Abstract
One goal in the post-genome-wide association study era is characterizing gene-environment interactions, including scanning for interactions with all available polymorphisms, not just those showing significant main effects. In recent years, several approaches to such “gene-environment-wide interaction studies” have been proposed. Two contributions in this issue of the American Journal of Epidemiology provide systematic comparisons of the performance of these various approaches, one based on simulation and one based on application to 2 real genome-wide association study scans for type 2 diabetes. The authors discuss some of the broader issues raised by these contributions, including the plausibility of the gene-environment independence assumption that some of these approaches rely upon, the need for replication, and various generalizations of these approaches.
Keywords: epidemiologic research design; genetic epidemiology; genome-wide association study; genotype-environment interaction; polymorphisms, single nucleotide
For decades, the pages of the Journal have been filled with philosophical debates over the meaning of words such as “interaction” and “synergism,” as well as distinctions among statistical, biologic, and public health contexts. Recently, there has been a resurgence of interest in this topic by a new cohort of genetic epidemiologists working on gene-environment (G-E) and gene-gene (G-G) interactions. Various authors have offered classifications of patterns of G-E interaction (1–3), although the concept of “epistasis” (G-G interaction) can be traced back to 1909 (4) and of G-E interaction to 1938 (5).
Most epidemiologic analyses of interactions have tested for a departure from some simple main effects model, most commonly a multiplicative one. Without belaboring the issue, we point out that this may not be interpretable as a biologic interaction or a synergistic public health impact (6). With the advent of genome-wide association studies, discussions have shifted from these philosophical topics to more practical concerns about study designs and analysis methods for discovering G-E interactions on a massive scale, what Khoury et al. called a “GEWIS” (gene-environment-wide interaction study(ies)) (6, 7). The advent of “EWAS” (environment-wide association studies) (8) and the “exposome” concept (9, 10) is likely to ratchet the importance of this topic up yet another notch. Two papers in this issue (11, 12) compare the performance of several novel approaches with GEWIS analysis.
Mukherjee et al. (11) used simulation to compare case-control, case-only, and several approaches that combine them in various ways. They found, as expected, that the case-only design generally yields the greatest power but is subject to substantial false positives in the presence of G-E associations. Empirical Bayes (13), Bayesian model averaging (14), and 2-step (15) methods all yield better power than the case-control method in most situations, with some performing better than the others for particular parameter combinations. The one notable exception is when a population-level G-E association goes in the opposite direction from the G-E interaction. Here, the case-only method also has low power because the G-E association among cases will tend to be small. Because any GEWIS is testing many different single nucleotide polymorphisms (SNPs) (and often several different exposure variables), there is no uniformly most powerful procedure across the full range of possible model parameters.
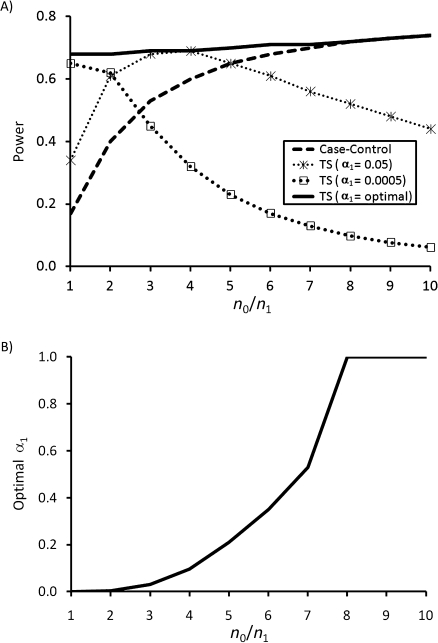
One observation reported by Mukherjee et al. (11) is that, for a fixed number of cases and a fixed screening threshold α1, the power of the 2-step method appears to decline with increasing number of controls—the only example we are aware of where having more data appears to be worse! However, the reason for this apparent “lack of coherence” is that the power of the 2-step design for a fixed α1 (Figure 1A at α1 = 0.05 or 0.0005) and the optimal first step critical value α1 (Figure 1B) depend strongly on the control:case ratio. If one chooses the optimal α1 for a given control:case ratio, the power does increase monotonically with increasing sample size (Figure 1A, optimal), as one might expect. In addition to the control:case ratio, the optimal choice for α1 depends strongly on the population disease prevalence and number of SNPs analyzed. Because all of these quantities are known or easily estimated prior to analysis, choosing an optimal or near-optimal α1 is possible in practice (16).
Figure 1.
Empirical power to detect a single causal single nucleotide polymorphism (SNP) for the case-control and 2-step (TS) analyses (for fixed α1 = 0.05, 0.0005, and optimal α1) as a function of the ratio of number of controls to number of cases (n0/n1) (A) and optimal α1 as a function of n0/n1 (B). Assumed parameter values are those used by Mukherjee et al. (11), specifically: M = 100,000 SNPs; n1 = 2,000 cases; Rg = Re = 1.0, Rge = 1.8; and Pr(E) = 0.5, minor allele frequency of dominantly coded casual SNP = 0.2.
In their discussion, Mukherjee et al. (11) claim that the 2-step procedure violates the likelihood principle (and that the other methods compared do not). Actually, any test of significance (whether multistep or not) violates the likelihood principle, as it does not rely exclusively on the likelihood for inferences but considers unobserved outcomes as well (more extreme ones than actually observed) (17). Multistep methods have a long tradition in statistics. For example, sequential and group sequential methods have been used in industrial applications to reduce costs and in clinical trials to minimize potential adverse side effects. Valid multistep methods have also been proposed in situations where the data are not collected sequentially, often leading to substantial improvements in power (18, 19).
We are aware of 2 applications to real data of the 2-step G-E approach. Ege et al. (20) applied the approach to data on asthma from genome-wide association studies from the GABRIELA consortium. They identified 15 genes showing evidence of interaction with farm-related variables, although none attained genome-wide significance. Figueiredo et al. (21) compared case-only, case-control, and the 2-step procedure on data from the Colorectal Cancer Family Registries for interactions with 14 established environmental risk factors. None attained genome-wide significance by any of the 3 methods. This work points out the difficulty in identifying G-E interactions for a complex trait and suggests the need for quite large sample sizes in addition to efficient analytical approaches. Programs to compute required sample size for interaction tests are available for several study designs (22, 23) including 2-step testing in a genome-wide association study (16).
Cornelis et al. (12) took a different approach, comparing similar methods on real data from 2 large GEWIS of type 2 diabetes. Here, the interacting variable was an “adipogenic environment,” as measured by a dichotomization of body mass index. What makes this an interesting application is that this “exposure” variable is also partially under genetic control, probably by some of the same genes that are involved in diabetes, so one might expect a substantial number of false positives due to G-E associations. Surprisingly, on the basis of an examination of the quantile-quantile (QQ) plot of P values, they found no evidence of an inflated type I error rate for any of these tests, even the simple case-only test. However, the loci that yielded the most significant interactions were generally those that were also most strongly associated with obesity, which should cause some concern. As the true state of nature is, of course, unknown, it isn’t possible to assess whether any of these are real interactions or simply reflect G-E associations.
One interesting observation in this paper was the lack of robustness of even the standard case-control test when the model for a continuous exposure variable was misspecified. This phenomenon was recently explored by Tchetgen Tchetgen and Kraft (24), who also proposed using a robust sandwich variance estimator that does not require dichotomization of the exposure variable (with its inherent loss of power) (25). Alternatively, more flexible modeling of the exposure using, e.g., generalized additive models (26) could also help to make the inferences on G-E interactions with quantitative exposures more robust.
So how plausible is the assumption of G-E independence? Most tag-SNPs used in genome-wide association studies are unlikely to be related to either the disease or exposure, so a screening tool using a case-only test seems reasonable. Even if a small proportion of the interactions discovered in this way are false positives, they will be weeded out by a second step that does not rely on this assumption. Nevertheless, there are a few circumstances where caution is warranted. One is for diseases with a strong behavioral component, such as lung cancer where many genes might be associated with nicotine addiction. Hormone-related cancers are another example where various genes could influence a woman’s age at menarche, menopause, or reproductive history as the “exposures” of interest. A third example is a nonrandomized study of treatment outcomes (e.g., second-cancer studies), where indications for treatment could relate to disease severity or other characteristics that are genetically influenced. Uncontrolled population stratification can easily induce spurious G-E associations due to confounding by genetic ancestry and cultural factors influencing exposures, emphasizing the importance of proper adjustment for ancestry covariates (27). Finally, differential survival over time can induce associations between genes and exposures so that both are risk factors even if the 2 are independent initially (28). Uncontrolled confounding of the G-E association will lead to inflated type I errors for the case-only or empirical Bayesian approach, but not for the case-control or 2-step approaches.
Although it is tempting to pretest for G-E independence among controls and on this basis decide whether to use the more powerful case-only test (which requires it for validity) or the more robust case-control test (which does not), Albert et al. (29) showed that this can be a seriously biased strategy. This bias arises because the pretesting is ignored when assessing significance in the follow-up test. Unlike the screening step in the Murcray et al. approach, the test of G-E independence using only controls is not independent of the standard case-control test. However, at least in principle, pretesting for G-E independence in controls would result in an acceptable 2-step test if one properly accounted for pretesting by either conditioning on the outcome of the pretest or by considering the true unconditional distribution of the resulting test statistic. This distribution is a weighted mixture of the case-only and case-control statistics with weights given by the probabilities of acceptance and rejection of the hypothesis of G-E independence in the pretest. The latter is similar to the empirical Bayes and the Bayes-model-averaging procedures, both of which are weighted averages of the 2 statistics. Nevertheless, one should be cautious about blindly disregarding concerns about the validity of the G-E independence assumption. Even if the various 2-step procedures (properly applied) ensure a valid test, power can be adversely affected if the first step passes too high a proportion of false positives to the second step.
As pointed out by Mukherjee et al. (11), the 2-step approach is the only alternative to the standard 1-step case-control test that guarantees asymptotic control of the type I error under departures from G-E independence. Both the empirical Bayes and Bayes model-averaging statistics, being weighted averages of the case-only and case-control statistics, are necessarily liberal under departures from G-E independence. Thus, because even a modest inflation of the type I error can translate into a sizable power increase, some of the power gain of the empirical Bayes and the Bayes model-averaging approaches over the standard 1-step case-control might be due to type I error inflation. One can argue that, among tests with similar power, it is preferable to use one that controls the type I error. After all, why trade an unknown increase in type I error (even if small) for extra power when one can simply increase the level of significance to achieve the same goal but with a known type I error?
However, perhaps a deeper question is whether we need be concerned about type I error at all in a climate that demands independent replication before publication in a top-tier journal. Won’t virtually all false positives be weeded out by the requirement of genome-wide significance in the discovery sample followed by significance at a conventional replication level such as α = 0.05? Perhaps yes, but this may be too conservative a requirement, tantamount to requiring a genome-wide significance level of α = 0.052 = 0.0025. The role of replication is more to rule out bias and to ensure generalizability by testing associations or interactions with different methods, by different investigators, in different populations, than to avoid chance statistical flukes (which can always be accomplished within a single study simply by adopting a more stringent significance level) (30, 31). If independent replication is planned anyway, then a good case could be made for always using the most powerful case-only test for the initial scan, provided the replication is performed with a case-control test in an independent data set. This, however, is not always an option in practice. In particular, for unique exposure situations, uniquely well-characterized cohorts, or consortia that comprise essentially the entirety of the world’s data to generate sufficient cases for studying rare diseases, replication may never be feasible (32). These situations put a premium on using a powerful testing procedure that maintains control of the type I error rate.
These 2 papers are certainly not the last word on this subject. Various extensions of 2-step procedures are possible. Kraft et al. (33) discussed the use of a 2 df joint test for gene main effects and G-E interactions, where the goal is not to detect the interaction per se but rather to identify genes that may be etiologically relevant either directly or through an interaction (this test is also evaluated in the paper by Cornelis et al. (12)). Our group recently described 2 different kinds of 2-step procedures, one for case-parent trios that exploits a between-family comparison of G-E association among the parents (19) and a hybrid approach (16) for case-control data that screens SNPs on the basis of both marginal association (34) and G-E association (15). Similar methods are applicable for G-G interactions, where the multiple comparisons burden is orders of magnitude more severe (half a trillion tests for an exhaustive scan of 1 million SNPs) and the power advantages of a 2-step method may be even larger than for G-E interaction scans (35). Tests that exploit Hardy-Weinberg equilibrium in the population (36), discussed in the contribution by Cornelis et al. (12), may also enhance power. As we move into the era of targeted, whole-exome, or even whole-genome sequencing, 2-step procedures for interaction testing may become even more necessary. Power for testing interactions with specific rare variants is likely to be miniscule, but interaction testing for aggregate indices of multiple rare variants in a gene or for discovering more complex pathways may be feasible. Exposure measurement error is a longstanding problem and can have unpredictable effects on G-E interactions, although in general it is likely to make their detection more difficult (37–39). The larger sample sizes available in a consortium setting may be necessary to achieve adequate power (40). Methods to analyze G-E interaction in the consortium setting have begun to appear (41), but this is an area of statistical research that requires more attention. All the GEWIS methods discussed so far are “agnostic” (with respect to the genes), but methods that incorporate external genetic and environmental information offer further potential to achieve substantial power gains (42, 43).
We commend Mukherjee et al. (11) for their rigorous comparison of several methods and Cornelis et al. (12) for their thoughtful application of methods to real data. Together, these papers raise a number of important issues, including the largely untapped potential of GEWIS to discover novel genetic variants in existing genome-wide association data sets. For nearly all complex human diseases, it is clear that neither genes nor environmental factors are exclusively to blame for increased risk. As we move forward, well-designed studies with careful measurement and efficient analysis of both genetic and environmental factors will likely hold the key to further understanding complex disease etiologies.
Acknowledgments
Author affiliation: Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California (Duncan C. Thomas, Juan Pablo Lewinger, Cassandra E. Murcray, W. James Gauderman).
This work was supported in part by the National Institute of Environmental Health Sciences (grants P30ES007048, R01ES019876, and P01ES011627); the National Heart, Lung, and Blood Institute (grants RO1HL087680 and RC2HL101651); and the National Institute of Child Health and Human Development (grant U01HD061968).
Conflict of interest: none declared.
Glossary
Abbreviations
- G-E
gene-environment
- GEWIS
gene-environment-wide interaction study(ies)
- G-G
gene-gene
- SNP
single nucleotide polymorphism
References
- 1.Lewontin RC. Annotation: the analysis of variance and the analysis of causes. Am J Hum Genet. 1974;26(3):400–411. [PMC free article] [PubMed] [Google Scholar]
- 2.Ottman R. An epidemiologic approach to gene-environment interaction. Genet Epidemiol. 1990;7(3):177–185. doi: 10.1002/gepi.1370070302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Q, Khoury MJ. Evolving methods in genetic epidemiology. III. Gene-environment interaction in epidemiologic research. Epidemiol Rev. 1997;19(1):33–43. doi: 10.1093/oxfordjournals.epirev.a017944. [DOI] [PubMed] [Google Scholar]
- 4.Bateson Discussion on the influence of heredity on disease, with special reference to tuberculosis, cancer, and diseases of the nervous system: introductory address. Proc R Soc Med. 1909;2(gen rep):22–30. doi: 10.1177/003591570900200404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haldane JBS. Heredity and Politics. New York, NY: W W Norton; 1938. [Google Scholar]
- 6.Thomas D. Gene–environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11(4):259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoury MJ, Wacholder S. Invited commentary: from genome-wide association studies to gene-environment-wide interaction studies—challenges and opportunities. Am J Epidemiol. 2009;169(2):227–230. doi: 10.1093/aje/kwn351. discussion 234–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel CJ, Bhattacharya J, Butte AJ. An environment-wide association study (EWAS) on type 2 diabetes mellitus. PLoS One. 2010;5(5):e10746. doi: 10.1371/journal.pone.0010746. (doi:10.1371/journal.pone.0010746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rappaport SM. Implications of the exposome for exposure science. J Expo Sci Environ Epidemiol. 2011;21(1):5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- 10.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 11.Mukherjee B, Ahn J, Gruber SB, et al. Testing gene-environment interaction in large scale case-control association studies: possible choices and comparisons. Am J Epidemiol. 2012;175(3):208–209. doi: 10.1093/aje/kwr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis MC, Tchetgen Tchetgen EJ, Liang L, et al. Gene-environment interactions in genome-wide association studies: a comparative study of tests applied to empirical studies of type 2 diabetes. Am J Epidemiol. 2012;175(3):191–202. doi: 10.1093/aje/kwr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee B, Chatterjee N. Exploiting gene-environment independence for analysis of case-control studies: an empirical Bayes-type shrinkage estimator to trade-off between bias and efficiency. Biometrics. 2008;64(3):685–694. doi: 10.1111/j.1541-0420.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- 14.Li D, Conti DV. Detecting gene-environment interactions using a combined case-only and case-control approach. Am J Epidemiol. 2009;169(4):497–504. doi: 10.1093/aje/kwn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murcray CE, Lewinger JP, Gauderman WJ. Gene-environment interaction in genome-wide association studies. Am J Epidemiol. 2009;169(2):219–226. doi: 10.1093/aje/kwn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murcray CE, Lewinger JP, Conti DV, et al. Sample size requirements to detect gene-environment interactions in genome-wide association studies. Genet Epidemiol. 2011;35(3):201–210. doi: 10.1002/gepi.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox DR, Hinkley DV. Theoretical Statistics. London, United Kingdom: Chapman and Hall; 1974. [Google Scholar]
- 18.O’Gorman TW. Using adaptive methods to select variables in case-control studies. Biometric J. 2004;46(5):595–605. [Google Scholar]
- 19.Gauderman WJ, Thomas DC, Murcray CE, et al. Efficient genome-wide association testing of gene-environment interaction in case-parent trios. Am J Epidemiol. 2010;172(1):116–122. doi: 10.1093/aje/kwq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ege MJ, Strachan DP, Cookson WO, et al. Gene-environment interaction for childhood asthma and exposure to farming in Central Europe. GABRIELA Study Group. J Allergy Clin Immunol. 2011;127(1):138–144. doi: 10.1016/j.jaci.2010.09.041. 144.e1–144.e4. [DOI] [PubMed] [Google Scholar]
- 21.Figueiredo JC, Lewinger JP, Song C, et al. Genotype-environment interactions in microsatellite stable/microsatellite instability-low colorectal cancer: results from a genome-wide association study. Cancer Epidemiol Biomarkers Prev. 2011;20(5):758–766. doi: 10.1158/1055-9965.EPI-10-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21(1):35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 23.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155(5):478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 24.Tchetgen Tchetgen EJ, Kraft P. On the robustness of tests of genetic associations incorporating gene-environment interaction when the environmental exposure is misspecified. Epidemiology. 2011;22(2):257–261. doi: 10.1097/EDE.0b013e31820877c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tchetgen Tchetgen E. Robust discovery of genetic associations incorporating gene-environment interaction and independence. Epidemiology. 2011;22(2):262–272. doi: 10.1097/EDE.0b013e318207ffc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood SN. Generalized Additive Models: An Introduction With R. London, United Kingdom: Chapman Hall; 2006. [Google Scholar]
- 27.Bhattacharjee S, Wang Z, Ciampa J, et al. Using principal components of genetic variation for robust and powerful detection of gene-gene interactions in case-control and case-only studies. Am J Hum Genet. 2010;86(3):331–342. doi: 10.1016/j.ajhg.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gauderman J, Millstein J. The case-only design to detect G × E interaction for a survival trait [abstract] Genet Epidemiol. 2002;23:282. [Google Scholar]
- 29.Albert PS, Ratnasinghe D, Tangrea J, et al. Limitations of the case-only design for identifying gene-environment interactions. Am J Epidemiol. 2001;154(8):687–693. doi: 10.1093/aje/154.8.687. [DOI] [PubMed] [Google Scholar]
- 30.Thomas DC, Siemiatycki J, Dewar R, et al. The problem of multiple inference in studies designed to generate hypotheses. Am J Epidemiol. 1985;122(6):1080–1095. doi: 10.1093/oxfordjournals.aje.a114189. [DOI] [PubMed] [Google Scholar]
- 31.Skol AD, Scott LJ, Abecasis GR, et al. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 32.Bookman EB, McAllister K, Gillanders E, et al. Gene-environment interplay in common complex diseases: forging an integrative model—recommendations from an NIH workshop. For the NIH G × E Interplay Workshop participants. Genet Epidemiol. 2011;35(4):217–225. doi: 10.1002/gepi.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraft P, Yen YC, Stram DO, et al. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63(2):111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 34.Kooperberg C, Leblanc M. Increasing the power of identifying gene x gene interactions in genome-wide association studies. Genet Epidemiol. 2008;32(3):255–263. doi: 10.1002/gepi.20300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewinger JP, Murcray CE, Gauderman WJ. Honolulu, HI: American Society of Human Genetics; 2009. Efficient two-step testing of gene-gene interactions in genomewide association studies [abstract] [Google Scholar]
- 36.Chatterjee N, Carroll RJ. Semiparametric maximum likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika. 2005;92(2):399–418. [Google Scholar]
- 37.Wong MY, Day NE, Luan JA, et al. The detection of gene-environment interaction for continuous traits: should we deal with measurement error by bigger studies or better measurement? Int J Epidemiol. 2003;32(1):51–57. doi: 10.1093/ije/dyg002. [DOI] [PubMed] [Google Scholar]
- 38.Lobach I, Fan R, Carroll RJ. Genotype-based association mapping of complex diseases: gene-environment interactions with multiple genetic markers and measurement error in environmental exposures. Genet Epidemiol. 2010;34(8):792–802. doi: 10.1002/gepi.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindström S, Yen YC, Spiegelman D, et al. The impact of gene-environment dependence and misclassification in genetic association studies incorporating gene-environment interactions. Hum Hered. 2009;68(3):171–181. doi: 10.1159/000224637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burton PR, Hansell AL, Fortier I, et al. Size matters: just how big is BIG?: quantifying realistic sample size requirements for human genome epidemiology. Int J Epidemiol. 2009;38(1):263–273. doi: 10.1093/ije/dyn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manning AK, LaValley M, Liu CT, et al. Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP × environment regression coefficients. Genet Epidemiol. 2011;35(1):11–18. doi: 10.1002/gepi.20546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewinger JP, Conti DV, Baurley JW, et al. Hierarchical Bayes prioritization of marker associations from a genome-wide association scan for further investigation. Genet Epidemiol. 2007;31(8):871–882. doi: 10.1002/gepi.20248. [DOI] [PubMed] [Google Scholar]
- 43.Roeder K, Devlin B, Wasserman L. Improving power in genome-wide association studies: weights tip the scale. Genet Epidemiol. 2007;31(7):741–747. doi: 10.1002/gepi.20237. [DOI] [PubMed] [Google Scholar]