Abstract
The rice disease resistance (R) gene Xa3/Xa26 (having also been named Xa3 and Xa26) against Xanthomonas oryzae pv. oryzae (Xoo), which causes bacterial blight disease, belongs to a multiple gene family clustered in chromosome 11 and is from an AA genome rice cultivar (Oryza sativa L.). This family encodes leucine-rich repeat (LRR) receptor kinase-type proteins. Here, we show that the orthologs (alleles) of Xa3/Xa26, Xa3/Xa26-2, and Xa3/Xa26-3, from wild Oryza species O. officinalis (CC genome) and O. minuta (BBCC genome), respectively, were also R genes against Xoo. Xa3/Xa26-2 and Xa3/Xa26-3 conferred resistance to 16 of the 18 Xoo strains examined. Comparative sequence analysis of the Xa3/Xa26 families in the two wild Oryza species showed that Xa3/Xa26-3 appeared to have originated from the CC genome of O. minuta. The predicted proteins encoded by Xa3/Xa26, Xa3/Xa26-2, and Xa3/Xa26-3 share 91–99% sequence identity and 94–99% sequence similarity. Transgenic plants carrying a single copy of Xa3/Xa26, Xa3/Xa26-2, or Xa3/Xa26-3, in the same genetic background, showed a similar resistance spectrum to a set of Xoo strains, although plants carrying Xa3/Xa26-2 or Xa3/Xa26-3 showed lower resistance levels than the plants carrying Xa3/Xa26. These results suggest that the Xa3/Xa26 locus predates the speciation of A and C genome, which is approximately 7.5 million years ago. Thus, the resistance specificity of this locus has been conserved for a long time.
Keywords: Bacterial blight, broad-spectrum resistance, durable resistance, Oryza officinalis, Oryza minuta, Oryza sativa
INTRODUCTION
Bacterial blight, caused by Xanthomonas oryzae pv. oryzae (Xoo), is one of the major diseases affecting rice production. Breeding rice with the quality of broad-spectrum and durable disease resistance is one of the principal goals of rice improvement (Yang et al., 2008; Kou and Wang, 2010). Exploitation and utilization of major resistance (R) genes is an effective way to control bacterial blight. Wild germplasm is always recognized as an important repository of useful allelic variation for crop improvement including resistance against Xoo.
Harlan and de Wet (1971) proposed a concept involving three levels of the gene pools for classifying crop species and their wild relatives, according to the crossability between them. The genus Oryza includes two cultivated rice species, Asian cultivated rice Oryza sativa (AA genome) and African cultivated rice Oryza glaberrima (AA genome), and 22 wild species, representing AA, BB, CC, BBCC, CCDD, EE, FF, GG, HHJJ, and KKLL genomes (Ge et al., 1999; Lu et al., 2009; Ammiraju et al., 2010). According to the crossability between O. sativa and other Oryza species, the wild species also have been categorized as the primary, secondary, and tertiary gene pools for the cultivars (Khush, 1997). All AA genome Oryza species are relatively easy to cross with O. sativa and are regarded as the primary gene pool, the wild species with BB, CC, BBCC, CCDD, EE, and FF genomes constitute the secondary gene pool, and the remaining wild species with the GG, HHJJ, and KKLL genomes belong to the tertiary gene pool (Khush, 1997; Lu et al., 2009).
The classic example of transfer of a disease resistance gene against Xoo from a wild species into cultivated rice is the introgression of Xa21 from the AA genome wild rice Oryza longistaminata into Asian cultivated rice (Khush et al., 1990). Xa21 has a broad-spectrum of resistance to nearly all races of Xoo in the Philippines (Cottyn and Mew, 2004); however, the activity of Xa21 is developmentally regulated (Century et al., 1999; Zhao et al., 2009). Another broad-spectrum bacterial blight resistance gene Xa23, from Oryza rufipogon (AA), was transferred into Asian cultivated rice and is active throughout the lifecycle of the plant (Zhang et al., 2000). Both Xa21 and Xa23 are located on chromosome 11 in cultivated rice (Song et al., 1995; Zhang et al., 2000). Wild Oryza species belonging to the secondary gene pool have also provided valuable genes that confer resistance to Xoo, such as Xa27 from Oryza minuta (BBCC) (Amante-Bordeos et al., 1992) and Xa29(t) from Oryza officinalis (CC) (Tan et al., 2004). The introgressed Xa27 and Xa29(t) genes are located on chromosomes 6 and 1 in cultivated rice, respectively (Gu et al., 2004; Tan et al., 2004). However, only limited gene transfer is possible from the secondary gene pool into the Asian cultivated rice by crossing, because there is limited homology between O. sativa and these wild Oryza species (Khush, 1997).
Rice Xa3/Xa26, encoding a leucine-rich repeat (LRR) receptor kinase-type protein, is an R gene conferring resistance against Xoo (Sun et al., 2004; Xiang et al., 2006). Asian-cultivated rice consists of two major subspecies, indica (O. sativa L. ssp. indica) and japonica (O. sativa L. ssp. japonica). Xa3/Xa26 was first identified in the indica rice cultivar Minghui 63 and named Xa26 (Yang et al., 2003). Further study revealed that Xa3, an R gene conferring resistance against Xoo, and Xa26 are actually the same gene and then it was renamed Xa3/Xa26 (Xiang et al., 2006). Xa3/Xa26 has been an important resistance gene for both indica and japonica rice production in China for a long time (Xu et al., 2004; Gao et al., 2010). Quantitative trail locus for disease resistance and defenses-responsive genes functioning and putatively functioning downstream of Xa3/Xa26 in defense signaling have been identified (Hu et al., 2008; Qiu et al., 2008, 2009; Kou et al., 2010). In this study, we used an ortholog (allele) mining strategy to isolate new R genes against Xoo in the Xa3/Xa26 family from wild Oryza species. The Xa3/Xa26 orthologs or alleles, Xa3/Xa26-2 from O. officinalis, and Xa3/Xa26-3 from O. minuta that encodes proteins different from Xa3/Xa26 protein can mediate a similar spectrum of resistance against Xoo. The Xa3/Xa26-carrying cultivars have been widely used in rice production for a long period of time. These results suggest that the Xa3/Xa26 locus may confer a durable resistance.
RESULTS
Isolation of Xa3/Xa26 Family Genes from O. officinalis and O. minuta
A total of nine and seven positive bacterial artificial chromosome (BAC) clones were identified after screening high-density hybridization filters from the O. officinalis and O. minuta BAC libraries, respectively, using a set of Xa3/Xa26 family DNA probes. Based on the finger printing analysis of these clones (Supplemental Figure 1), BAC clones OO_Ba0120J21 from O. officinalis and OM_Ba0293H21 from O. minuta, which had the greatest degree of overlap with other positive clones from the same genome, were sequenced.
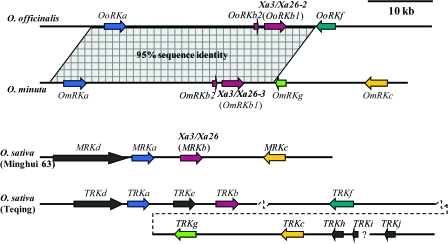
Analysis of approximately 113-kb sequences of clone OO_Ba0120J21 identified four putative genes that belonged to the Xa3/Xa26 gene family. Three of the four paralogs in O. officinalis were orthologs of the Xa3/Xa26 gene family TRKa, TRKb, and TRKf in Asian rice cultivar Teqing (O. sativa L. ssp. indica; Sun et al., 2006), respectively, according to their sequence similarity, corresponding locations within the family, and transcriptional orientation; they were thus named OoRKa, OoRKb1, and OoRKf (Figure 1). The fourth paralog in O. officinalis showed very high sequence similarity with OoRKb1 (94%) and TRKb (94%); it could be generated through tandem duplication or unequal crossover and thus was named OoRKb2.
Figure 1.
Comparison of Organization of the Xa3/Xa26 Gene Family in Two Wild Oryza Species, O. officinalis and O. minuta, with that in Cultivated Rice Varieties, Minghui 63 and Teqing.Members of the Xa3/Xa26 family and their transcription orientation are indicated by arrows. Regions showing extreme sequence identity (95% DNA identity overall) between the two wild species are shadowed. Organization of the Xa3/Xa26 family in Minghui 63 and Teqing is drawn according to published data (Sun et al., 2004, 2006); the dotted slant indicates the border between two sequence contigs; the question mark indicates that the neighborhood of two adjacent contigs is deduced according to sequence similarity and the Xa3/Xa26 family organization in other cultivated rice varieties.
Analysis of approximately 149 kb of sequence of clone OM_Ba0293H21 identified five putative genes that showed sequence homology with the Xa3/Xa26 family orthologs. The five paralogs spanned an approximately 50-kb region (Figure 1) and four of them showed a high degree of sequence similarity respective to the Xa3/Xa26 family orthologs TRKa, TRKb, TRKg, and TRKc in rice cultivar Teqing; they were designated OmRKa, OmRKb1, OmRKg, and OmRKc. The fifth paralog in O. minuta showed very high sequence similarity with OoRKb2 (99%) and OmRKb1 (94%); it was named OmRKb2. These results suggest that the origin of member b2 must have occurred before polyploidization.
OoRKb1, OmRKa, and OmRKb1 were predicted to be intact genes, using Xa3/Xa26 as the reference (Supplemental Figure 2). OoRKa and OmRKc were pseudogenes, with each containing a frame-shift mutation. OoRKb2, OoRKf, OmRKb2, and OmRKg were truncated as compared to Xa3/Xa26 (Supplemental Figure 2). OoRKb2 and OmRKb2 appeared to encode only an incomplete kinase domain. OoRKf lost the predicted intron and the following sequence of this family and putatively encoded an intact LRR domain, but an incomplete kinase domain. OmRKg contained an in-frame stop codon and putatively encoded an incomplete LRR domain. The orthologs a, b, and c in rice cultivar Teqing are intact genes; the orthologs f and g in Teqing are pseudogenes with insertion, deletion, or in-frame stop codon, which are different from the mutations in OoRKf and OmRKg (Sun et al., 2006) (Supplemental Figure 2).
Comparing the sequences of Xa3/Xa26 family orthologs in the two wild Oryza species to four Asian rice cultivars, Minghui 63 (O. sativa L. ssp. indica), Teqing, 93–11 (O. sativa L. ssp. indica), and Nipponbare (O. sativa L. ssp. japonica) (Sun et al., 2006), it was easy to recognize that they had very low sequence similarity (<49% sequence identity) between cultivated rice and wild Oryza species in the intergenic regions. However, nearly 34-kb regions with 95% sequence identity in the Xa3/Xa26 family were identified between O. officinalis and O. minuta, which harbored orthologs a, b1, and b2 (Figure 1). These results suggest that the Xa3/Xa26 families in the two wild Oryza species are less diverse between each other than as compared to the diversity found between them and cultivars.
Xa3/Xa26 Orthologs of the Wide Oryza Species Were Functional Disease Resistance Genes
OoRKb1 and OmRKb1 were the orthologs of the R gene Xa3/Xa26. To determine whether OoRKb1 and OmRKb1 were functional in the rice–Xoo interaction, genomic fragments containing OoRKb1 and OmRKb1 with their native promoters were individually transformed into rice cultivar Mudanjiang 8, which is susceptible to Xoo (Supplemental Figure 3). Thirty-one independent positive transformants carrying OoRKb1 (named D101OM) and 18 independent positive transformants carrying OmRKb1 (named D103OM) were obtained. Eleven of the T0 plants transformed with OoRKb1 construct showed significantly (P < 0.01) enhanced resistance to Xoo strain PXO61, with lesion areas ranging from 3.4 to 35.0%, compared to 50.3% for wild-type Mudanjiang 8; another 10 of the T0 plants transformed with the same construct showed significantly (P < 0.01) enhanced resistance to Xoo strain PXO341, with lesion areas ranging from 1.6 to 44.4%, compared to 61.2% for wild-type Mudanjiang 8 (Supplemental Table 1). All 18 T0 plants transformed with OmRKb1 construct showed significantly (P < 0.01) enhanced resistance to Xoo strain PXO61, with lesion areas ranging from 6.7 to 27.5%, compared to 55.2% for wild-type Mudanjiang 8 (Supplemental Table 1).
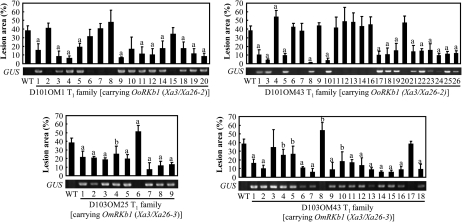
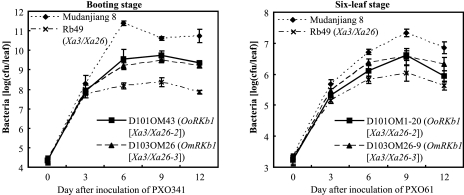
Two T1 families from resistant T0 (D101OM1 and D101OM43) and Two T1 families from resistant T0 (D103OM25 and D103OM43) were further analyzed for resistance to PXO61, and for the existence of the transgenic marker gene GUS that was tightly linked to the targeted transgene. The results showed that the enhanced resistance was associated with the presence of OoRKb1/GUS or OmRKb1/GUS in the T1 families (Figure 2). The bacterial growth rates in OoRKb1- and OmRKb1-carrying plants were 7–51-fold and 13–153-fold lower than that in wild-type at the booting (panicle development) stage and 4–8-fold and 2–5-fold lower than that in wild-type at six-leaf stage at 6–12 d after infection, respectively (Figure 3). These results suggest that OoRKb1 and OmRKb1 confer Xoo resistance. Thus, we designated them Xa3/Xa26-2 (OoRKb1; GenBank accession number: HQ148674) and Xa3/Xa26-3 (OmRKb1; GenBank accession number: HQ148675) based on the naming system of rice R genes against Xoo.
Figure 2.
Enhanced Resistance to Xoo Strain PXO61 Associated with the Presence of a GUS Marker Gene that Was Tightly Linked to OoRKb1 or OmRKb1 in T1 Families at Booting Stage.Bars represent mean (three to five replicates) ± standard deviation. The ‘a’ or ‘b’ indicates that a significant difference between transgenic and wild-type (WT) Mudanjiang 8 was detected at P < 0.01 or P < 0.05, respectively.
Figure 3.
Growth of Xoo Strains PXO341 and PXO61 in Leaves of OoRKb1-Carrying (D101OM) and OmRKb1-Carrying (D103OM) Plants at Adult (Booting) Stage (T1 Plants) and Seedling (Six-Leaf) Stage (T3 Homozygous Lines).Bacterial populations were determined from three leaves at each time point by counting colony-forming units (cfu). Mudanjiang 8 was wild-type. Rb49 was a transgenic line carrying Xa3/Xa26 regulated by its native promoter in Mudanjiang 8 background.
Xa3/Xa26-2 and Xa3/Xa26-3 Mediated Broad-Spectrum Resistance
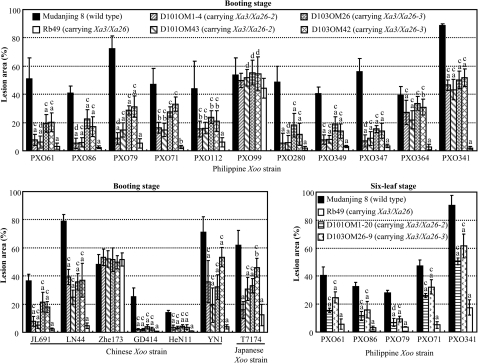
Transgenic plants carrying a single copy of Xa3/Xa26-2 or Xa3/Xa26-3 were examined for their responses to different Xoo strains. The resistance of the transgenic plants to 16 of the 18 strains was significantly enhanced as compared to the wild-type Mudanjiang 8 at adult stage (Figure 4). The lesion areas of the transgenic plants carrying Xa3/Xa26-2 and plants carrying Xa3/Xa26-3 to the 16 Xoo strains were 31–92% and 15–91% smaller than those of the wild-type plants, respectively. The plants carrying Xa3/Xa26-2, and the plants carrying Xa3/Xa26-3, showed a similar resistance spectrum to the transgenic rice line Rb49, which carried a single copy of Xa3/Xa26 driven by its native promoter with the same genetic background as the transgenic plants carrying Xa3/Xa26-2 or Xa3/Xa26-3 (Cao et al., 2007a). However, the resistance level of plants carrying Xa3/Xa26 was significantly higher than plants carrying Xa3/Xa26-2 or Xa3/Xa26-3 to all 16 Xoo strains, which showed incompatible reactions with all the transgenic plants (Figure 4). Although the Rb49 line was compatible with Xoo strain PXO99, plants carrying Xa3/Xa26-2 and plants carrying Xa3/Xa26-3 appeared to be more susceptible to PXO99 than the Rb49. In addition, plants carrying Xa3/Xa26-2 appeared to have a higher level of resistance to some of the Xoo strains than plants carrying Xa3/Xa26-3 (Figure 4). Xa3/Xa26-2- and Xa3/Xa26-3-carrying plants were also showed enhanced resistance to different Xoo strains at seedling stage, but the resistance level was significantly lower than that of Xa3/Xa26-carrying plants (Figure 4). Consistent with these results, bacterial growth rates in Xa3/Xa26-2- and Xa3/Xa26-3-carrying plants were also higher than that in Xa3/Xa26-carrying plants (Figure 3). These results suggest that Xa3/Xa26-2 and Xa3/Xa26-3 can mediate a broad-spectrum resistance to Xoo as their ortholog Xa3/Xa26.
Figure 4.
Resistance Spectrum of Xa3/Xa26-2 and Xa3/Xa26-3 to Xoo at Adult (Booting) and Seedling (Six-Leaf) Stages.All the transgenic plants carried a single copy of transgene. D101OM43, D103OM26, and D103OM42 are resistance T1 transgenic plants. D101OM1-4 is resistance T2 transgenic plants. D101OM1-20 and M103OM26-9 are homozygous resistance transgenic lines (T3 generation). Rb49 is a resistance transgenic line carrying Xa3/Xa26 regulated by its native promoter with the genetic background of Mudanjiang 8 (Cao et al. 2007a). Bars represent mean (6–10 replicates for D101OM and D103OM and 3–5 replicates for Rb49 and wild-type at booting stage and 3–6 replicates at six-leaf stage) ± standard deviation. The ‘a’ or ‘b’ indicates that a significant difference between transgenic and wild-type was detected at P < 0.01 or P < 0.05, respectively. The ‘c’ or ‘d’ indicates that a significant difference between the Xa3/Xa26-2-carrying or Xa3/Xa26-3-carrying plants and Xa3/Xa26-carrying plants was detected at P < 0.01 or P < 0.05, respectively.
The two wild Oryza accessions used for constructing the BAC libraries were resistance to Xoo (Brar and Khush, 2002). We inoculated O. officinalis (accession 100896) and O. minuta (accession 101141), which were used for construction of the BAC libraries, with three Philippine Xoo races, PXO61, PXO99, and PXO341. They both present a high-level resistance to these Xoo strains as compared to susceptible rice variety Mudanjiang 8 (Table 1). Transgenic plants carrying Xa3/Xa26-2 or Xa3/Xa26-3 showed enhanced resistance to PXO61 and PXO341 but susceptibility to PXO99 as the transgenic line (Rb49) carrying Xa3/Xa26 (Figure 4). These results suggest that the resistance of the two wild Oryza species to Xoo is at least partly contributed by Xa3/Xa26-2 or Xa3/Xa26-3. However, they may also carry other R genes against Xoo in addition to Xa3/Xa26-2 or Xa3/Xa26-3.
Table 1.
Resistance of Two Wild Oryza Species to Xoo as Compared to Cultivated Rice Mudanjiang 8.1
| Xoo strain | Mudanjiang 8 | O. officinalis (accession 100896)2 | O. minuta (accession 101141)2 | Mudanjiang 8 | Rb49 (Xa3/Xa26, Mudanjiang 8 background)3 |
| PXO61 | 55.3 ± 16.0 | 2.7 ± 0.9a | 1.4 ± 0.5a | 50.9 ± 14.7 | 3.1 ± 2.1a |
| PXO99 | 57.7 ± 14.8 | 8.6 ± 2.1a | 2.4 ± 0.2a | 53.8 ± 11.8 | 44.5 ± 7.0 |
| PXO341 | 81.6 ± 6.0 | 9.9 ± 3.4a | 1.5 ± 1.0a | 88.7 ± 1.0 | 2.1 ± 0.8a |
Three to five uppermost fully expanded leaves of each plant were inoculated. Data represent mean (three to five replicates) ± standard deviation. The ‘a’ indicates that a significant difference between wild species or rice transgenic line Rb49 and susceptible rice Mudanjiang 8 (control) was detected at P < 0.01.
The lesion areas (%) were measured 23 d after inoculation.
The lesion areas (%) were measured 14 d after inoculation of PXO61 and PXO99 and 12 d after inoculation of PXO341.
Variations of Amino-Acid Sequences Encoded by Xa3/Xa26 Orthologs
Aligning the genomic and cDNA sequences of Xa3/Xa26-2 and Xa3/Xa26-3 showed that the two orthologs had similar structures to Xa3/Xa26: two exons and one intron that was inserted in the region encoding the kinase domain (Figure 5). Xa3/Xa26-2 and Xa3/Xa26-3 had nearly the same structure: the same sizes of exons and 5'- and 3'-untranslated regions. The only difference between the two orthologs was that they had different sizes of introns (103 versus 104 nucleotides). Comparative sequence analyses of the coding regions and introns of Xa3/Xa26, Xa3/Xa26-2, and Xa3/Xa26-3 revealed 82–99% identity among the three orthologs.
Figure 5.
Comparison of the Structures of Xa3/Xa26-2 and Xa3/Xa26-3 with Xa3/Xa26.The coding regions (black boxes) of the genes are interrupted by one intron (line). The positions of 5' and 3' UTR (white boxes), translation start codon (ATG), and translation stop codon (TGA) are also indicated. The numbers indicate the nucleotides of each substructure.
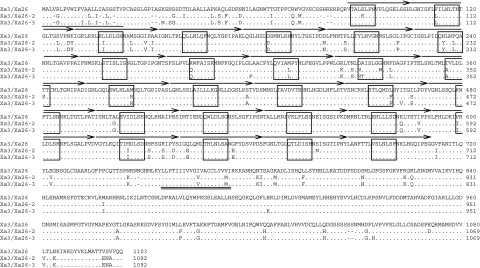
Both Xa3/Xa26-2 and Xa3/Xa26-3 encode proteins that consisted of 1092 amino acids as compared to 1 103 amino acids encoded by Xa3/Xa26. The predicted proteins encoded by the three orthologs share 91–99% sequence identity and 94–99% sequence similarity. Xa3/Xa26-2 and Xa3/Xa26-3 proteins share 98% sequence identity and have only a total of 14 amino-acid residues distinct from each other (Figure 6). Of these 14 amino-acid changes, four are located in the region in front of the LRR domain, seven in the LRR domain, and three in the kinase domain.
Figure 6.
Alignment of Xa3/Xa26-2, Xa3/Xa26-3, and Xa3/Xa26 Proteins.All sequences are compared with the reference Xa3/Xa26. The predicted signal peptide sequence is underlined. The predicted transmembrane region is double underlined. The arrows above the amino-acid residues indicate the LRR repeats and the xxLxLxx motifs of the LRR domain are boxed. Dots represent identical amino acids of Xa3/Xa26-2 and Xa3/Xa26-3 to Xa3/Xa26. The dash represents the single amino acid absent in Xa3/Xa26-2 and Xa3/Xa26-3.
Both Xa3/Xa26-2 and Xa3/Xa26-3 proteins share 91% sequence identity to Xa3/Xa26 protein. The two proteins have 88 and 90 amino-acid difference from Xa3/Xa26, respectively (Figure 6). Of these 88 and 90 amino-acid changes, 11 are amino-acid deletions (eight in the regions in front of the LRR domain, two in the kinase domains, and one in the juxtamembrane regions) in the same sites of Xa3/Xa26-2 and Xa3/Xa26-3. Of the 77 and 79 total amino-acid substitutions compared to Xa3/Xa26, approximately half of them (39 and 38) are in the LRR domains of Xa3/Xa26-2 and Xa3/Xa26-3, respectively. Most of the substitution sites are the same in the LRR domains of the two proteins. Compared to Xa3/Xa26, the distributions of the amino-acid changes in the LRR regions of Xa3/Xa26-2 and Xa3/Xa26-3 are dispersed. Nearly one-third of the variants in the LRR regions lie within the xxLxLxx (‘x’ indicates any amino-acid residue) motifs. Xa3/Xa26-2 and Xa3/Xa26-3, compared to Xa3/Xa26, have the same nine substitutions in the transmembrane and juxtamembrane regions. Nearly half of the remaining variants are concentrated in the regions in front of the LRR domain, within an approximate a range of 80 amino acids. The rest of the variants are dispersed in the kinase domains of Xa3/Xa26-2 and Xa3/Xa26-3 proteins. These results suggest that Xa3/Xa26-2 and Xa3/Xa26-3 are more evolutionarily related to each other than their evolutionary relationship with Xa3/Xa26.
DISCUSSION
The Xa3/Xa26 family is a potential disease resistance gene reservoir. In addition to Xa3/Xa26, the MRKa, a paralog of Xa3/Xa26 in rice cultivar Minghui 63, can mediate Xoo resistance when enhancing its expression (Cao et al., 2007b). Other paralogs of this family in different Asian rice cultivars are expressed in rice leaves, which are one of the major invasion sites of pathogens, suggesting their potential role in rice–pathogen interactions (Xu et al., 2007). The present results further support the assumption that the Xa3/Xa26 family is rich in genes for disease resistance.
It is generally accepted that the LRR domains of R proteins function directly or indirectly in recognition of pathogen effectors and play an important role in race-specific resistance (Rivas and Thomas, 2005; Ellis et al., 2007). Comparative sequence analysis of Xa3/Xa26 family paralogs in four rice cultivars reveals that positive selection of point mutations in the LRR domains is a major force for the evolution of this family, suggesting the important roles of LRR domains in the functions of these family members (Sun et al., 2006). Domain swapping analysis further supports this hypothesis; this analysis has revealed that the LRR domain of Xa3/Xa26 protein is a major determinant of race-specific recognition during rice–Xoo interaction (Zhao et al., 2009). Furthermore, the juxtamembrane region of Xa3/Xa26 protein also appears to contribute to resistance specificity (Zhao et al., 2009). In addition, genetic background also influences the resistance spectrum and resistance level conferred by Xa3/Xa26 (Cao et al., 2007a; Zhou et al., 2009). Analysis of the predicted amino-acid sequences of Xa3/Xa26-2 and Xa3/Xa26-3 revealed the same known motif (transmembrane region) and domains (LRR and kinase) with Xa3/Xa26 (Figure 6). Xa3/Xa26-2 and Xa3/Xa26-3 also harbor 26 imperfect LRRs with consensus sequence of L/IxxLxxLxxLxLxxNxLxGxIPxx for LRR (‘x’ indicating any amino acid) as Xa3/Xa26. Although more than 40% of the amino-acid diversity residues between Xa3/Xa26-2 or Xa3/Xa26-3 and Xa3/Xa26 occur in the LRR domain, even in the xxLxLxx motif that forms the solvent-exposed surface for pathogen recognition in the LRR domain (Padmanabhan et al., 2009), plants carrying Xa3/Xa26-2 or Xa3/Xa26-3 showed a similar resistance spectrum to plants carrying Xa3/Xa26. These results suggest that these polymorphic amino-acid residues in Xa3/Xa26-2 and Xa3/Xa26-3 proteins, compared to Xa3/Xa26 protein, are not at the core region for pathogen recognition specificity. However, further study is required to determine whether some of these amino-acid changes may influence the level of resistance.
O. officinalis and O. minuta belong to the O. officinalis complex, and both species carry the C genome. O. minuta is a BBCC genome allotetraploid and is thought to have arisen from a hybridization between O. officinalis, a CC genome diploid, and an extinct BB diploid species (Wang et al., 2009). Comparative sequence analysis revealed a high degree of sequence homology of Xa3/Xa26 families between diploid O. officinalis and tetraploid O. minuta, suggesting that the analyzed BAC clone containing Xa3/Xa26 family in O. minuta might have originated from the CC genome. Thus, both Xa3/Xa26-2 and Xa3/Xa26-3 belong to the CC genome. Xa3/Xa26 was first isolated from rice cultivar Minghui 63 (AA genome) (Sun et al., 2004). Minghui 63 has the pedigree of O. nivara, an AA genome wild species (Xie, 1998; Khush and Virk, 2005). Comparison of the Xa3/Xa26 family sequences in Minghui 63 (Sun et al., 2006) and O. nivara (Li and Wang, unpublished data) revealed that Xa3/Xa26 and its paralog MRKc as well as the intergenic region between Xa3/Xa26 and MRKc in Minghui 63 share 98, 98, and 97% sequence identify to their orthologs and the corresponding intergenic region in O. nivara, respectively. However, the ortholog of Xa3/Xa26 in the O. nivara accession used for sequencing is a pseudogene with an in-frame stop codon. This comparison suggests that the region harboring Xa3/Xa26 in Minghui 63 might have been introduced from an O. nivara accession that was similar to the accession used for sequencing. The AA and CC genome lineages diversified ∼7.5 million years ago (Ammiraju et al., 2008; Lu et al., 2009; Ammiraju et al., 2010; Sanyal et al., 2010). The orthologs at Xa3/Xa26 locus in different Oryza species confers a similar resistance spectrum, which may imply that this resistance locus appeared earlier than the divergence of the AA and CC genomes. Thus, the resistance function of Xa3/Xa26 locus appears to be relatively conserved during evolution.
Xa3/Xa26 family proteins belong to the same type of LRR-receptor kinase proteins as rice Xa21 protein, which also mediates resistance to Xoo (Sun et al., 2004). Xa21 functions both as an R protein and as a pattern recognition receptor (PRR) by recognition of an evolutionarily conserved pathogen-associated molecular pattern, a sulfated peptide (Lee et al., 2009). Another well-studied PRR is Arabidopsis FLS2, which also encodes a LRR-receptor kinase (Gómez-Gómez and Boller, 2000; Ali and Reddy, 2008). The Xa3/Xa26, Xa3/Xa26-2, and Xa3/Xa26-3 from the two diverged genomes mediate similar resistance spectrums. It remains to be elucidated whether the encoding proteins of Xa3/Xa26 and its alleles perceive the same conserved pathogen component in rice–Xoo interaction.
Durable resistance refers to resistance that remains effective during its prolonged and widespread use in environments favorable to pathogen or disease spread (Johnson, 1981). The indica rice cultivar Minghui 63, carrying Xa3/Xa26, is a parent of a set of hybrids that account for more than 20% of total rice production area in China for the last two decades. In addition, Xa3/Xa26 is also an important resistance gene in japonica cultivar breeding in China (Xu et al., 2004). The two wild Oryza accessions used both presented a high level of resistance to Xoo strains PXO61, PXO99, and PXO341. However, transgenic plants carrying Xa3/Xa26-2 or Xa3/Xa26-3 showed enhanced resistance to PXO61 and PXO341 but susceptibility to PXO99 as the transgenic line (Rb49) carrying Xa3/Xa26. These results indicated that there is another R gene(s) in the two wild Oryza species against PXO99. Although the CC and BBCC wild rice species have not been cultivated in large areas, the orthologs, originated at least 7.5 million years ago, still remain functionally in the same genomes as other R genes together by the long natural selection. The wide use of Xa3/Xa26-carrying cultivars in rice production and the similar resistance specificity of the ancient Xa3/Xa26-2 and Xa3/Xa26-3 as the present Xa3/Xa26 suggest that this R gene locus may confer durable resistance in addition to conferring a relative broad-spectrum resistance.
METHODS
Selection of BAC Clone
Mixed probes of DNA segments of Xa3/Xa26 and its paralog MRKa from rice cultivar Minghui 63 (Oryza sativa ssp. indica; Cao et al., 2007b) were used to screen the O. officinalis and O. minuta BAC libraries (Ammiraju et al., 2006). DNA segments corresponding to the LRR domain of MRKa and the kinase domain of Xa3/Xa26 were amplified using gene-specific primers (Supplemental Table 2). Positive BACs were digested with restriction enzyme HindIII, transferred to nylon membranes, and hybridized separately using Xa3/Xa26 and MRKa DNA probes. BAC that has the most hybridizing bands in each genome was sequenced.
BAC Sequencing and Sequence Assembly
BAC clones were sequenced using a shotgun strategy. To construct a subclone library for sequencing, DNA from each BAC clone was randomly sheared by sonication. DNA fragments in the 2–4-kb size frame were size-selected, blunt-ended, and then ligated into the pUC19 vector. Clones were sequenced from both directions using M13 universal forward and reverse primers and BigDye Terminator v3.0 Cycle Sequencing Kits (Applied Biosystems, Foster City, CA, USA). Sequence reads were assembled with Sequencher 4.5 (Gene Codes Corporation, Ann Arbor, MI, USA). Gaps were filled by a combination of primer walking and shotgun sequencing of subclones with extremes at both sides of the sequencing gaps.
Sequence Annotation and Computational Analysis
Assembled BAC sequences were annotated by using the gene prediction program Fgenesh (http://morissardjerome.free.fr/infobiogen/www.softberry.com/berry.html) (Salamov and Solovyev, 2000) and BLAST (Blastn, Blastx and Blastp) analyses against different databases (Altschul et al., 1997). Pair-wise sequence comparisons were carried out using YASS program (http://bioinfo.lifl.fr; Noe and Kucherov, 2005) and the Global Sequence Alignment Tool (www.ncbi.nlm.nih.gov/blast/Blast.cgi; Needleman and Wunsch, 1970). ClustalX (Thompson et al., 1997) was used for multiple sequence alignment.
Gene Cloning and Rice Transformation
BAC clones OO_Ba0120J21 from O. officinalis and OM_Ba0293H21 from O. minuta were digested with restriction enzymes EcoRV and SamI, respectively. An 11.9-kb DNA fragment harboring OoRKb1 and its native promoter from OO_Ba0120J21 and a 13.7-kb DNA fragment harboring OmRKb1 and its native promoter from OM_Ba0293H21 were individually ligated with the vector pCAMBIA1301 digested with restriction enzyme SmaI (Supplemental Figure 2). The constructs containing OoRKb1 and OmRKb1 were transferred into Agrobacterium tumefaciens strain EHA105 by electroporation. Agrobacterium-mediated transformation was performed using calli derived from mature embryos of susceptible rice cultivars Mudanjiang 8 (O. sativa L. ssp. japonica) according to a published procedure (Lin and Zhang, 2005). Positive transgenic plants were identified by PCR amplification of the marker gene β-glucuronidase (GUS) using gene-specific primers (Supplemental Table 2).
Pathogen Inoculation and Disease Scoring
Three to five uppermost fully expanded leaves of each plant were inoculated with different Xoo strains using the leaf-clipping method (Chen et al., 2002) at six-leaf and booting stages. Xoo strains included Chinese strains GD414, HeN11, JL691, LN44, YN1, and ZHE173, Japanese strain T7174, and Philippine strains PXO61 (race 1), PXO86 (race 2), PXO79 (race 3), PXO71 (race 4), PXO112 (race 5), PXO99 (race 6), PXO280 (race 8), PXO349 (race 9b), PXO347 (race 9c), PXO364 (race 9d), and PXO341 (race 10). Because cultivated rice and wild Oryza species had different leaf length, disease was scored by measuring percent lesion area (lesion length/leaf length) 2–3 weeks after inoculation. The bacterial growth rate in rice leaves was determined by counting colony-forming units (Sun et al., 2004).
Gene Structure Analysis
Total RNA extracted from the leaves of resistant T1 transgenic plants was used to analyze the structures of the transgenes. The 5' and 3' end cDNA sequences of target genes were determined by rapid amplification of cDNA ends (RACE) using the SMARTer RACE cDNA Amplification Kit (Clontech) according to the manufacturer's instructions (Qiu et al., 2007). Intermediate cDNA fragments of the transgenes were obtained by reverse transcription (RT)–PCR. The primers used for RACE and RT–PCR analyses are listed in Supplemental Table 1. The RACE and RT–PCR products were ligated into the pGEM–T Easy vector (Promega, Madison, WI, USA) and sequenced.
Statistical Analyses
The significant differences between the samples of transgenic and wild-type plants were analyzed by the pair-wise t-test installed in the Microsoft Office Excel program.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by grants from the National Program on the Development of Basic Research in China (2011CB100700) and the National Natural Science Foundation of China (30930063, 30921091).
Supplementary Material
Acknowledgments
We thank Dr Meizhong Luo of Huazhong Agricultural University (originally Arizona Genomics Institute, University of Arizona) for helping obtaining BAC clones, Dr Mingsheng Chen of Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for providing the O. officinalis and O. minuta accessions, and Drs Jetty S.S. Ammiraju, Chuanzhu Fan, and Fusheng Wei for critical reading of the manuscript. No conflict of interest declared.
References
- Ali GS, Reddy A. PAMP-triggered immunity: early events in the activation of FLAGELLIN SENSITIVE2. Plant Signal Behav. 2008;3:423–426. doi: 10.4161/psb.3.6.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amante-Bordeos A, Sitch LA, Nelson R, Dalmacio RD, Oliva NP, Aswidinnoor H, Leung H. Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa. Theor. Appl. Genet. 1992;84:345–354. doi: 10.1007/BF00229493. [DOI] [PubMed] [Google Scholar]
- Ammiraju JS, et al. The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 2006;16:140–147. doi: 10.1101/gr.3766306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammiraju JS, et al. Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell. 2008;20:3191–3209. doi: 10.1105/tpc.108.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammiraju JS, et al. Spatio-temporal patterns of genome evolution in allotetraploid species of the genus Oryza. Plant J. 2010;63:430–442. doi: 10.1111/j.1365-313X.2010.04251.x. [DOI] [PubMed] [Google Scholar]
- Brar DS, Khush GS. Transferring genes from wild species into rice. In Quantitative Genetics, Genomics and Plant Breeding. Kang, M.S., ed. (Wallingford, UK: CAB International) [Google Scholar]
- Cao Y, Ding X, Cai M, Zhao J, Lin Y, Li X, Xu C, Wang S. The expression pattern of a rice disease resistance gene Xa3/Xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics. 2007a;177:523–533. doi: 10.1534/genetics.107.075176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Duan L, Li H, Sun X, Zhao Y, Xu C, Li X, Wang S. Functional analysis of Xa3/Xa26 family members in rice resistance to Xanthomonas oryzae pv. Oryzae. Theor. Appl. Genet. 2007b;115:887–895. doi: 10.1007/s00122-007-0615-0. [DOI] [PubMed] [Google Scholar]
- Century KS, Lagman RA, Adkisson M, Morlan J, Tobias R, Schwartz K, Smith A, Love J, Ronald PC, Whalen MC. Developmental control of Xa21-mediated disease resistance in rice. Plant J. 1999;20:231–236. doi: 10.1046/j.1365-313x.1999.00589.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Wang S, Zhang Q. New gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, an elite restorer line. Phytopathology. 2002;92:750–754. doi: 10.1094/PHYTO.2002.92.7.750. [DOI] [PubMed] [Google Scholar]
- Cottyn B, Mew T. In: Bacterial blight of rice. In Encyclopedia of Plant and Crop Science. Goodman RM, editor. Abingdon, UK: Taylor and Francis; 2004. [Google Scholar]
- Ellis JG, Dodds PN, Lawrence GJ. Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu. Rev. Phytopathol. 2007;45:289–306. doi: 10.1146/annurev.phyto.45.062806.094331. [DOI] [PubMed] [Google Scholar]
- Gao J, Zhao J, Xu C, Li X, Wang S. Development of rice germplasms conferring high-level and broad-spectrum resistance to Xanthomonas oryzae pv. oryzae at both seedling and adult stages. Mol. Plant Breed. 2010;8:420–525. [Google Scholar]
- Ge S, Sang T, Lu BR, Hong DY. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc. Natl Acad. Sci. U S A. 1999;96:14400–14405. doi: 10.1073/pnas.96.25.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Gu K, Tian D, Yang F, Wu L, Sreekala C, Wang D, Wang GL, Yin Z. High-resolution genetic mapping of Xa27(t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor. Appl. Genet. 2004;108:800–807. doi: 10.1007/s00122-003-1491-x. [DOI] [PubMed] [Google Scholar]
- Harlan JR, de Wet JMJ. Toward a rational classification of cultivated plants. Taxon. 1971;20:509–517. [Google Scholar]
- Hu K, Qiu D, Shen X, Li X, Wang S. Isolation and manipulation of quantitative trait loci for disease resistance in rice using a candidate gene approach. Mol. Plant. 2008;1:786–793. doi: 10.1093/mp/ssn039. [DOI] [PubMed] [Google Scholar]
- Johnson R. Durable resistance: definition of, genetic control, and attainment in plant breeding. Phytopathology. 1981;71:567–568. [Google Scholar]
- Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997;35:25–34. [PubMed] [Google Scholar]
- Khush GS, Virk PS. 2005. IR Varieties and Their Impact. International Rice Research Institute, Los Baños, Philippines. [Google Scholar]
- Khush GS, Bacalangco E, Ogawa T. A new gene for resistance to bacterial blight from O. longistaminate. Rice Genet. Newslett. 1990;7:121–122. [Google Scholar]
- Kou Y, Wang S. Broad-spectrum and durability: understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 2010;13:181–185. doi: 10.1016/j.pbi.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Kou Y, Li X, Xiao J, Wang S. Identification of genes contributing to quantitative disease resistance in rice. Sci. China Life Sci. 2010;53:1263–1273. doi: 10.1007/s11427-010-4081-6. [DOI] [PubMed] [Google Scholar]
- Lee SW, Han SW, Sririyanum M, Park CJ, Seo YS, Ronald PC. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science. 2009;326:850–853. doi: 10.1126/science.1173438. [DOI] [PubMed] [Google Scholar]
- Lin Y, Zhang Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005;23:540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- Lu F, et al. Comparative sequence analysis of MONOCULM1-orthologous regions in 14 Oryza genomes. Proc. Natl Acad. Sci. U S A. 2009;106:2071–2076. doi: 10.1073/pnas.0812798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Noe L, Kucherov G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 2005;33(Web Server issue):W540–W543. doi: 10.1093/nar/gki478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan M, Cournoyer P, Dinesh-Kumar SP. The leucine-rich repeat domain in plant innate immunity: a wealth of possibilities. Cell Microbiol. 2009;11:191–198. doi: 10.1111/j.1462-5822.2008.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant–Microbe Interact. 2007;20:492–499. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Cheng H, Li X, Wang S. Exploring transcriptional signaling mediated by OsWRKY13, a potential regulator of multiple physiological processes in rice. BMC Plant Biol. 2009;9:74. doi: 10.1186/1471-2229-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S. Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol. Plant. 2008;1:538–551. doi: 10.1093/mp/ssn012. [DOI] [PubMed] [Google Scholar]
- Rivas S, Thomas CM. Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 2005;43:395–436. doi: 10.1146/annurev.phyto.43.040204.140224. [DOI] [PubMed] [Google Scholar]
- Salamov A, Solovyev V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, et al. Orthologous comparisons of the Hd1 region across genera reveal Hd1 gene lability within diploid Oryza species and disruptions to microsynteny in Sorghum. Mol. Biol. Evol. 2010;27:2487–2506. doi: 10.1093/molbev/msq133. [DOI] [PubMed] [Google Scholar]
- Song WY, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;279:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Sun X, Cao Y, Wang S. Point mutations with positive selection were a major force during the evolution of a receptor-kinase resistance gene family of rice. Plant Physiol. 2006;140:998–1008. doi: 10.1104/pp.105.073080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313x.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- Tan GX, Ren X, Weng QM, Shi ZY, Zhu LL, He GC. Mapping of a new resistance gene to bacterial blight in rice line introgressed from Oryza officinalis. Acta Genetica Sinica. 2004;31:724–729. [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Ding Z, Liu W, Pan J, Li C, Ge S, Zhang D. Polyploid evolution in Oryza officinalis complex of the genus Oryza. BMC Evol. Biol. 2009;9:250. doi: 10.1186/1471-2148-9-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Cao Y, Xu C, Li X, Wang S. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor. Appl. Genet. 2006;113:1347–1355. doi: 10.1007/s00122-006-0388-x. [DOI] [PubMed] [Google Scholar]
- Xie H. Selection and utilization of Minghui 63. Fujian Journal of Agricultural Sciences. 1998;4:1–6. [Google Scholar]
- Xu S, Cao Y, Li X, Wang S. Expressional and biochemical characterization of rice disease resistance gene Xa3/Xa26 family. J. Integr. Plant Biol. 2007;49:852–862. [Google Scholar]
- Xu Z, Sun Q, Liu F, Chen Z, Hu B, Guo Y, Liu Y, Liu H. Race monitoring of rice bacterial blight (Xanthomonas oryzae pv. oryzae) in China. Chin. J. Rice Sci. 2004;18:469–472. [Google Scholar]
- Yang D, Li Q, Deng Y, Lou Y, Wang M, Zhou G, Zhang Y, He Z. Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol. Plant. 2008;1:528–537. doi: 10.1093/mp/ssn021. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sun X, Wang S, Zhang Q. Genetic and physical mapping of a new gene for bacterial blight resistance in rice. Theor. Appl. Genet. 2003;106:1467–1472. doi: 10.1007/s00122-003-1205-4. [DOI] [PubMed] [Google Scholar]
- Zhang Q, et al. Identifying and mapping a new gene Xa-23(t) for resistance to bacterial blight (Xanthomonas oryzae pv. oryzae) from O. rufipogon. Acta Agronomica Sinica. 2000;26:536–542. [Google Scholar]
- Zhao J, Fu J, Li X, Xu C, Wang S. Dissection of the factors affecting development-controlled and race-specific disease resistance conferred by leucine-rich repeat receptor kinase-type R genes in rice. Theor. Appl. Genet. 2009;119:231–239. doi: 10.1007/s00122-009-1032-3. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Cao Y, Huang Y, Xie W, Xu Cai, Li X, Wang S. Multiple gene loci affecting genetic background-controlled disease resistance conferred by R gene Xa3/Xa26 in rice. Theor. Appl. Genet. 2009;120:127–138. doi: 10.1007/s00122-009-1164-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.