Abstract
Conventional DNA ladder assay has certain shortcomings such as loss of DNA fragments during sample processing, involvement of multiple steps and requirement of expensive reagents. The present study demonstrates a rapid, easy-to-perform cost-effective method for detection of apoptotic DNA fragments with considerable improvement in the sensitivity by avoiding loss of DNA fragments. It involves a few minutes of procedure involving direct lysis of cells with dimethyl sulphoxide (DMSO), brief vortexing, addition of 2% SDS–TE buffer, and a single step of centrifugation. This cost- and time-efficient method reduces the assay time considerably and can be used for a large number of samples with excellent sensitivity.
Keywords: Cell death, Apoptosis, Sf9, DNA ladder, DNA fragmentation
Introduction
Apoptosis is a highly regulated cellular process that involves the activation of distinct biochemical/molecular cascades and can manifest into morphological features characterized by cell shrinkage, chromatin condensation, nuclear fragmentation and formation of apoptotic bodies. Cellular/molecular assays for detecting externalization of phosphatidylserine moieties (using annexin-V staining), cytosolic cytochrome-c release from mitochondria (using western blotting or immunofluorescence microscopy) and caspase activity are used routinely for monitoring apoptosis, besides routine morphological assessment. Fragmentation of DNA and chromatin is an integral process during apoptosis, and has been reported to occur in more than one distinct stage. During initial stages, high molecular weight DNA fragments of 50 kbp or longer size have been observed in morphologically normal cells committed to undergo apoptosis. The low molecular weight DNA fragments are associated with late events such as formation of apoptotic bodies (Czene et al. 2002), although the phenomenon of nucleosome excision (ladder formation) is also reported to initiate before any obvious apoptotic changes in cell morphology (Tanuma et al. 1993).
The extensive DNA fragmentation induced during apoptosis can be detected using techniques such as DNA ladder assay (agarose gel electrophoresis), terminal deoxynucleotidyl transferase mediated dUTP-biotin nick end labeling (TUNEL assay) and comet assay (Otsuki 2000; Chandna 2004). Out of these methods, DNA fragmentation assay using agarose gel electrophoresis is the most frequent technique used for the detection of apoptosis and can easily discriminate between apoptotic and non-apoptotic (necrotic) modes of cell death, as in most cases the inter-nucleosomal cleavage of genomic DNA yielding the characteristic DNA ladder is a molecular hallmark of apoptotic cells (Kerr et al. 1972; Pandey et al. 1994). In the typical DNA fragment ladder obtained, molecular weights of the genome fragments are integer multiples of 180 base-pairs length associated with a nucleosome subunit. On the other hand, genomic fragments of irregular sizes are generally induced during necrotic cells, and a DNA smear is obtained during agarose gel electrophoresis.
The conventionally used DNA fragmentation assay involves separation of DNA fragments following “phenol–chloroform” method of DNA isolation, which has been the method of choice. However, it is associated with certain drawbacks such as requirement of relatively large amount of starting material (e.g., in vitro cultured cells or tissue pieces), loss of smaller fragments during the precipitation step and longer time-consumption. In addition, it is cumbersome to use for large number of samples due to the long processing time involved. In recent past extensive efforts has been made to develop simpler assay methodology for detection of apoptosis (Singh 2000; Rosl 1992; Basnakian and James 1994; Lamm et al. 1997; Willingham 1999; Saraste and Pulkki 2000; Sgonc and Guber 1998; Compton 1992; Nicoletti et al. 1991; Umansky et al. 1981). A simple DNA fragmentation method developed by Gong et al. (1994) is although rapid and has few advantages as well but it may have reduced sensitivity because of ethanol fixation, which is known to remove smaller DNA fragments during processing (Telford et al. 1991, 1992) and also require enzymatic removal of proteins and RNA (Gong et al. 1994). In addition to these modifications in DNA ladder assay other methods of apoptosis detections such as radioisometric, fluorimetric, flow cytometric and comet assay based methods were also tried for rapid detection of apoptosis (Willingham 1999), but despite these efforts the conventional DNA ladder method is still popular and is being widely used indicating the need of simplification, which can be easily adapted by the researchers.
In this study, we report a much simpler and shorter alternative method for isolating DNA with increased sensitivity for DNA fragment detection. We used Sf9 insect cells that display classical apoptotic morphology when treated with apoptogenic agents such as Actinomycin-D (a transcriptional inhibitor), Etoposide (topoisomerase inhibitor) and ionizing radiation (Chandna et al. 2004; Suman et al. 2009; Kumarswamy et al. 2009). In order to demonstrate the advantages of this new method, we validated the results with conventional phenol–chloroform method, and monitored morphological apoptotic alterations using the differential image contrast (DIC) microscopy. Our results show that the proposed method not only detects DNA fragments with improved sensitivity, it can also facilitate assessment of apoptosis in a large number of samples due to ease of processing.
Materials and methods
Cell-culture
Sf9 cells were maintained as monolayers in 25 cm2 culture flasks (T-25) at 28 °C in Grace’s insect cell medium (Sigma, St. Louis, MO, USA) supplemented with 3.33 g/L lactalbumin hydrolysate, 3.33 g/L yeastolate, 0.35 g/L NaHCO3 and antibiotics (Penicillin-sodium salt 50,000 units/L, Streptomycin sulphate 50,000 μg/L, Nystatin 2,000 μg/L from 500,000 USP units/mg; all Sigma USA). Growth medium (pH 6.2) was prepared by adding 10% heat inactivated FBS (Fetal Bovine Serum, Sigma, USA) and stored at 2–8 °C. Sf9 cells were regularly sub-cultured twice a week in exponential phase by seeding at a density of 40,000–50,000 cells/cm2 area.
Irradiation and treatment with apoptotic agents
Cells were irradiated in exponential growth phase at dose-rate of 36.66 Gy/min in a 60Co gamma chamber (Gamma Chamber 5000, Board of Radiation and Isotope Technology, Department of Atomic Energy, Mumbai, India). Actinomycin-D (0.5 μg/mL; Sigma USA), Etoposide (100 μM, Dabur, India) were directly added to the culture media of cells. Cells were harvested at 8, 24 and 48 h time-points and for analysis of DNA fragmentation.
Live cell morphology analysis
Cells undergoing apoptosis were observed by Nomarski-DIC time-lapse imaging using the Axiovert 200 Zeiss inverted microscope (Carl Zeiss, Germany) and Axiovision software (version 4.0).
Detection of DNA fragmentation (ladder assay)
DNA isolation was done using following methods:
The conventional “phenol–chloroform” DNA isolation method: 3–4 million Sf9 cells were taken as starting material for DNA isolation. Cells were dislodged in the culture medium, and centrifuged at 5,000g to obtain the pellet containing both intact and apoptotic cells. Cells were further washed with PBS (phosphate buffer saline, pH 7.4) and again centrifuged at 5,000g before overnight lysis of the cell pellet with 150 μL of lysis buffer (10 mM EDTA, 0.5% SDS, 10 mM Trizma Base and 0.5 μg/mL Proteinase K; pH 7.5). Proteinase-K was deactivated by heating at 75 °C for 15 min followed by addition of RNAse-A (0.5 μg/mL) at 37 °C for 2 h. DNA was isolated by phenol–chloroform-isoamyl alcohol method and DNA was dissolved in 40 μL TE buffer (pH 7.4). The total 40 μL DNA solution was loaded on 2% agarose gel (Hermann and Frischauf 1987; Zhuang et al. 2008).
The new DMSO (dimethyl sulphoxide)–SDS (sodium dodecyl sulphate)–TE (Tris-EDTA) method: 1.8–2 million Sf9 cells were taken as starting material for DNA isolation. Cells were dislodged and washed with PBS in the same manner as used in conventional method. Instead of overnight lysis used in the conventional method, 100 μL of DMSO was added directly to the cell pellet and mixed well followed immediately by vortexing. Equal volume (100 μL) of TE buffer (pH 7.4) with 2% SDS was added, followed by mixing and vortexing. The resulting solution was centrifuged at 12,000g at 4 °C and 40 μL of the supernatant (equal volume as used for the conventional assay) was loaded on agarose gel. The isolated DNA was analyzed using UV-spectroscopy by recording the absorbance at 260, 280 and 230 nm respectively. Further, the A260/A280 and A260/A230 was calculated to analyze the purity of the isolated DNA and the isolated DNA was found to be of good quality. Stability of the isolated DNA was also analyzed by running the same sample after few days. When samples were stored at 4 °C, DNA degradation was not observed up to 3 days of storage.
Results and discussion
The DNA fragmentation assay involves two basic steps, viz., isolation of genomic DNA and separation of DNA fragments using agarose gel electrophoresis. The untreated normal cells yielded a very high molecular weight DNA in the range of 5–7 kbp but a ladder could be seen in apoptotic cells using the phenol–chloroform (PC) method of DNA isolation. Although this method yields reasonably good quality of DNA, it involves multiple time-consuming steps and is cumbersome to use for a large number of samples. Besides, it is prone to loss of low molecular weight fragments during centrifugation and phase-separation steps, and DNA may also get degraded due to mishandling during these multiple steps. Therefore, good level of skill is required to isolate DNA using the PC method.
In the new protocol detailed herein, we modified the DNA isolation steps in such a manner as to retain good quality DNA while considerably reducing the overall time of processing and minimizing chances of losing smaller DNA fragments. For this method we selected DMSO as the component of lysis buffer because of being a polar-aprotic solvent (Wang et al. 2008; Di and Kerns 2006). Lipids and proteins have limited solubility in DMSO, which removes proteins through its denaturing and precipitating action and also inhibits the activity of nucleases (Arakawa et al. 2007), which helps reduce false DNA fragmentation during sample processing. In addition, DMSO is known to inhibit the formation of folded DNA structure (Kang et al. 2005), which results in increased band intensity on the agarose gel following staining with fluorochromes such as ethidium bromide.
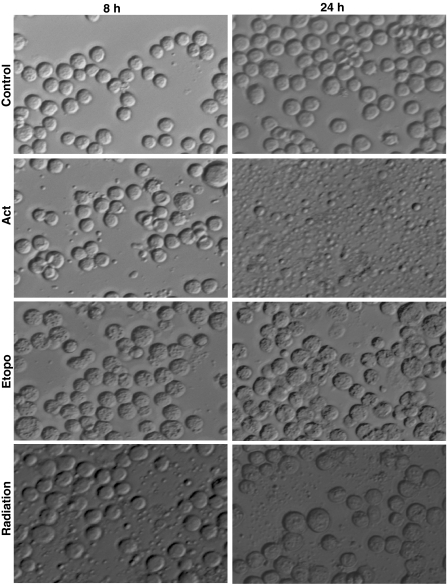
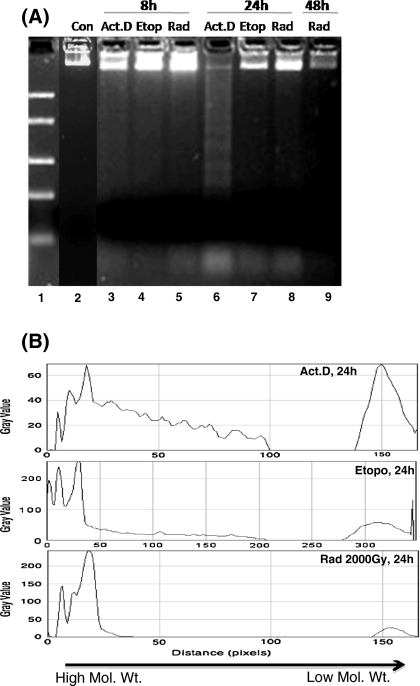
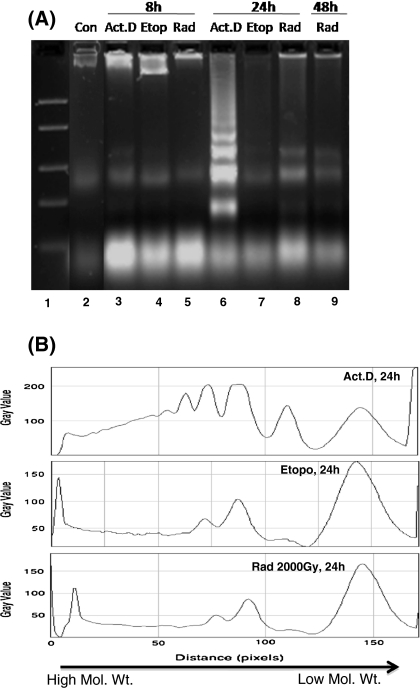
Since the DNA fragmentation is known to proceed before the onset of morphological changes during apoptosis, we studied morphological changes and DNA fragmentation at an early time-point (8 h) when very few apoptotic bodies were visible (Fig. 1). No DNA fragmentation could be detected using the PC method (lane 3, 4, 5 in Fig. 2), whereas the DMSO-SDS-TE method clearly shows the presence of fragmented DNA (lane 3, 4, 5 in Fig. 3). This ability of the new DMSO-SDS-TE method to detect DNA fragmentation at early time point was accompanied by a better yield of low molecular weight fragments at later time-points, while high molecular weight band was predominant in the PC method especially after etoposide treatment or irradiation (lane-7, 8 in Figs. 2b, 3b). Therefore, our results show that the DMSO-SDS-TE method can detect the low molecular weight DNA fragments with much better sensitivity compared to the conventional PC method.
Fig. 1.
Morphological analysis of stress-induced apoptotic body formation in Sf9 cells. Cells were treated with known apoptogenic agents; Actinomycin D (0.5 μg/mL), Etoposide (100 μM) and γ-radiation (2,000 Gy). Images were acquired with an Axiovert 200 Zeiss inverted microscope (Carl Zeiss, Germany) using Nomarski-DIC mode and Axiovision (version 4.0) software
Fig. 2.
DNA ladders of treated Sf9 cells obtained using the conventional phenol–chloroform method. a 3 × 106 cells were used and 40 μL of final sample was loaded in all lanes. Electrophoresis was performed on 2% agarose gel. b Densitometric analysis of ladder lanes using Image-J software, with distinct peaks representing a single band on the gel, ranging from high to low molecular weight
Fig. 3.
DNA ladders of treated Sf9 cells obtained using the new DMSO-SDS-TE method. a Equal volume (40 μL) of lysate was loaded in all lanes. Electrophoresis was performed on 2% agarose gel. Relative abundance of small DNA fragments is evident in the new assay compared to the conventional method. b Densitometric analysis of ladder lanes using Image-J software, with distinct peaks representing a single band on the gel, ranging from high to low molecular weight
Although DNA ladder is seemingly a simple assay, yet the extensive use of this technique prompted us to enhance its sensitivity and reliability through this super-short and highly simplified procedure. The obvious multiple advantages of the new DMSO–SDS–TE method are: (1) negligible loss of DNA fragments that increases the yield of DNA fragments and enhances sensitivity of detection, (2) early detection of DNA fragmentation due to increased sensitivity, (3) extremely short (less than 30 min) processing of cells as compared to the conventional (overnight) isolation method, (4) suitability for use in large number of samples, (5) cost-effectiveness (requiring fewer reagents and chemicals), and (6) higher stability of the isolated sample (up to 3 days when stored at 4 °C). Limitation of this method is that the DNA isolated using this protocol cannot be used for other purposes such as PCR, because of presence of high amount of DMSO. Therefore, experiments only aiming at detection or monitoring of DNA fragmentation should be performed using this technique, for which the new technique definitely offers an edge over the existing phenol–chloroform method. We believe that this short and sensitive technique will be very useful for numerous labs that routinely study cell death or carry out routine experimental/clinical screening of drugs and chemotherapeutics.
Acknowledgment
This work is an offshoot from the routine cellular radiobiology studies being carried out under project INM-311.1.5 funded by DRDO, Government of India. Constant support received in our studies from Dr. R. P. Tripathi, Director INMAS, is duly acknowledged.
References
- Arakawa T, Kita Y, Timasheff SN. Protein precipitation and denaturation by dimethyl sulfoxide. Biophys Chem. 2007;131:62–70. doi: 10.1016/j.bpc.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Basnakian AG, James SJ. A rapid and sensitive assay for detection of DNA fragmentation during early phases of apoptosis. Nucleic Acids Res. 1994;22:2714–2715. doi: 10.1093/nar/22.13.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandna S. Single-cell gel electrophoresis assay monitors precise kinetics of DNA fragmentation induced during programmed cell death. Cytometry Part A. 2004;61A:127–133. doi: 10.1002/cyto.a.20071. [DOI] [PubMed] [Google Scholar]
- Chandna S, Dwarakanath BS, Seth RK, Khaitan D, Adhikari JS, Jain V. Radiation responses of Sf9, a highly radioresistant lepidopteran insect cell line. Int J Radiat Biol. 2004;80:301–315. doi: 10.1080/09553000410001679794. [DOI] [PubMed] [Google Scholar]
- Compton MM. A biochemical hallmark of apoptosis: intranucleosomal degradation of the genome. Cancer Metast Rev. 1992;11:105–119. doi: 10.1007/BF00048058. [DOI] [PubMed] [Google Scholar]
- Czene S, Testa E, Nygren J, Belyaev I, Harms-Ringdahl MH. DNA fragmentation and morphological changes in apoptotic human lymphocytes. Biochem Biophys Res Commun. 2002;294:872–878. doi: 10.1016/S0006-291X(02)00588-0. [DOI] [PubMed] [Google Scholar]
- Di L, Kerns EH. Biological assay challenges from compound solubility: strategies for bioassay optimization. Drug Discov Today. 2006;11:446–451. doi: 10.1016/j.drudis.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Gong J, Traganos F, Darzynkiewicz Z. A selective procedure for DNA extraction from apoptotic cells applicable for gel electrophoresis and flow cytometry. Anal Biochem. 1994;218:314–319. doi: 10.1006/abio.1994.1184. [DOI] [PubMed] [Google Scholar]
- Hermann BG, Frischauf AM. Isolation of genomic DNA. Methods Enzymol (US) 1987;152:180–183. doi: 10.1016/0076-6879(87)52018-3. [DOI] [PubMed] [Google Scholar]
- Kang J, Lee MS, Gorenstein DG. The enhancement of PCR amplification of a random sequence DNA library by DMSO and betaine: application to in vitro combinatorial selection of aptamers. J Biochem Biophys Methods. 2005;64:147–151. doi: 10.1016/j.jbbm.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarswamy R, Seth RK, Dwarakanath BS, Chandna S. Mitochondrial regulation of insect cell apoptosis: evidence for permeability transition pore-independent cytochrome-c release in the Lepidopteran Sf9 cells. Int J Biochem Cell Biol. 2009;41:1430–1440. doi: 10.1016/j.biocel.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Lamm GM, Steinlein P, Cotton M, Christofori G. A rapid, quantitative and inexpensive method for detecting apoptosis by flow cytometry in transiently transfected cells. Nucleic Acids Res. 1997;25:4855–4857. doi: 10.1093/nar/25.23.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods. 1991;139:271–280. doi: 10.1016/0022-1759(91)90198-O. [DOI] [PubMed] [Google Scholar]
- Otsuki Y. Various methods of apoptosis detection. Acta Histochem Cytochem. 2000;33:235–241. doi: 10.1267/ahc.33.235. [DOI] [Google Scholar]
- Pandey S, Walker PR, Sikorska M. Separate pools of endonuclease activity are responsible for inter-nucleosomal and high-molecular-mass DNA fragmentation during apoptosis. Biochem cell Biol (biochimie et biologie cellulaire) 1994;72:625–629. doi: 10.1139/o94-082. [DOI] [PubMed] [Google Scholar]
- Rosl F. A simple and rapid method for detection of apoptosis in human cells. Nucleic Acids Res. 1992;20:5243. doi: 10.1093/nar/20.19.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste A, Pulkki K. Morphological and biochemical hallmarks of apoptosis. Cardiovasc Res. 2000;45:528–537. doi: 10.1016/S0008-6363(99)00384-3. [DOI] [PubMed] [Google Scholar]
- Sgonc R, Guber J. Apoptosis detection: an overview. Exp Gerontol. 1998;33:525–533. doi: 10.1016/S0531-5565(98)00031-X. [DOI] [PubMed] [Google Scholar]
- Singh NP. A simple method for accurate estimation of apoptotic cells. Exp Cell Res. 2000;256:328–337. doi: 10.1006/excr.2000.4810. [DOI] [PubMed] [Google Scholar]
- Suman S, Khaitan D, Pati U, Seth RK, Chandna S. Stress response of a p53 homologue in the radioresistant Sf9 insect cells. Int J Radiat Biol. 2009;85:238–249. doi: 10.1080/09553000902748591. [DOI] [PubMed] [Google Scholar]
- Tanuma S, Shiokawa D, Tanimoto Y, Ikekita M, Sakagami H, Takada M, Fukuda S, Kochi M. Benzylideneascorbate induces apoptosis in L029 tumor cells. Biochem Biophys Res Commun. 1993;15:29–35. doi: 10.1006/bbrc.1993.1780. [DOI] [PubMed] [Google Scholar]
- Telford WG, King LE, Fraker PJ. Evaluation of glucocorticoid-induced DNA fragmentation in mouse thymocytes by flow-cytometry. Cell Prolif. 1991;24:447–459. doi: 10.1111/j.1365-2184.1991.tb01173.x. [DOI] [PubMed] [Google Scholar]
- Telford WG, King LE, Fraker PJ. Comparative evaluation of several DNA binding dyes in the detection of apoptosis-associated chromatin degradation by flow cytometry. Cytometry. 1992;13:137–143. doi: 10.1002/cyto.990130205. [DOI] [PubMed] [Google Scholar]
- Umansky SR, Korol BR, Nelipaovich PA. In vivo DNA degradation in the thymocytes of gamma-irradiated or hydrocortisone-treated rats. Biochim Biophys Acta. 1981;655:281–290. doi: 10.1016/0005-2787(81)90060-5. [DOI] [PubMed] [Google Scholar]
- Wang G, Gong Y, Burczynski FJ, Hasinoff BB. Cell lysis with dimethyl sulphoxide produces stable homogeneous solutions in the dichlorofluorescein oxidative stress assay. Free Radic Res. 2008;42:435–441. doi: 10.1080/10715760802074462. [DOI] [PubMed] [Google Scholar]
- Willingham MC. Cytochemical methods for the detection of apoptosis. J Histochem Cytochem. 1999;47:1101–1109. doi: 10.1177/002215549904700901. [DOI] [PubMed] [Google Scholar]
- Zhuang Z, Fei F, Chen Y, Jin W. Suberoyl bis-hydroxamic acid induces p53-dependent apoptosis of MCF-7 breast cancer cells. Acta Pharmacol Sin. 2008;29:1459–1466. doi: 10.1111/j.1745-7254.2008.00906.x. [DOI] [PubMed] [Google Scholar]