisomes are described as acidic, polyphosphate-enriched cell compartments, which are enclosed by a lipid membrane possessing ATPases, pyrophosphatases and different cation and anion transporters (Docampo et al. 2005). In contrast to the properties of acidocalcisomes, we could not detect significant acidic conditions within the polyphosphate inclusions (Fig. 3). A specific fluorescence increase at pH values between 4 and 5 which is typical for LysoSensor Green DND-189 as shown for polyphosphate granules in Chlamydomonas reinhardtii (Komine et al. 2000) was not observed. Also Acridine Orange, which would be indicative for a pronounced pH difference between cytoplasm and polyphosphate inclusions (Ramos et al. 2010), showed no corresponding accumulation in the inclusions. Nevertheless, we observed smaller acidic compartments within the cell (Fig. 3), which are not associated with polyphosphate but indicate that the staining for acidic regions was successful. Thus, we conclude that the large polyphosphate inclusions are not notably acidic, at least under our experimental conditions.
Among large sulfide-oxidizing bacteria, the investigated polyphosphate inclusions constitute a new type of vacuolar compartment. They are unique intracellular components in addition to the nitrate-storing vacuoles, which are known from the large marine genera such as “Candidatus Marithioploca”, “Candidatus Maribeggiatoa” and Thiomargarita and from smaller freshwater Thioploca species. In these genera, the stored nitrate serves as electron acceptor for sulfide oxidation (McHatton et al. 1996; Otte et al. 1999; Schulz et al. 1999; Zemskaya et al. 2009). For the genus “Candidatus Marithrix”, vacuoles without nitrate have been observed but the function of these vacuoles is still under debate (Kalanetra and Nelson 2010). Therefore, it seems plausible that vacuolar structures in sulfide-oxidizing bacteria can also serve other purposes than nitrate storage. The only genus in which both a large nitrate-storing vacuole and polyphosphate inclusions have been unambiguously detected is Thiomargarita. In contrast to the genera Beggiatoa and Thiomargarita, polyphosphate inclusions seem to be absent in “Candidatus Marithioploca” (Holmkvist et al. 2010).
In an earlier study (Brock and Schulz-Vogt 2011), we showed for Beggiatoa strain 35Flor that decomposition of polyphosphate enables the filaments to survive short periods of exposure to sulfide without an electron acceptor such as oxygen or nitrate. Although the cellular mechanism behind this physiological correlation remains unknown we assume that polyphosphate-filled vacuoles in narrow Beggiatoa spp. might have a similar purpose as the nitrate-filled vacuoles in the larger species of the family Beggiatoaceae. In the latter, the vacuoles contain nitrate concentrations of up to 800 mmol l−1 (Schulz et al. 1999), which is used for sulfide oxidation under diminished oxygen concentrations (Otte et al. 1999; McHatton et al. 1996; Schulz et al. 1999).
The narrow Beggiatoa spp. define the phylogenetic root of the family Beggiatoaceae as shown by Ahmad et al. (2006) and by our 16S rRNA data (Fig. 5). Since the polyphosphate inclusions in Beggiatoa strain 35Flor are enclosed by a lipid layer, probably a vacuolar membrane, it is likely that they are related to the aqueous, mostly nitrate-storing vacuoles found in the other genera of the Beggiatoaceae. This implies that the ability to form vacuoles is a basic evolutionary feature of the family Beggiatoaceae and that the modulation of the vacuolar size and function might reflect adaptations to different environmental conditions. Within the genus Beggiatoa, a large central vacuole for nitrate storage has so far only been found in two strains (‘Chiprana’ and 1124, Fig. 5), which are the widest representatives of this genus (6–9 μm). Likewise, the Beggiatoa strain 35Flor is among the widest of its phylogenetic group (6 μm) and features a compartmentalization by polyphosphate inclusions. This observation suggests that the overall expression of vacuoles in a filament strongly depends on the filament diameter, as only organisms beyond 5 μm so far contain intracellular vacuoles. This conclusion is in congruence with an increased diffusion limitation in cells with diameters exceeding a few μm (Schulz and Jørgensen 2001). However, already filaments of around 4.4 μm possess small polyphosphate inclusions (around 150 nm) that even compartmentalize these thin filaments (de Albuquerque et al. 2010).
In conclusion, the large polyphosphate inclusions of Beggiatoa strain 35Flor share similarities with acidocalcisomes and vacuoles in general. They possibly constitute an adaptation of narrow sulfide-oxidizing bacteria to their habitat and to their increasing cell size comparable to the aqueous, nitrate-storing vacuoles known from other genera of Beggiatoaceae. The habitats of Beggiatoa spp. and its relatives include organic-rich sediments, vent systems and cyanobacterial mats, which are characterized by frequently changing gradients of sulfide, oxygen and nitrate. The storage of compounds such as energy reserves is therefore a crucial life-strategy of gradient organisms and now includes the large polyphosphate inclusions identified here as one more extraordinary feature among these unusual bacteria.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (through MARUM Center for Marine Environmental Sciences) and the Max Planck Society. We thank M. Meyer for technical help.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Ahmad A, Kalanetra KM, Nelson DC. Cultivated Beggiatoa spp. define the phylogenetic root of morphologically diverse, noncultured, vacuolate sulfur bacteria. Can J Microbiol. 2006;52(6):591–598. doi: 10.1139/w05-154. [DOI] [PubMed] [Google Scholar]
- Ault-Riche D, Fraley CD, Tzeng CM, Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol. 1998;180(7):1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock J, Schulz-Vogt HN. Sulfide induces phosphate release from polyphosphate in cultures of a marine Beggiatoa strain. ISME J. 2011;5:497–506. doi: 10.1038/ismej.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque JP, Keim CN, Lins U. Comparative analysis of Beggiatoa from hypersaline and marine environments. Micron. 2010;41(5):507–517. doi: 10.1016/j.micron.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Docampo R, Ulrich P, Moreno SNJ. Evolution of acidocalcisomes and their role in polyphosphate storage and osmoregulation in eukaryotic microbes. Phil Trans R Soc B. 2005;365(1541):775–784. doi: 10.1098/rstb.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossing H, Gallardo VA, Jørgensen BB, Hüttel M, Nielsen LP, Schulz H, et al. Concentration and transport of nitrate by the mat-forming sulfur bacterium Thioploca. Nature. 1995;374(6524):713–715. doi: 10.1038/374713a0. [DOI] [Google Scholar]
- Greenspan P, Fowler SD. Spectrofluorometric studies of the lipid probe, Nile Red. J Lipid Res. 1985;26(7):781–789. [PubMed] [Google Scholar]
- Han JY, Burgess K. Fluorescent indicators for intracellular pH. Chem Rev. 2010;110(5):2709–2728. doi: 10.1021/cr900249z. [DOI] [PubMed] [Google Scholar]
- Hinck S, Neu TR, Lavik G, Mussmann M, De Beer D, Jonkers HM. Physiological adaptation of a nitrate-storing Beggiatoa sp. to diel cycling in a phototrophic hypersaline mat. Appl Environ Microbiol. 2007;73(21):7013–7022. doi: 10.1128/AEM.00548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmkvist L, Arning ET, Küster-Heins K, Vandieken V, Peckmann J, Zabel M, et al. Phosphate geochemistry, mineralization processes, and Thioploca distribution in shelf sediments off central Chile. Mar Geol. 2010;277(1–4):61–72. doi: 10.1016/j.margeo.2010.08.011. [DOI] [Google Scholar]
- Jørgensen BB. Distribution of colorless sulfur bacteria (Beggiatoa spp.) in a coastal sediment. Mar Biol. 1977;41(1):19–28. doi: 10.1007/BF00390577. [DOI] [Google Scholar]
- Kalanetra KM, Nelson DC. Vacuolate-attached filaments: highly productive Ridgeia piscesae epibionts at the Juan de Fuca hydrothermal vents. Mar Biol. 2010;157(4):791–800. doi: 10.1007/s00227-009-1362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalanetra KM, Huston SL, Nelson DC. Novel, attached, sulfur-oxidizing bacteria at shallow hydrothermal vents possess vacuoles not involved in respiratory nitrate accumulation. Appl Environ Microbiol. 2004;70(12):7487–7496. doi: 10.1128/AEM.70.12.7487-7496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp A, Stief P, Schulz-Vogt HN. Anaerobic sulfide oxidation with nitrate by a freshwater Beggiatoa enrichment culture. Appl Environ Microbiol. 2006;72(7):4755–4760. doi: 10.1128/AEM.00163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Teske A, Fukui M. Morphological and phylogenetic characterizations of freshwater Thioploca species from Lake Biwa, Japan, and Lake Constance, Germany. Appl Environ Microbiol. 2003;69(1):390–398. doi: 10.1128/AEM.69.1.390-398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y, Eggink LL, Park HS, Hoober JK. Vacuolar granules in Chlamydomonas reinhardtii: polyphosphate and a 70-kDa polypeptide as major components. Planta. 2000;210(6):897–905. doi: 10.1007/s004250050695. [DOI] [PubMed] [Google Scholar]
- Kornberg A. Inorganic polyphosphate—toward making a forgotten polymer unforgettable. J Bacteriol. 1995;177(3):491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Kulaev IS, Vagabov VM, Kulakovskaja TV. The biochemistry of inorganic polyphosphates. 2. New York: Wiley; 2004. [Google Scholar]
- Lawry NH, Jensen TE. Deposition of condensed phosphate as an effect of varying sulfur deficiency in the cyanobacterium Synechococcus sp. (Anacystis nidulans) Arch Microbiol. 1979;120(1):1–7. doi: 10.1007/BF00413264. [DOI] [Google Scholar]
- Lin HJ, Herman P, Kang JS, Lakowicz JR. Fluorescence lifetime characterization of novel low-pH probes. Anal Biochem. 2001;294(2):118–125. doi: 10.1006/abio.2001.5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucl Acids Res. 2004;32(4):1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier S, Murray RGE. Fine structure of Thioploca ingrica and a comparison with Beggiatoa. Can J Microbiol. 1965;11(4):645–655. doi: 10.1139/m65-087. [DOI] [PubMed] [Google Scholar]
- McHatton SC, Barry JP, Jannasch HW, Nelson DC. High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl Environ Microbiol. 1996;62(3):954–958. doi: 10.1128/aem.62.3.954-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mußmann M, Schulz HN, Strotmann B, Kjaer T, Nielsen LP, Rossello-Mora RA, et al. Phylogeny and distribution of nitrate-storing Beggiatoa spp. in coastal marine sediments. Environ Microbiol. 2003;5(6):523–533. doi: 10.1046/j.1462-2920.2003.00440.x. [DOI] [PubMed] [Google Scholar]
- Mußmann M, Hu FZ, Richter M, de Beer D, Preisler A, Jørgensen BB, et al. Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 2007;5(9):1923–1937. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Teske A, Wirsen CO, Jannasch HW. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164(3):165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- Nelson DC, Jannasch HW. Chemoautotrophic growth of a marine Beggiatoa in sulfide-gradient cultures. Arch Microbiol. 1983;136(4):262–269. doi: 10.1007/BF00425214. [DOI] [Google Scholar]
- Nelson DC, Waterbury JB, Jannasch HW. Nitrogen fixation and nitrate utilization by marine and freshwater Beggiatoa. Arch Microbiol. 1982;133(3):172–177. doi: 10.1007/BF00414997. [DOI] [Google Scholar]
- Nelson DC, Revsbech NP, Jørgensen BB. Microoxic-anoxic niche of Beggiatoa spp.: microelectrode survey of marine and freshwater strains. Appl Environ Microbiol. 1986;52(1):161–168. doi: 10.1128/aem.52.1.161-168.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Wirsen CO, Jannasch HW. Characterization of large, autotrophic Beggiatoa spp. abundant at hydrothermal vents of the Guaymas basin. Appl Environ Microbiol. 1989;55(11):2909–2917. doi: 10.1128/aem.55.11.2909-2917.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte S, Kuenen JG, Nielsen LP, Paerl HW, Zopfi J, Schulz HN, et al. Nitrogen, carbon, and sulfur metabolism in natural Thioploca samples. Appl Environ Microbiol. 1999;65(7):3148–3157. doi: 10.1128/aem.65.7.3148-3157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplies J, Kottmann R, Ludwig W, Glöckner FO. A standard operating procedure for phylogenetic inference (SOPPI) using (rRNA) marker genes. Syst Appl Microbiol. 2008;31(4):251–257. doi: 10.1016/j.syapm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res. 2007;35(21):7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos IB, Miranda K, Ulrich P, Ingram P, LeFurgey A, Machado EA, et al. Calcium- and polyphosphate-containing acidocalcisomes in chicken egg yolk. Biol Cell. 2010;102(7):421–434. doi: 10.1042/BC20100011. [DOI] [PubMed] [Google Scholar]
- Rosenberg R, Diaz RJ. Sulfur bacteria (Beggiatoa spp.) mats indicate hypoxic conditions in the inner Stockholm archipelago. AMBIO. 1993;22(1):32–36. [Google Scholar]
- Salman V, Amann R, Girnth A-C, Polerecky L, Bailey JV, Høgslund S, et al. A single-cell sequencing approach to the classification of large, vacuolated sulfur bacteria. Syst Appl Microbiol. 2011;34(4):243–259. doi: 10.1016/j.syapm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Schmaljohann R, Drews M, Walter S, Linke P, von Rad U, Imhoff JF. Oxygen-minimum zone sediments in the northeastern Arabian Sea off Pakistan: a habitat for the bacterium Thioploca. Mar Ecol Prog Ser. 2001;211:27–42. doi: 10.3354/meps211027. [DOI] [Google Scholar]
- Schulz HN, Jørgensen BB. Big bacteria. Annu Rev Microbiol. 2001;55:105–137. doi: 10.1146/annurev.micro.55.1.105. [DOI] [PubMed] [Google Scholar]
- Schulz HN, Schulz HD. Large sulfur bacteria and the formation of phosphorite. Science. 2005;307(5708):416–418. doi: 10.1126/science.1103096. [DOI] [PubMed] [Google Scholar]
- Schulz HN, Brinkhoff T, Ferdelman TG, Marine MH, Teske A, Jørgensen BB. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science. 1999;284(5413):493–495. doi: 10.1126/science.284.5413.493. [DOI] [PubMed] [Google Scholar]
- Schulz HN, Strotmann B, Gallardo VA, Jørgensen BB. Population study of the filamentous sulfur bacteria Thioploca spp. off the Bay of Concepcion, Chile. Mar Ecol Prog Ser. 2000;200:117–126. doi: 10.3354/meps200117. [DOI] [Google Scholar]
- Seufferheld M, Vieira MCF, Ruiz FA, Rodrigues CO, Moreno SNJ, Docampo R. Identification of organelles in bacteria similar to acidocalcisomes of unicellular eukaryotes. J Biol Chem. 2003;278(32):29971–29978. doi: 10.1074/jbc.M304548200. [DOI] [PubMed] [Google Scholar]
- Seufferheld MJ, Alvarez HM, Farias ME. Role of polyphosphates in microbial adaptation to extreme environments. Appl Environ Microbiol. 2008;74(19):5867–5874. doi: 10.1128/AEM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohl WR. Genus III. Beggiatoa. In: Garrity G, Brenner DJ, Krieg NR, Staley JR, editors. Bergey’s manual of systematic bacteriology. New York: Springer; 2005. pp. 148–161. [Google Scholar]
- Strohl WR, Larkin JM. Enumeration, isolation and characterization of Beggiatoa from freshwater sediments. Appl Environ Microbiol. 1978;36(5):755–770. doi: 10.1128/aem.36.5.755-770.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohl WR, Geffers I, Larkin JM. Structure of the sulfur inclusion envelopes from four beggiatoas. Curr Microbiol. 1981;6(2):75–79. doi: 10.1007/BF01569007. [DOI] [Google Scholar]
- Sweerts JPRA, De Beer D, Nielsen LP, Verdouw H, Vandenheuvel JC, Cohen Y, et al. Denitrification by sulphur oxidizing Beggiatoa spp. mats on freshwater sediments. Nature. 1990;344(6268):762–763. doi: 10.1038/344762a0. [DOI] [Google Scholar]
- Teske A, Nelson DC. The genera Beggiatoa and Thioploca. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes. New York: Springer; 2006. pp. 784–810. [Google Scholar]
- Tijssen JPF, Beekes HW, Vansteveninck J. Localization of polyphosphates in Saccharomyces fragilis, as revealed by 4′, 6-diamidino-2-phenylindole fluorescence. Biochim Biophys Acta. 1982;721(4):394–398. doi: 10.1016/0167-4889(82)90094-5. [DOI] [PubMed] [Google Scholar]
- Zemskaya TI, Chernitsyna SM, Dul’tseva NM, Sergeeva VN, Pogodaeva TV, Namsaraev BB. Colorless sulfur bacteria Thioploca from different sites in Lake Baikal. Microbiology. 2009;78(1):117–124. doi: 10.1134/S0026261709010159. [DOI] [PubMed] [Google Scholar]