Abstract
The growth, metabolism, and productivity of five Chinese hamster ovary (CHO) clones were explored in response to stimulation with insulin (5 mg/L) and LONG®R3IGF-I (20 μg/L or 100 μg/L). All five clones were derived from the same parental CHO cell line (DG44) and produced the same recombinant monoclonal antibody, with varying specific productivities. There was no uniform response among the clones to stimulation with the different trophic factors. One of the high productivity clones (clone D) exhibited significantly better growth in response to LONG®R3IGF-I; whereas the other clones showed equivalent or slightly better growth in the presence of insulin. Three out of the five clones had higher specific productivities in the presence of insulin (although not statistically significant); one was invariant, and the final clone exhibited slightly higher specific productivity in the presence of LONG®R3IGF-I. Total product titers exhibited moderate variation between culture conditions, again with neither trophic factor being clearly superior. Overall product titers were affected by variations in both integrated viable cell density and specific productivity. Nutrient uptake and metabolite generation patterns varied strongly between clones and much less with culture conditions. These results point to the need for careful clonal analysis when selecting clones, particularly for platform processes where media and culture conditions are predetermined.
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-011-9388-z) contains supplementary material, which is available to authorized users.
Keywords: CHO cells, Monoclonal antibody, Insulin, LONG®R3IGF-I, Metabolism
Introduction
Currently over 165 recombinant therapeutic proteins are approved for human use, with another 500 in preclinical and clinical development (Durocher and Butler 2009), accounting for approximately 25% of all new drugs approved in the United States and Europe (Walsh 2006, 2007). The 2009 protein therapeutics market was approximately $92 billion (La Merie Business Intelligence 2010), approximately one-third of the total pharmaceutical sales, with expectations that by 2014 therapeutic proteins will comprise half of the top 100 pharmaceuticals (Strohl and Knight 2009). Equally importantly, therapeutic proteins have provided new treatments for many previously untreatable conditions including immunological and metabolic disorders, have eliminated the risks associated with purification of proteins from human and animal tissues for conditions such as hemophilia and diabetes, and have provided entirely new, targeted approaches to cancer treatment. While these statistics are encouraging, it is important to note that the success rate of pharmaceuticals in clinical trials, even biopharmaceuticals, is still relatively low. For example, between 1990 and 1997, 134 small molecule candidates for cancer therapeutics entered clinical trials; 77% of those trials were completed during that period, resulting in 12 newly approved compounds, a 12% success rate. During that same period, 16 humanized monoclonal antibodies entered clinical trials as cancer therapeutics; 62.5% of those trials were completed, resulting in 3 new biopharmaceuticals, a 30% success rate (Reichert and Wenger 2008). This relatively low success rate increases the pressure on companies to maintain a large volume of products in their pipeline, increasing demands on process development groups to reduce time-to-clinic and time-to-market.
More than two-thirds of the marketed therapeutic proteins are glycoproteins (Li and d’Anjou 2009), produced predominantly in mammalian cell culture systems, with Chinese hamster ovary (CHO) cell systems being the most widely used production host (Durocher and Butler 2009). CHO cells have emerged as the preferred host due to their extensive characterization and human-like glycosylation, particularly in comparison with murine cell lines. Two primary selection systems are used to generate stable cell lines with high levels of expression, the dihydrofolate reductase (DHFR) system (Kaufman et al. 1983; Kaufman 1990; Urlaub and Chasin 1980), and the glutamine synthetase (GS) system (Bebbington et al. 1992). While there are advantages and disadvantages to both systems, extensive clonal selection and screening are still required in both systems. This selection and screening is time consuming and labor intensive and is usually focused on identifying clones that generate high titers. To improve throughput and reduce time-to-clinic, a number of biotechnology companies have adopted platform processes in which a fairly limited number of process parameters are varied in process development, regardless of the therapeutic moiety of interest. There are limited numbers of publications addressing platform processes for downstream processes or product purification (Eppink et al. 2009; Shukla et al. 2007) and surprisingly, none in the upstream process-development literature, despite the topic being extensively discussed in oral presentations (Leonard and Onadipe 2010; Seeworster 2010) and trade journals (DePalma 2008). A critical issue in clone selection is the compatibility of the selected clone with the platform process. This can become an even greater issue when a change is made in the platform process, e.g. transitioning to chemically defined media.
In this study, we examined the growth, productivity, and metabolic activity of five different CHO cell clones, all of which were derived from the same parental cell line and produce the same recombinant monoclonal antibody. Clones were grown in the presence of either insulin or LONG®R3IGF-I, a potent substitute for insulin that has been reported to increase growth and productivity in some cell lines (Morris and Schmid 2000). In addition to differences in growth and specific productivity between the clones, we found substantial differences in metabolic behavior and responsiveness to insulin or LONG®R3IGF-I, despite all the cell clones being grown in the same, serum-free medium. Our results highlight the need for a thorough analysis of the behavior of different cell clones before a clone is selected and a master cell bank generated, as well as suggesting the value of maintaining several “backup” clones that might be revived if a working clone does not respond well to process changes.
Materials and methods
Cell lines and cell culture
Chinese hamster ovary cell clones that produce a recombinant monoclonal humanized IgG with different specific productivities (qP) were provided by Biogen Idec (Cambridge, MA). These cell lines were developed by cotransfecting two plasmids, one containing IgG heavy chain (HC) and dihydrofolate reductase (DHFR) genes and the other containing IgG light chain (LC) and neomycin phosphotransferase (Neo) genes. Transfected cell lines were initially selected in medium containing 400 μg/mL neomycin (G418). After selection, the neomycin was removed, and all subsequent cultures were performed in the absence of neomycin. Subsequently, gene amplification was performed by stepwise selection with increasing methotrexate (MTX) concentrations. Based upon previously determined specific productivities, higher producers A1, C1, C2, and D and lower producer G were selected for the studies. Cell clones A1, C1, and C2 have been previously described (Jiang et al. 2006; Jiang and Sharfstein 2009). Cells were cultured in a serum-free modification of DME-F12 and alpha MEM medium supplemented with MTX (Sigma) as needed. The complete medium formulation is available in supplementary information (On-line resource 1). Frozen cell stocks were thawed into fresh medium and seeded at 0.2 × 106 cells/mL. Cell suspensions (10 mL) were cultured on an orbital shaker in upright T-25 flasks (Corning, Corning, NY) at 36°C, 5% CO2, and 125 rpm. Cells were routinely sub-cultured every 3 days. After two passages, all cell viabilities exceeded 95%, indicating cells were fully recovered from the frozen state. Cell culture samples were taken every 24 h for biochemical assays and determination of IgG levels.
Cell clones were seeded at ~0.2 × 106 cells/mL in duplicate with either 5 mg/L insulin (SAFC Biosciences (91077C)) or LONG®R3IGF-I (20 μg/L or 100 μg/L, Novozymes Biopharma DK A/S, available from SAFC Biosciences, product number 85580C or 91590C). Cell-culture samples were taken every 24 h for cell counts, biochemical assays, and determination of IgG levels. Samples for biochemical and antibody analysis were centrifuged and the supernatant removed. Sodium azide was added to a final concentration of 0.01% and supernatant samples were stored at −20 °C until analyzed. Cultures were monitored for a total of 7 days.
Determination of cell counts
Cell density and viability were measured using trypan-blue exclusion. Samples were taken and cell counts determined every 24 h.
Determination of glucose, glutamine and lactate levels
Cell-culture supernatants were analyzed for glucose and lactate using a YSI 2700 biochemical analyzer (Yellow Springs, OH, USA). The instrument was standardized using 2.5 g/L glucose and the lactate standard from YSI (YSI 2776). 20 μL injections were used. Glutamine and glutamate concentration were analyzed using a YSI 7100 analyzer with 5 mM glutamine and glutamate standards.
Amino acid analysis
Amino acid consumption in spent media was measured using the optimized Waters AccQ·Tag amino acid analysis method and kit based on pre-column derivatisation with AQC [6-aminoquinolyl-N-hydroxysuccinimidyl carbamate. Prior to analysis, spent medium samples were diluted 1 in 5 using DPBS (Dulbecco’s phosphate buffered saline) and heated for 10 min at 55 °C. Samples were analyzed by rp-HPLC and the elution of 17 amino acids monitored by UV detector at 254 nm. The amount of amino acids consumed or synthesized by different clones was calculated against an amino acid standard H solution (PIERCE, Prod# NCI0180).
Determination of IgG levels using protein A chromatography
The productivity of cells was determined by measuring the concentration of the IgG secreted into the sampled medium by affinity chromatography. Briefly, a Millipore ProSep® Protein A column, 4.6 × 50 mm (or equivalent) was used to separate IgG from each sample. 100% binding buffer (150 mM NaCl, 4.5 mM NaH2PO4, 15.5 mM Na2HPO4, pH 7.4) was used for the first 2 min followed by a gradient of 0–100% elution buffer (18.9 mM Na3C6H5O7·2H2O, 81.1 mM C6H8O7·H2O, pH 3.2) over 3 min, with a total analysis time of 10 min. The flow gradient program entailed a step of 0.5–1.0 mL/min from 0 to 2 min to capture IgG and wash off unbound material, a step of 1.0–1.5 mL/min from 2 to 8 min to elute IgG and a step of 2 mL/min from 8-10 min to equilibrate with binding buffer. The elution of IgG was monitored by a UV detector at 280 nm and fluorescence detection with excitation at 275 nm and emission at 303 nm. The amount of IgG recovered from each medium sample was calculated against a human IgG reference (BETHYL, Cat# E88-104).
Statistical analysis
Statistical analysis was performed using GraphPad Prism, version 5.03 for Windows, GraphPad Software, San Diego, CA, USA, www.graphpad.com.
Results
Cell growth
Five cell clones were selected from a population of 13 cell clones that had previously been provided by Biogen Idec. Three of these cell clones (A1, C1, C2) have been characterized with respect to gene copy number, localization, and mRNA levels (Jiang et al. 2006; Jiang and Sharfstein 2009); the other two cell clones D (a high producer) and G (a low producer) were selected based on productivity and differential responses to insulin and LONG®R3IGF-I in preliminary studies. Insulin (5 mg/L) or LONG®R3IGF-I at either a low (20 μg/L) or high (100 μg/L) concentration were added at the beginning of batch cultures, and samples were taken daily for cell density, immunoglobulin, and metabolic assays.
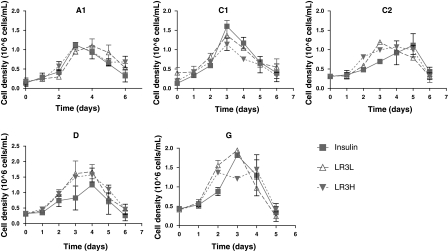
Cell clones exhibited significantly different growth profiles, with D and G clones reaching maximum viable cell densities of nearly 2 × 106 cells/mL, while A1 and C2 clones reached maximum viable cells densities of only slightly more than 1 × 106 cells/mL (Fig. 1). Only the D cell clone showed a marked preference for one of the trophic factors over the other, preferring the LONG®R3IGF-I (this difference was statistically significant over days 0–5, p < 0.05 by two-way ANOVA). In contrast, the other clones were either indifferent or showed a slight (not statistically significant) preference for insulin.
Fig. 1.
Viable cell densities for all cell clones. Error bars represent the standard deviation of duplicate biological replicates. Viability was maintained at greater than 90% through day 3 in all cultures. Insulin (squares)-human recombinant insulin 5 mg/L; LR3L (open triangles)- LONG®R3IGF-I 20 μg/L; LR3H (filled inverted triangles)- LONG®R3IGF-I 100 μg/L
Antibody productivity
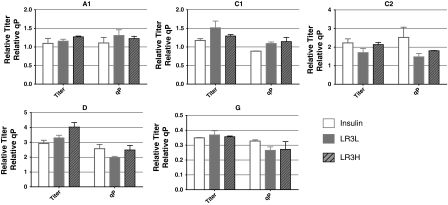
The total product titer and specific productivity, qP (over days 0–4 for all clones except G, over days 0–3 for clone G) are shown in Fig. 2. As expected, the higher productivity clones showed much higher titers than the lower productivity clone. In general, the qP was slightly higher in the cultures stimulated with insulin than those stimulated with LONG®R3IGF-I, with the exception of the C1 cell clone, which showed a slight preference for LONG®R3IGF-I; however, this preference was not statistically significant (p = 0.06 by one-way ANOVA). The total product titers also showed only small differences between the different trophic factors, with only the D cell clone exhibiting a statistically significant difference between the culture conditions (p < 0.05 by one-way ANOVA). In this case, the highest titer was observed in the culture supplemented with 100 μg/L of LONG®R3IGF-I, but the difference in titers was largely due to differences in integrated viable cells density (IVCD) rather than specific productivity. There was no apparent dose dependence in the effects of LONG®R3IGF-I. Cultures supplemented with 20 μg/L behaved similarly to those supplemented with 100 μg/L.
Fig. 2.
Product titer and specific productivity (qP) for all clones. All values are relative to the A1 insulin culture, replicate 1. Actual product titers were between 5 and 100 μg/mL and specific productivities were between 5 and 100 picograms/cell/day. Values were determined at day 4 in all cultures except G clone which was determined at day 3 due to a rapid decrease in viable cell density after day 3. Error bars represent the standard deviation of duplicate biological replicates. Insulin (open bars)-human recombinant insulin 5 mg/L; LR3L (filled bars)- LONG®R3IGF-I 20 μg/L; LR3H (hashed bars)- LONG®R3IGF-I 100 μg/L
Metabolism
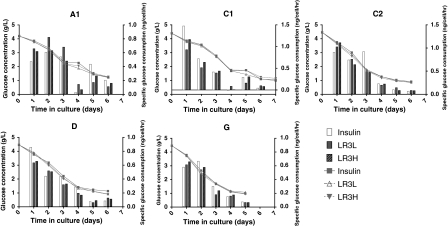
Significant differences in metabolic profiles were also observed between the different clones and the differences between the clones were, in general, much more significant than the differences between the different trophic factors. The glucose concentrations and glucose uptake rates are shown in Fig. 3. Regardless of clone or trophic factor stimulation, all cultures showed a final glucose concentration of ~1 g/L upon culture termination, indicating that the cultures were not glucose limited. The different clones exhibited fairly significant differences in glucose uptake rates, with the highest initial rates observed in the C1 and C2 (>1 ng/cell/h) and the lowest rates observed in the A1 and G cell lines (~0.5–0.6 ng/cell/h). While there was some variation in glucose uptake between the insulin-stimulated and LONG®R3IGF-I-stimulated cultures, no clear pattern emerged. A priori, one might expect a higher glucose consumption in the cells stimulated with insulin than in those stimulated with LONG®R3IGF-I, but that appears not to be the case.
Fig. 3.
Glucose concentration (lines) and specific glucose consumption rates (bars) for all cell clones. Values are the mean of duplicate biological replicates. Insulin-human recombinant insulin 5 mg/L; LR3L- LONG®R3IGF-I 20 μg/L; LR3H- LONG®R3IGF-I 100 μg/L
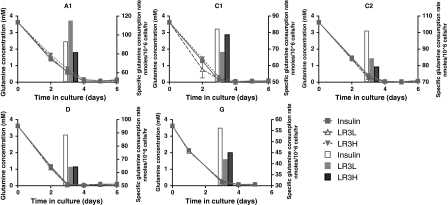
As in the case of glucose, the different cell clones exhibited much larger variations in glutamine consumption patterns than the clones stimulated with different trophic factors (Fig. 4). All cell lines exhibited glutamine depletion by day 4, presumably leading to the cessation of growth. While there was no statistically significant difference in the glutamine concentrations between the different trophic factors, cell lines C2 and D exhibited statistically significant (p < 0.05 by one-way ANOVA) differences in specific glutamine consumption rates between cultures supplemented with insulin and those stimulated with LONG®R3IGF-I, with lower specific consumption rates in the LONG®R3IGF-I cultures. The G cell clone appears to follow a similar pattern, although the difference in consumption rates was not statistically significant. One might hypothesize that the rapid decline in viable cell density of the G cell clone might have been due to more rapid depletion of the glutamine, but that appears not to be the case; in fact, the G cell clone actually exhibited the lowest specific glutamine consumption rates of all the clones, suggesting that glutamine supplementation would not have extended its growth significantly.
Fig. 4.
Glutamine concentration (lines) and specific glutamine consumption rates bars for all cell lines. Specific glutamine consumption rates were calculated from day 0 to day 3. Values are the mean of duplicate biological replicates. Insulin-human recombinant insulin 5 mg/L; LR3L- LONG®R3IGF-I 20 μg/L; LR3H- LONG®R3IGF-I 100 μg/L
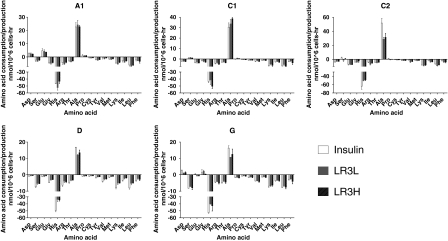
Some of the most striking variations in metabolic profiles can be seen in the amino acid metabolic patterns (Fig. 5). The greatest differences occurred in glycine and proline and the small polar amino acids, aspartic acid, serine and glutamic acid. As shown in Table 1, in some clones there was a net consumption of these amino acids, and in others, a net production. Histidine showed the highest net consumption of any amino acid (other than glutamine) and was nearly depleted in all cultures despite additional supplementation of histidine after preliminary experiments. As before, there was no clear distinction between cultures supplemented with insulin and those supplemented with LONG®R3IGF-I. Interestingly, there was an apparent correlation between the amount of histidine consumed and the amount of alanine produced when comparing the different trophic factors in any one cell clone.
Fig. 5.
Amino acid production (positive values) and consumption (negative values) for all clones. Error bars represent the standard deviation of duplicate biological measurements. All values were obtained at day 4 in culture expect for clone G which was obtained at day 3. Insulin-human recombinant insulin 5 mg/L; LR3L- LONG®R3IGF-I 20 μg/L; LR3H- LONG®R3IGF-I 100 μg/L
Table 1.
Variation in amino acid consumption and production between different cell clones
| Amino acid | |||||
|---|---|---|---|---|---|
| Clone | Aspartate | Serine | Glutamate | Glycine | Proline |
| A1 | ++ | –– | ++ | –– | + |
| C1 | –– | ± | –– | –– | ± |
| C2 | –– | ± | –– | –– | ± |
| D | –– | –– | –– | –– | –– |
| G | ++ | –– | ± | + | –– |
All data from day 4 in culture except for clone G which was determined at day 3
++ More than 50% increase over initial concentration; + less than 50% increase of starting concentration; ± mixed responses between culture conditions-all less than 50% change from starting values; –– decrease from initial conditions with more than 50% of initial concentration remaining; –– decrease from initial conditions with less than 50% of initial concentration remaining
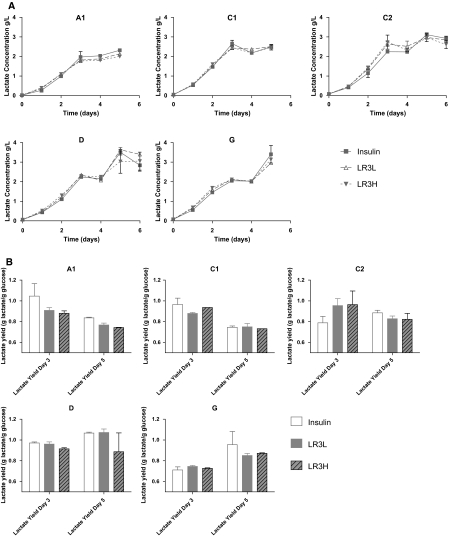
The lactate levels and the yields of lactate from glucose are shown in Fig. 6a, b, respectively. In all cell clones, the lactate levels reached a plateau around day 3, corresponding to the transition into stationary (or decline) phase. The actual lactate concentrations varied substantially between the clones. After this plateau, a variety of behaviors were observed with some clones consuming lactate, and others resuming lactate production. These differences can be seen by comparing the yields of lactate from glucose at day 3 and day 5 (Fig. 6b). In all cases the lactate yields were relatively high, ranging from a low of ~0.7 g lactate/g glucose to a high of slightly greater than 1 g lactate/g glucose.
Fig. 6.
a Lactate concentrations for all clones. Error bars represent the standard deviation of duplicate biological cultures. b Yield of lactate from glucose at day 3 and day 5 of culture. Error bars represent the standard deviation of duplicate biological replicates. Insulin-human recombinant insulin 5 mg/L; LR3L- LONG®R3IGF-I 20 μg/L; LR3H- LONG®R3IGF-I 100 μg/L
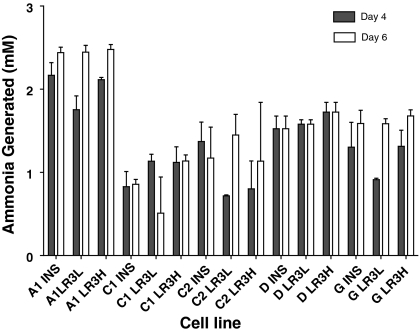
The ammonia generated for each cell clone under the varying growth conditions is shown in Fig. 7. Approximately 1–2.5 mM ammonia was generated in each clone. As the starting ammonia level in the medium (presumably due to spontaneous deamination of glutamine) was approximately 1.5 mM, final ammonia concentrations ranged from ~2 to 4 mM. This level is within the range that has been reported to be toxic to some cell lines (Schneider et al. 1996), although in some systems, no toxicity was reported below 6 mM. However, no correlation was observed between the ammonia levels and the final cell density (data not shown). As was the case for most of the metabolic parameters, greater variation was seen between the different cell clones than between the different trophic factors, with the C1 and C2 cell clones generating the lowest levels of ammonia and the A1 cell clone the highest levels.
Fig. 7.
Ammonia production at day 4 (filled bars) and day 6 (open bars) of culture (day 3 and day 5 for clone G). Error bars represent the standard deviation of duplicate biological replicates. Insulin-human recombinant insulin 5 mg/L; LR3L- LONG®R3IGF-I 20 μg/L; LR3H- LONG®R3IGF-I 100 μg/L
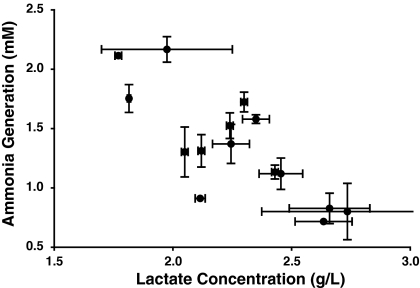
The lactate and ammonia production are compared in Fig. 8. As can be seen, they exhibit a strong inverse correlation (Pearson r = −0.7818, p = 0.0006, r2 = 0.6113, Pearson correlation), indicating that the cells that produce high levels of lactate produce lower levels of ammonia and vice versa, suggesting that some cell clones have a more glycolytic metabolic pattern, i.e. more lactate production (particularly C1 and C2), whereas other cell clones (particularly A1) have a metabolic pattern that favors generation of ammonia. Interestingly, the A1 cell clone did not deplete the glutamine more rapidly (see Fig. 4); however it did have one of the higher rates of specific glutamine consumption, particularly for the LONG®R3IGF-I culture supplemented at 20 μg/L. The C2 clone exhibited similar rates of glutamine consumption as the A1 although there was some variation between the trophic factors. The A1 clone did exhibit the lowest specific rate of glucose consumption (Fig. 3) and a slightly higher final glucose concentration than the other cell clones. Notably, the A1 cell clone is the only clone that produced significant amounts of aspartate, glutamate, and proline as shown in Table 1 (possible shunts for carbon that are not converted into lactate); however, its production of alanine was somewhat lower than the C1 and C2 clones (Fig. 5); hence C1 and C2 may have reduced their ammonia generation by excreting some of the ammonia as alanine.
Fig. 8.
Lactate concentrations (day 3) versus ammonia production (day 4, except for clone G which is reported at day 3). Error bars represent the standard deviation of duplicate biological experiments
Discussion
Use of insulin and insulin-like growth factors (IGF) in cell culture media
Insulin has been a routine component of serum-free media formulations from many of the earliest studies, being widely reported to stimulate cell growth and essential for many cell lines (Barnes and Sato 1980). Insulin has also been reported to stimulate protein synthesis, both in adipocytes and muscle cells (Pause et al. 1994) as well as in CHO cells (Zhang et al. 1997). However, the concentrations of insulin required in cell culture (typically 1–10 mg/L) are much higher than the physiological concentrations of insulin, suggesting a variety of possible explanations for its effect. One suggestion was that insulin was rapidly inactivated by cysteine in the basal medium; however, when cysteine was replaced by cystine, the necessary concentration of insulin was decreased tenfold, but this level was still outside of the physiological range (Hayashi et al. 1978). Other possibilities are that the preparations of insulin were contaminated by other factors or much more likely, that insulin is acting through other receptors, such as the insulin-like growth factor type 1 receptor (or hybrid heterodimers of the insulin and IGF receptors). The development of recombinant insulin in response to a desire for animal-derived component free media (ADCF) has eliminated concerns about contamination of pancreatic-derived insulin with other proteins, leaving questions about the mechanisms of insulin action remaining.
Insulin-like growth factor I (IGF-I) is a 70 amino acid polypeptide hormone with a molecular weight of 7.6 kDa with high homology to insulin. Insulin and IGF-1 both signal by binding to their respective receptors. Both receptors are composed of two extracellular α-subunits and two transmembrane β-subunits. The receptors have high homology in their tyrosine kinase domains (84%) but differ markedly in the transmembrane (~25% homology) and carboxy-terminal domain (Laviola et al. 2007). Binding of the respective hormones leads to a conformational change causing autophosphorylation on cytoplasmic tyrosine residues. Previous work at Novozymes determined that the number of IGF-1 receptors in CHO cells is approximately tenfold greater than the number of insulin receptors (Yandell et al. 2004); however, literature reports indicate that in the presence of a large excess of one type of receptor, insulin and IGF-1 receptors tend to heterodimerize (Laviola et al. 2007). The IGF-1 receptor is reported to have 100–1,000-fold less affinity for insulin than IGF. Interestingly, the heterodimeric receptors show very little loss of affinity for IGF-1, but fairly low affinity for insulin (Pandini et al. 2002; Slaaby et al. 2006; Benyoucef et al. 2007), suggesting that the heterodimers would also require super-physiological concentrations of insulin to stimulate growth.
A number of authors have suggested that IGF-I could substitute for insulin in cell culture medium, either by exogenous addition (Rasmussen et al. 1998), or by engineering the cell lines to produce their own IGF-I (Hunt et al. 1997; Li et al. 2000; Sunstrom et al. 1998; Sunstrom et al. 2000). However, exogenous addition of IGF-I is complicated by the presence of IGF binding proteins (IGFBP) secreted by the cells. In vivo, virtually all of the secreted IGF-I appears to be bound by one of at least six IGFBPs which have significantly higher affinity for IGF-I than the IGF receptors (Holly 2004), and in one of the CHO cell studies in which IGF-I was exogenously expressed, IGFBP-3 expression was induced in response to the presence of IGF-I, potentially limiting the effectiveness of IGF-I at stimulating growth (Sunstrom et al. 1998). To alleviate the effects of IGFBPs, an IGF-I analog (LONG®R3IGF-I) was developed by Novozymes (previously Gropep) in which the third amino acid, glutamate was replaced by arginine (Francis et al. 1992), reducing the affinity of LONG®R3IGF-I for IGFBPs by > 100-fold without compromising its biological potency.
A limited number of published studies have compared growth (Chun et al. 2003) or viability and productivity (Morris and Schmid 2000) in CHO cells grown either in the presence of insulin and/or LONG®R3IGF-I. Additional studies have examined receptor activation (Voorhamme and Yandell 2006) and the signaling pathways activated by the two mitogens (Yandell et al. 2004). LONG®R3IGF-I was found to be effective at much lower concentrations (5–100 μg/L) than insulin, as would be expected. In the study performed by Morris and Schmid, two different CHO cell lines, producing different recombinant proteins from the same parental cell line were examined. Both cell lines exhibited extended viability in the presence of LONG®R3IGF-I compared to insulin, and this extended viability presumably led to the increase in product titer that was reported in both cell lines. Glucose consumption and lactate production were unaffected by the choice of trophic factor in both cell lines. In the study performed by Chun et al., a factorial design study was applied to identify stimulatory proteins that would have a beneficial effect on serum-free CHO cell cultures. They examined the effects of insulin, LONG®R3IGF-I, transferrin, and basic fibroblast growth factor (bFGF). LONG®R3IGF-I had the most beneficial effect on growth, followed by insulin; bFGF had a minor positive effect on growth, and transferrin showed little to no benefit. Interestingly, supplying both insulin and LONG®R3IGF-I had an antagonistic affect, possibly due to competition between the insulin and LONG®R3IGF-I for the IGF-I receptors. This result is somewhat surprising considering that the IGF-I receptors (and any insulin/IGF-1 receptor heterodimers) would be expected to have a much higher affinity for LONG®R3IGF-I than for insulin. It is possible that this outcome is an artifact of the factorial design study, as other investigators have reported synergistic effects of combining insulin and LONG®R3IGF-I (personal communication).
In contrast to the previous studies, we found that only one cell clone (clone D) exhibited improved growth in response to LONG®R3IGF-I in comparison with insulin; the other clones were largely indifferent, although clone A1 did exhibit a slight extension of viability (Fig. 1). It is possible that had our cultures not become glutamine depleted, we might have seen a similar extension of viability to that observed by Moriss and Schmid. However, we also did not see a general improvement in viable density at day 3 as observed in the studies by Chun et al. (whose initial glutamine level was 4 mM, the same as ours). We did observe that in addition to the D clone, which showed an overall improvement in growth in the LONG®R3IGF-I over the insulin cultures, the C2 line showed a higher cell density at day 3 (the study endpoint in the Chun paper). However, the insulin culture did ultimately surpass the LONG®R3IGF-I cultures in its maximum cell density; moreover, the titers were reduced in the LONG®R3IGF-I cultures (particularly the 20 μg/L supplemented culture) due to a reduction in the specific productivities. Neither of the previous studies reported specific productivities, although the study by Morris and Schmid did report changes in product titers. As they did not report IVCD values, one can only surmise that the increase in productivity was due primarily to the extension of growth and not to a change in qP. We observed that the specific productivities were in generally slightly higher in the insulin supplemented cultures (although not statistically significant) with the one exception of the C1 clone, which showed a slightly higher (though not statistically significant) specific productivity in the cultures supplemented by LONG®R3IGF-I. Hence, when considering medium supplementation with different trophic factors, it is clearly important to examine the effects on both growth and product titer. It is likely that these observations would also hold for fed-batch cultures, but this would need to be verified.
Clonal variation in cellular metabolism
While the initial goal of these studies was to investigate the effects of insulin and LONG®R3IGF-I on growth and antibody productivity for a collection of cell clones with industrial productivity levels, a somewhat surprising outcome of these studies was the significant variability in metabolic behavior between different cell clones. Much of the early work on industrially relevant mammalian cell culture systems, particularly hybridoma cells, focused on understanding metabolic behavior (Bonarius et al. 1996; Leno et al. 1992; Mancuso et al. 1994, 1998; Ozturk and Palsson 1991; Savinell and Palsson 1992; Sharfstein et al. 1994; Xie and Wang 1994; Zupke and Stephanopoulos 1995). However, it became apparent that unlike microbial systems, there was not a direct link between metabolic behavior and productivity (Sharfstein et al. 1994). As production of monoclonal antibodies moved from hybridoma cells to CHO cells, with the introduction of strong viral promoters and amplified gene copy numbers, the focus shifted to designing media and feeding strategies to achieve higher viable cell densities and extended growth phases (Wurm 2004). To achieve that goal, industrial studies focused primarily on factorial experimental design and spent medium analysis (Chun et al. 2003; Ganne and Mignot 1991; Kim and Lee 2009; Kontoravdi et al. 2005; Liu et al. 2001; Sandadi et al. 2006). However, with the development of genome-scale models that can address all of the metabolic pathways in cells (Selvarasu et al. 2009; Sheikh et al. 2005) and the development of microarray technologies that can evaluate gene expression levels (Dorai et al. 2007; Griffin et al. 2007; Shen and Sharfstein 2006; Swiderek and Al-Rubeai 2007; Shen et al. 2010), there has been renewed interest in understanding cellular metabolism and its relationship with protein productivity (Baughman et al. 2010a, b).
In these studies, the specific glucose and glutamine consumption rates differed sharply between different cell clones. The C2 clone showed the highest specific rate of glucose consumption and one of the highest rates of glutamine consumption, but not the highest cell titers. The G cell line, which had the lowest titers, did not exhibit the lowest consumption rate of glucose, but the LONG®R3IGF-I supplemented cultures of the G clone did exhibit the lowest specific glutamine consumption rates. The amino acid utilization patterns were also quite striking (Table 1). The A1 clone, which had the lowest overall maximum viable cell density and the G clone, which had the highest overall viable cell density, both secreted aspartic acid, while all of the other clones consumed it. The A1 clone also secreted glutamate and proline, the only clone to do so. The G clone produced glycine, while all other clones consumed it. Similar variation was seen in the metabolite patterns. While all cell clones produced lactate and ammonia, the amount produced and the yields differed sharply. A strong inverse correlation was observed between ammonia production and lactate production (Fig. 8) with the A1 clone favoring ammonia production and both C clones, lactate production. As lactate is generally believed to be much less toxic than ammonia, this suggests that clones could be screened for lactate and ammonia production, and those that exhibit more beneficial phenotypes selected to go forward. One might argue when comparing the C2 and D clones, both of which exhibited quite high productivities and titers that the C2 clone with its slightly lower titer might perform better in fed-batch cultures due to its somewhat lower production of lactate and ammonia. This reduced production of lactate and ammonia may be due to the high rate of alanine synthesis (Fig. 5), serving as both a repository for carbon that is not consumed by the TCA cycle as well as a sink for nitrogen removed from glutamine. Interestingly, the two C clones, which were derived from the same parental transfection, both exhibit fairly high rates of alanine production when compared with the other clones. The relationship between productivity and metabolism is still unclear as the two highly productive clones (C2 and D) exhibit quite different metabolic behaviors, suggesting that there are a variety of ways for cells to utilize nutrients that can still lead to optimal growth and productivity patterns. Current work in our laboratory is attempting to address the relationships between these metabolic patterns and growth and productivity using a genome-scale reconstruction based on our previous state-space approach to modeling cellular metabolism (Baughman et al. 2010a, b). However, our experimental results strongly suggest that several highly productive clones be screened when attempting to optimize medium formulations or “fit” a new cell line and product into an existing platform process. While the results presented here are for batch cultures and do not address fed-batch conditions, the metabolic differences between clones are apparent long before nutrients become depleted, suggesting that the clonal variation might be exacerbated rather than diminished under fed-batch conditions.
Conclusions
Five CHO cell clones producing the same recombinant monoclonal antibody with industrial levels of productivity were examined for growth, productivity and metabolic behavior in cultures stimulated with either insulin or LONG®R3IGF-I. Neither trophic factor improved growth or productivity in all clones, and in general, the differences in growth and productivity between the different culture conditions were fairly modest. The least productive culture conditions produced more than 70% as much antibody as the most productive conditions, and the culture conditions that exhibited the highest growth had an IVCD that was ~1.5-fold higher than the lowest conditions for the same cell clone. Most strikingly, the different clones exhibited quite different metabolic behaviors with respect to nutrient uptake and metabolite production. These behaviors were not correlated with growth or productivity either between the clones or in response to stimulation with trophic factors. The substantial differences in response to trophic factors and cellular metabolism between different clones, all from the same parental CHO cell line and all producing the same recombinant monoclonal antibody strongly suggests that multiple clones be brought forward during process development in order to identify clones that will both provide high levels of productivity and good compatibility with platform processes.
Electronic supplementary material
Acknowledgments
The authors would like to thank Drs. Sally Grosvenor, Larissa Chirkova, Anthony Simula, and Geoffrey Francis for advice on experimental design and comments on the manuscript and Dr. Thomas Kiehl, Christian Schenkelberg, and Yong Jun An for technical assistance with the cell counting. This work was supported by Novozymes Biopharma AU.
References
- Barnes D, Sato G. Methods for growth of cultured cells in serum-free medium. Anal Biochem. 1980;102(2):255–270. doi: 10.1016/0003-2697(80)90151-7. [DOI] [PubMed] [Google Scholar]
- Baughman AC, Sharfstein ST, Martin LL. A flexible state-space approach for the modeling of metabolic networks I: development of mathematical methods. Metab Eng. 2010;13:125–137. doi: 10.1016/j.ymben.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Baughman AC, Sharfstein ST, Martin LL. A flexible state-space approach for the modeling of metabolic networks II: advanced interrogation of hybridoma metabolism. Metab Eng. 2010;13:138–149. doi: 10.1016/j.ymben.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Bebbington CR, Renner G, Thomson S, King D, Abrams D, Yarranton GT. High-level expression of a recombinant antibody from myeloma cells using a glutamine synthetase gene as an amplifiable selectable marker. Biotechnology (NY) 1992;10:169–175. doi: 10.1038/nbt0292-169. [DOI] [PubMed] [Google Scholar]
- Benyoucef S, Surinya KH, Hadaschik D, Siddle K. Characterization of insulin/IGF hybrid receptors: contributions of the insulin receptor L2 and Fn1 domains and the alternatively spliced exon 11 sequence to ligand binding and receptor activation. Biochem J. 2007;403:603–613. doi: 10.1042/BJ20061709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonarius HPJ, Hatzimanikatis V, Meesters KPH, deGooijer CD, Schmid G, Tramper J. Metabolic flux analysis of hybridoma cells in different culture media using mass balances. Biotechnol Bioeng. 1996;50:299–318. doi: 10.1002/(SICI)1097-0290(19960505)50:3<299::AID-BIT9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Chun C, Heineken K, Szeto D, Ryll T, Chamow S, Chung JD. Application of factorial design to accelerate identification of CHO growth factor requirements. Biotechnol Prog. 2003;19:52–57. doi: 10.1021/bp025575+. [DOI] [PubMed] [Google Scholar]
- DePalma A (2008) Platform technologies ease scale-up pain. Genet Eng Biotechnol News 28:44+
- Dorai H, Li K, Huang CC, Bittner A, Galindo J, Carmen A. Genome-wide analysis of mouse myeloma cell lines expressing therapeutic antibodies. Biotechnol Prog. 2007;23:911–920. doi: 10.1021/bp0700051. [DOI] [PubMed] [Google Scholar]
- Durocher Y, Butler M. Expression systems for therapeutic glycoprotein production. Curr Opin Biotechnol. 2009;20:700–707. doi: 10.1016/j.copbio.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Eppink MHM, Schreurs R, Gusen A, Verhoeven K. Platform technology for developing purification processes. Biopharm Int. 2009;20:44. [Google Scholar]
- Francis GL, Ross M, Ballard FJ, Milner SJ, Senn C, McNeil KA, Wallace JC, King R, Wells JR. Novel recombinant fusion protein analogues of insulin-like growth factor (IGF)-I indicate the relative importance of IGF-binding protein and receptor binding for enhanced biological potency. J Mol Endocrinol. 1992;8:213–223. doi: 10.1677/jme.0.0080213. [DOI] [PubMed] [Google Scholar]
- Ganne V, Mignot G. Application of statistical design of experiments to the optimization of factor VIII expression by CHO cells. Cytotechnology. 1991;6:233–240. doi: 10.1007/BF00624762. [DOI] [PubMed] [Google Scholar]
- Griffin TJ, Seth G, Xie HW, Bandhakavi S, Hu WS. Advancing mammalian cell culture engineering using genome-scale technologies. Trends Biotechnol. 2007;25:401–408. doi: 10.1016/j.tibtech.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Hayashi I, Larner J, Sato G. Hormonal growth control of cells in culture. In Vitro. 1978;14:23–30. doi: 10.1007/BF02618171. [DOI] [PubMed] [Google Scholar]
- Holly J (2004) Physiology of the IGF system. In: Bock G, Goode J (eds) Biology of IGF-1: its interaction with insulin in health and malignant states, Novartis Foundation Symposium 262, pp 19–26; discussion pp 26–35, pp 265–268, Wiley, Chichester, UK. doi:10.1002/0470869976.ch3 [PubMed]
- Hunt SMN, Pak SCO, Bridges MW, Gray PP, Sleigh MJ. Chinese hamster ovary cells produce sufficient recombinant insulin-like growth factor I to support growth in serum-free medium—serum-free growth of IGF-I producing CHO cells. Cytotechnology. 1997;24:55–64. doi: 10.1023/A:1007969502256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Sharfstein ST. Characterization of gene localization and accessibility in DHFR-amplified CHO cells. Biotechnol Prog. 2009;25:296–300. doi: 10.1002/btpr.82. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Huang Y, Sharfstein ST. Regulation of recombinant monoclonal antibody production in Chinese hamster ovary cells: a comparative study of gene copy number, mRNA level, and protein expression. Biotechnol Prog. 2006;22:313–318. doi: 10.1021/bp0501524. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Selection and coamplification of heterologous genes in mammalian cells. Methods Enzymol. 1990;185:537–566. doi: 10.1016/0076-6879(90)85044-O. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ, Sharp PA, Latt SA. Evolution of chromosomal regions containing transfected and amplified dihydrofolate reductase sequences. Mol Cell Biol. 1983;3:699–711. doi: 10.1128/mcb.3.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lee GM. Development of serum-free medium supplemented with hydrolysates for the production of therapeutic antibodies in CHO cell cultures using design of experiments. Appl Microbiol Biotechnol. 2009;83:639–648. doi: 10.1007/s00253-009-1903-1. [DOI] [PubMed] [Google Scholar]
- Kontoravdi C, Asprey SP, Pistikopoulos EN, Mantalaris A. Application of global sensitivity analysis to determine goals for design of experiments: an example study on antibody-producing cell cultures. Biotechnol Prog. 2005;21:1128–1135. doi: 10.1021/bp050028k. [DOI] [PubMed] [Google Scholar]
- La Merie Business Intelligence (2010) Top 20 Biologics 2009. R&D Pipeline News, vol March 10, 2009
- Laviola L, Natalicchio A, Giorgino F. The IGF-I signaling pathway. Curr Pharm Des. 2007;13:663–669. doi: 10.2174/138161207780249146. [DOI] [PubMed] [Google Scholar]
- Leno M, Merten OW, Hache J. Kinetic-analysis of hybridoma growth and monoclonal-antibody production in semicontinuous culture. Biotechnol Bioeng. 1992;39:596–606. doi: 10.1002/bit.260390603. [DOI] [PubMed] [Google Scholar]
- Leonard M, Onadipe K (2010) Practical challenges in biotherapeutic development: when and how do rapidly-developed, platform phase 1 processes “Transform Themselves” into optimized, robust commercial ones? Paper presented at the cell culture engineering XII, Banff, Alberta, Canada, April 25–30
- Li H, d’Anjou M. Pharmacological significance of glycosylation in therapeutic proteins. Curr Opin Biotechnol. 2009;20:678–684. doi: 10.1016/j.copbio.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Li DJ, Hettle S, McLean J, MacDonald C. Survival of 3T3 cells expressing or co-expressing bFGF and/or IGF-I and/or IGF-II in low serum and serum free media. Cytotechnology. 2000;32:209–218. doi: 10.1023/A:1008151302358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Chu I, Hwang S. Factorial designs combined with the steepest ascent method to optimize serum-free media for CHO cells. Enzyme Microb Technol. 2001;28:314–321. doi: 10.1016/S0141-0229(00)00346-X. [DOI] [PubMed] [Google Scholar]
- Mancuso A, Sharfstein ST, Tucker SN, Clark DS, Blanch HW. Examination of primary metabolic pathways in a murine hybridoma with carbon-13 nuclear magnetic resonance spectroscopy. Biotech Bioeng. 1994;44:563–585. doi: 10.1002/bit.260440504. [DOI] [PubMed] [Google Scholar]
- Mancuso A, Sharfstein ST, Fernandez EJ, Clark DS, Blanch HW. Effect of extracellular glutamine concentration on primary and secondary metabolism of a murine hybridoma: an in vivo 13C nuclear magnetic resonance study. Biotech Bioeng. 1998;57:172–186. doi: 10.1002/(SICI)1097-0290(19980120)57:2<172::AID-BIT6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Morris AE, Schmid J. Effects of insulin and LongR(3) on serum-free Chinese hamster ovary cell cultures expressing two recombinant proteins. Biotechnol Prog. 2000;16:693–697. doi: 10.1021/bp0000914. [DOI] [PubMed] [Google Scholar]
- Ozturk SS, Palsson BO (1991) Growth, metabolic, and antibody production kinetics of hybridoma cell culture: 2. Effects of serum concentration, dissolved oxygen concentration, and medium pH in a batch reactor. Biotechnol Prog 7:481–494 [DOI] [PubMed]
- Pandini G, Frasca F, Mineo R, Sciacca L, Vigneri R, Belfiore A. Insulin/insulin-like growth factor I hybrid receptors have different biological characteristics depending on the insulin receptor isoform involved. J Biol Chem. 2002;277:39684–39695. doi: 10.1074/jbc.M202766200. [DOI] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras A-C, Donze O, Lin T-A, Lawrence JC, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5’-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Rasmussen B, Davis R, Thomas J, Reddy P. Isolation, characterization and recombinant protein expression in Veggie-CHO: a serum-free CHO host cell line. Cytotechnology. 1998;28:31–42. doi: 10.1023/A:1008052908496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert JM, Wenger JB. Development trends for new cancer therapeutics and vaccines. Drug Discov Today. 2008;13:30–37. doi: 10.1016/j.drudis.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Sandadi S, Ensari S, Kearns B. Application of fractional factorial designs to screen active factors for antibody production by Chinese hamster ovary cells. Biotechnol Prog. 2006;22:595–600. doi: 10.1021/bp050300q. [DOI] [PubMed] [Google Scholar]
- Savinell JM, Palsson BO (1992) Network analysis of intermediary metabolism using linear optimization. II. Interpretation of hybridoma cell metabolism. J Theor Biol 154:455–473 [DOI] [PubMed]
- Schneider M, Marison IW, Stockar U. The importance of ammonia in mammalian cell culture. J Biotechnol. 1996;46:161–185. doi: 10.1016/0168-1656(95)00196-4. [DOI] [PubMed] [Google Scholar]
- Seeworster T (2010) Where are you in the platform funnel? Paper presented at the cell culture engineering XII, Banff, Alberta, Canada, April 25–30
- Selvarasu S, Wong VV, Karimi IA, Lee DY. Elucidation of metabolism in hybridoma cells grown in fed-batch culture by genome-scale modeling. Biotech Bioeng. 2009;102:1494–1504. doi: 10.1002/bit.22186. [DOI] [PubMed] [Google Scholar]
- Sharfstein ST, Tucker SN, Mancuso A, Blanch HW, Clark DS. Quantitative in vivo nuclear magnetic resonance studies of hybridoma metabolism. Biotech Bioeng. 1994;43:1059–1074. doi: 10.1002/bit.260431109. [DOI] [PubMed] [Google Scholar]
- Sheikh K, Förster J, Nielsen LK. Modeling hybridoma cell metabolism using a generic genome-scale metabolic model of Mus musculus. Biotechnol Prog. 2005;21:112–121. doi: 10.1021/bp0498138. [DOI] [PubMed] [Google Scholar]
- Shen D, Sharfstein ST. Genome-wide analysis of the transcriptional response of murine hybridomas to osmotic shock. Biotechnol Bioeng. 2006;93:132–145. doi: 10.1002/bit.20691. [DOI] [PubMed] [Google Scholar]
- Shen D, Kiehl TR, Khattak SF, Li ZJ, He A, Kayne PS, Patel V, Neuhaus IM, Sharfstein ST. Transcriptomic responses to sodium chloride-induced osmotic stress: a study of industrial fed-batch CHO cell cultures. Biotechnol Prog. 2010;26:1104–1115. doi: 10.1002/btpr.398. [DOI] [PubMed] [Google Scholar]
- Shukla AA, Hubbard B, Tressel T, Guhan S, Low D. Downstream processing of monoclonal antibodies—application of platform approaches. J Chromatogr B Anal Technol Biomed Life Sci. 2007;848:28–39. doi: 10.1016/j.jchromb.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Slaaby R, Schaffer L, Lautrup-Larsen I, Andersen AS, Shaw AC, Mathiasen IS, Brandt J. Hybrid receptors formed by insulin receptor (IR) and insulin-like growth factor I receptor (IGF-IR) have low insulin and high IGF-1 affinity irrespective of the IR splice variant. J Biol Chem. 2006;281:25869–25874. doi: 10.1074/jbc.M605189200. [DOI] [PubMed] [Google Scholar]
- Strohl WR, Knight DM. Discovery and development of biopharmaceuticals: current issues. Curr Opin Biotechnol. 2009;20:668–672. doi: 10.1016/j.copbio.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Sunstrom NA, Baig M, Cheng L, Sugyiono DP, Gray P. Recombinant insulin-like growth factor-I (IGF-I) production in super-CHO results in the expression of IGF-I receptor and IGF binding protein 3. Cytotechnology. 1998;28:91–99. doi: 10.1023/A:1008073513948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunstrom NA, Gay RD, Wong DC, Kitchen NA, DeBoer L, Gray PP. Insulin-like growth factor-I and transferrin mediate growth and survival of Chinese hamster ovary cells. Biotechnol Prog. 2000;16:698–702. doi: 10.1021/bp000102t. [DOI] [PubMed] [Google Scholar]
- Swiderek H, Al-Rubeai M. Functional genome-wide analysis of antibody producing NS0 cell line cultivated at different temperatures. Biotechnol Bioeng. 2007;98:616–630. doi: 10.1002/bit.21445. [DOI] [PubMed] [Google Scholar]
- Urlaub G, Chasin LA. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. PNAS. 1980;77:4216–4220. doi: 10.1073/pnas.77.7.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhamme D, Yandell CA. LONG R3IGF-I as a more potent alternative to insulin in serum-free culture of HEK293 cells. Mol Biotechnol. 2006;34:201–204. doi: 10.1385/MB:34:2:201. [DOI] [PubMed] [Google Scholar]
- Walsh G. Biopharmaceutical benchmarks 2006. Nat Biotechnol. 2006;24:769–776. doi: 10.1038/nbt0706-769. [DOI] [PubMed] [Google Scholar]
- Walsh G (2007) Approval trends in 2006. Biopharm Int 20:56+
- Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- Xie L, Wang DI. Applications of improved stoichiometric model in medium design and fed-batch cultivation of animal cells in bioreactor. Cytotechnology. 1994;15:17–29. doi: 10.1007/BF00762376. [DOI] [PubMed] [Google Scholar]
- Yandell C, Lawson J, Butler I, Wade B, Sheehan A, Grosvenor S, Goddard C, Simula T (2004) An analogue of IGF-I. Bioprocess Int, March:2–7
- Zhang YP, Katakura Y, Seto P, Shirahata S. Evidence that phosphatidylcholine-specific phospholipase C is a key molecule mediating insulin-induced enhancement of gene expression from human cytomegalovirus promoter in CHO cells. Cytotechnology. 1997;23:193–196. doi: 10.1023/A:1007955332526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupke C, Stephanopoulos G. Intracellular flux analysis in hybridomas using mass balances and in vitro (13)C NMR. Biotech Bioeng. 1995;45:292–303. doi: 10.1002/bit.260450403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.