Abstract
Staphylococcus aureus possesses three MsrA enzymes (MsrA1, MsrA2, MsrA3) that reduce the S-epimer of methionine sulfoxide (MetO) and an MsrB enzyme that reduces R-MetO. The four msr genes are expressed from three different promoters. The msrA1/msrB genes are coexpressed. To determine the expression pattern of msr genes, three independent reporter strains were constructed where msr promoter was cloned in front of a promoterless lacZ and the resulting construct was integrated in the chromosome. Using these strains, it was determined that the msrA1/B expression is significantly higher in S. aureus compared to msrA2 or msrA3. Expression of msrA1/B was highest during stationary phase growth, but the expression of msrA2 and msrA3 was highest during the early to midexponential growth phase. Expression of msrA1/B was induced by oxacillin and the expression of msrA3 was upregulated by salt. Expression of msrA2 remained unchanged under all tested conditions.
1. Introduction
Staphylococcus aureus is a versatile and aggressive pathogen responsible for causing a wide array of diseases ranging from mild skin infections such as folliculitis and carbuncles to life-threatening conditions such as bacteremia, pneumonia, and endocarditis [1–3]. In response to S. aureus invasion, the host immune system recruits neutrophils and macrophages that trigger the release of highly reactive oxygen species such as hydrogen peroxide, hydroxyl radical, singlet oxygen, and hypochlorous acid. These highly reactive species lead to the oxidation of DNA, lipids, and proteins [4].
In proteins, oxidative damage usually leads to a loss of protein function that disturbs cellular processes and metabolism [5, 6]. Such oxidative damage includes oxidation of the sulfur atom of methionine producing methionine sulfoxide. Oxidation of methionine results in two diastereomic forms of MetO (R-MetO and S-MetO). These two stereoisomeric MetO products are reduced by two different kinds of Msr enzymes—MsrA and MsrB. MsrA specifically reduces S-MetO, whereas MsrB specifically reduces R-MetO [7–9]. The MsrA and MsrB proteins share no homology at the primary sequence or structural levels. Orthologs of msrA and msrB are present in most organisms [10, 11]. In bacterial species, the genetic organization of msrA and msrB shows great variation. In numerous cases, msrA and msrB are transcribed as independent units and are located in different regions of the chromosome [12]. However, in many bacterial species, these two genes are located adjacent to each other and are cotranscribed [12–14]. In a few cases like Neisseria, msrA and msrB are transcriptionally fused [12, 15–17]. The copy number of msrA and msrB orthologs also varies widely in bacterial species. For example, Escherichia coli contains one copy each of msrA and msrB; S. aureus, 3 msrA and 1 msrB; Vibrio cholerae, 2 msrA and 3 msrB; all present in the chromosome [12, 15, 16]. Rhizobium meliloti possesses 3 msrA and 3 msrB genes and one of each is located on a plasmid [12]. Genetic redundancy is considered a strategy where organisms express specific genes under specific environments [12]. Additionally, MsrA and MsrB proteins are some of the most conserved proteins across prokaryotic and eukaryotic organisms suggesting important cellular functions [11, 12, 18]. In many studies, bacterial species deficient in Msr proteins have been shown to be sensitive to oxidative stress and in the cases of many pathogenic bacteria, their Msr knockout derivatives were shown to be attenuated in virulence [12, 15–17, 19–22].
Previously, in proteomic studies, upon exposure of actively growing S. aureus cells to oxacillin (a cell wall-active antibiotic), MsrA1 and MsrB proteins were observed to be produced in elevated amounts [23]. Subsequent gene fusion, Northern analysis, and transcriptional profiling experiments demonstrated an increased expression of msrA1 and msrB genes in the presence of oxacillin as well as several other cell wall-active antibiotics such as cephalothin, D-cycloserine, and bacitracin [24, 25].
It was speculated that the expression of different msr genes in S. aureus are regulated differently under different growth conditions as part of better survival strategy. Three independent reporter strains were constructed to test this assumption. In these reporter strains, the msrA1/B, msrA2, and msrA3 promoters were cloned in front of a promoterless lacZ gene and the resulting constructs were introduced into the chromosome of the S. aureus strain SH1000. Findings of this study suggest that the msr gene loci are differentially regulated in S. aureus that may have important physiological significances.
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Antibiotics, and Growth Conditions
The bacterial strains and plasmids used in this study are shown in Table 1. S. aureus cells were grown in tryptic soy broth (TSB) or tryptic soy agar (TSA), and E. coli cells were grown in Luria-Bertani broth (LB) or Luria-Bertani agar (LBA). Plasmids in E. coli cells were maintained by adding ampicillin at 100 μg mL−1 and erythromycin at 15 μg mL−1, when required. Overnight cultures of S. aureus msr reporter strains were prepared in the presence of erythromycin at 10 μg mL−1.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Characteristics | Reference |
|---|---|---|
| Strains | ||
| S. aureus SH1000 | S. aureus strain 8325-4 with functional RsbU | [41] |
| S. aureus RN4220 | A restriction minus derivative of S. aureus 8325-4 | [42] |
| VKS1009 | SH1000 with msrA1/B-lacZ integration (Ermr) | This study |
| VKS1010 | SH1000 with msrA2-lacZ integration (Ermr) | This study |
| VKS1011 | SH1000 with msrA3-lacZ integration (Ermr) | This study |
| E. coli JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1thi Δ(lac-proAB) F′(traD36proAB+laclqΔM15) | [43] |
|
| ||
| Plasmids | ||
| pGEMT | Cloning vector for E. coli (Ampr) | Promega |
| pAZ106 | lacZ fusion vector (Ampr, E. coli; Ermr, S. aureus) | [27] |
| pGEMT-msrA2P | pGEMT containing the msrA2 promoter fragment | This study |
| pGEMT-msrA3P | pGEMT containing the msrA3 promoter fragment | This study |
| pAZ-msrA2P | pAZ106 containing msrA2 promoter-lacZ fusion | This study |
| pAZ-msrA3P | pAZ106 containing msrA3 promoter-lacZ fusion | This study |
Ermr: erythromycin resistant, Ampr: ampicillin resistant.
2.2. DNA Manipulations
Plasmid DNA was isolated using the QIAprep Miniprep kit (Qiagen). All restriction and modification enzymes were purchased from Promega (Promega). PCR was performed using a Peltier Thermal Cycler-200 system (MJ research). DNA manipulations were carried out as described [26]. Oligonucleotide primers were obtained from Sigma Genosys.
2.3. Construction of an msrA1/B Promoter-lacZ Reporter Strain in S. aureus Strain SH1000
An msrA1/B promoter-lacZ reporter constructed previously in S. aureus strain RN450 [13, 23] was transferred to S. aureus strain SH1000 using a phage transduction procedure as described previously [13, 23] and the resulting construct was verified by PCR.
2.4. Construction of an msrA2 Promoter-lacZ Reporter Strain in S. aureus Strain SH1000
Primers MsrA2P-1 (5′-TCTAGACAAGCAATTCACGTTG-3′) and MsrA2P-2 (5′-GAATTCCTTTCATTAGACCTTAG-3′) were used to amplify a 1281 bp DNA fragment using genomic DNA from S. aureus SH1000 as template. This amplicon represents the upstream and 8 nt of the 5′-end of the msrA2 gene. The amplicon was cloned in the correct orientation upstream of a promoterless lacZ gene of vector pAZ106 [27] and was introduced into the chromosome of S. aureus RN4220 by electroporation with selection on erythromycin. Phage 80α lysate of the resulting transformant was used to transduce the msrA2 promoter-lacZ fusion into strain S. aureus SH1000. A single copy integration of the msrA2P-lacZ in the chromosome was confirmed by Southern blot analysis.
2.5. Construction of an msrA3 Promoter-lacZ Reporter Strain in S. aureus Strain SH1000
To construct the msrA3 reporter strain, two primers MsrA3P-1 (5′-GATCCAGCGACACCTCATCATTTGC-3′) and MsrA3P-2 (5′-GAATTCACCCTCCTGCTACATAAAC-3′) and genomic DNA from S. aureus strain SH1000 as template were used to PCR amplify a 1427 bp DNA fragment. The amplicon represents the upstream and 39 nt of the 5′-end of the msrA3 gene. The amplicon was cloned in the correct orientation upstream of the promoterless lacZ gene of vector pAZ106, introduced into the chromosome of S. aureus RN4220 by electroporation, and subsequently into strain S. aureus SH1000 using a phage transduction procedure. A single copy integration of the msrA3P-lacZ in the chromosome was confirmed by Southern blot analysis.
2.6. Growth Kinetics of msr Reporter Strains and Expression of msr Genes in S. aureus
Overnight cultures of msr(A1/B)P-lacZ, msrA2P-lacZ, and msrA3P-lacZ reporter constructs in S. aureus strain SH1000 were diluted 100-fold in fresh TSB with a flask-to-medium volume ratio of 6 : 1 and grown at 37°C with aeration at 220 rpm. Growth of these cultures was recorded by measuring OD600 every 30 min. The expression of individual msr gene locus was determined in these reporter constructs at different time points by assaying β-galactosidase using O-nitrophenyl-β-D-galactopyranoside (ONPG) as the substrate as described previously [13, 23].
2.7. Expression of msr Genes in S. aureus under Stress Conditions
Overnight cultures of msr(A1/B)P-lacZ, msrA2P-lacZ, and msrA3P-lacZ reporter constructs were diluted 100-fold in fresh TSB and allowed to grow at 37°C with aeration and shaking. At OD600 = 0.3, cells from 10.0 mL of culture were harvested and resuspended in 10.0 mL fresh TSB or TSB modified to impose a variety of different stress conditions. Antibiotic stress used oxacillin at 1.2 μg mL−1; oxidative stress, H2O2 at 15 mM; alkaline stress, TSB at pH 9.0; acidic stress, TSB at pH 5.0; osmotic stress, TSB supplemented with 1.5 M NaCl. Cells were allowed to grow for 1 h. Subsequently, the bacterial cells were harvested and the β-galactosidase activity was measured. The msr reporter constructs pre-grown to OD600 = 0.3 were also exposed for 1 h to following chemical agents: diamide (5 mM), N-ethylmaleimide (0.05 mM), methyl viologen (paraquat, 20 mM), menadione (0.05 mM), cumene hydroperoxide (0.0125%), and sodium nitroprusside (5 mM). The bacterial cells were subsequently used to determine β-galactosidase activity.
2.8. Statistical Analysis
All results are reported as the mean ± SD of at least three trials. Data were analyzed with Dunnett's Method in one-way analysis of variance or with Student-Newman-Keuls Method in two-way analysis of variance using a statistical analysis computer programs (SigmaPlot for Windows, version 11.0, Systat Software, Inc.). Statistical significance was set at P < 0.05.
3. Results
3.1. Growth Kinetics of msr(A1/B)P-lacZ, msrA2P-lacZ, and msrA3P-lacZ Reporter Constructs in S. aureus
Fusion of individual msrA1/B, msrA2, and msrA3 promoters with the promoterless lacZ gene and their subsequent integration into S. aureus chromosome was verified by PCR and Southern blot analysis (data not shown). Subsequently, the growth rates of the above constructed reporter strains were analyzed to see if this promoter-lacZ integration in the staphylococcal chromosome caused any impact on growth. The results showed that the S. aureus msrA2 and msrA3 reporter strains grew almost at the same rate, whereas the msrA1/B reporter strain demonstrated a slightly slower growth rate (Figure 1).
Figure 1.
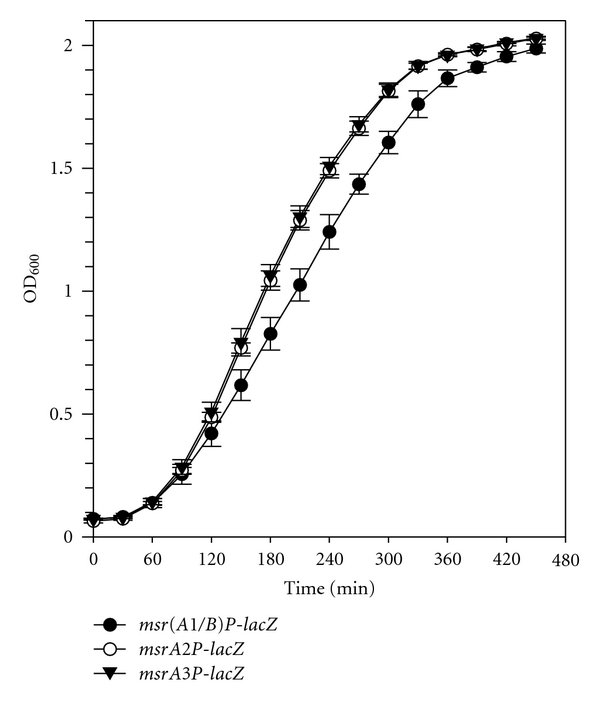
Growth comparison of the msr(A1/B)P-lacZ, msrA2P-lacZ, and msrA3P-lacZ reporter constructs in S. aureus strain SH1000. Growth was measured by recording OD600 periodically. Values indicate averages of data from three independent experiments ± standard deviation (SD). The msr(A1/B)P-lacZ, msrA2P-lacZ, and msrA3P-lacZ reporter strains are represented by closed circles, open circles, and closed triangles, respectively.
3.2. Expression of msr Genes during Various Growth Stages in S. aureus
Bacterial cells from the cultures of msr(A1/B)P-lacZ, msrA2P-lacZ, and msrA3P-lacZ reporter strains were collected during different stages of growth to investigate if the expression of these genes is growth phase dependent. In these studies, the expression of the msrA1/B gene locus was low during the early- and mid-exponential growth phases, but was significantly higher during the late exponential and stationary growth phases (Figure 2(a)). Expression of msrA2 and msrA3 genes, on the other hand, was more pronounced during the early- and mid-exponential phases of growth and was much lower during the stationary growth phase (Figures 2(b) and 2(c)). These experiments also showed that the msrA2 and msrA3 genes are expressed at significantly lower levels compared to the expression of the msrA1/B locus at all stages of growth (Figures 2(a), 2(b), and 2(c)).
Figure 2.
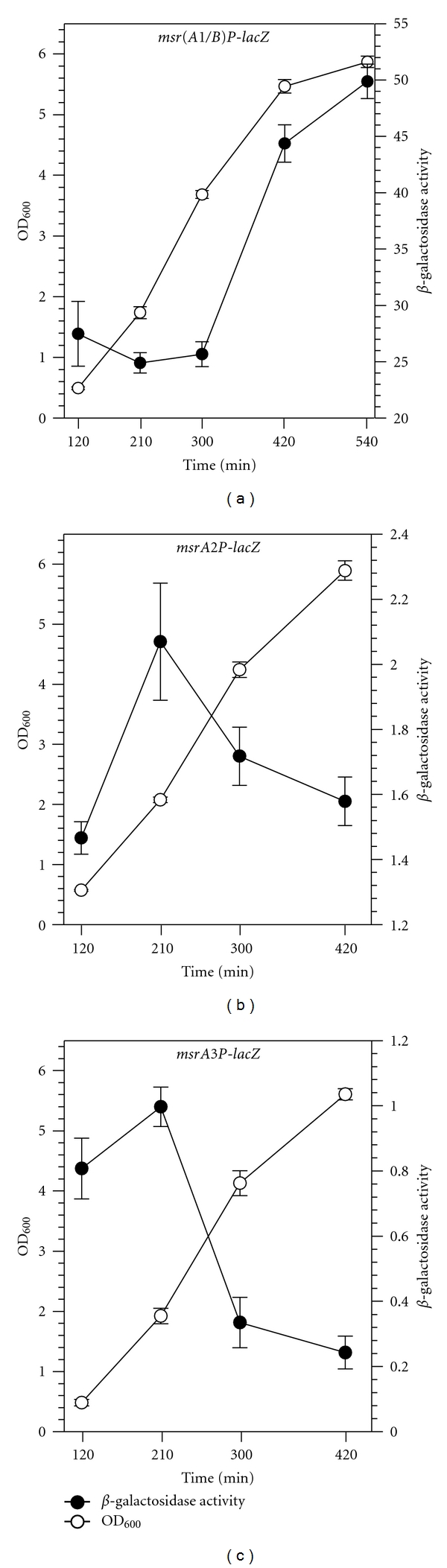
β-galactosidase activity levels in msr(A1/B)P-lacZ (a), msrA2P-lacZ (b), and msrA3P-lacZ (c) reporter strains during different stages of growth under standard growth conditions. Growth and β-galactosidase activity were measured at different time points spectrophotometrically. For precise OD600 determination, the late-stage cultures were diluted appropriately to bring cell density in measurable range of the spectrophotometer. OD600 is indicated by open circles, and β-galactosidase activity (OD420) is indicated by closed circles. Values indicate averages of data from three independent experiments ± standard deviation (SD).
3.3. Expression of S. aureus msr Genes under Stress Conditions
Expression of msr genes in S. aureus was investigated under various stress conditions. In these studies, the expression of msrA1/B gene locus was significantly increased (~6.5-fold) (Figure 3(a)) in the presence of oxacillin, an observation consistent with prior findings [23–25]. No significant change in the expression of msrA1/B gene locus was observed under oxidative, alkaline, acidic, or osmotic stress conditions (Figure 3(a)). None of these stress conditions caused any increase in msrA2 expression (Figure 3(b)). Expression of msrA2, in fact, was significantly repressed under acidic pH (Figure 3(b)). Studies utilizing msrA3 reporter strains demonstrated an approximately 5.5-fold increase in msrA3 expression under osmotic stress. Other stress conditions did not significantly affect msrA3 expression in S. aureus (Figure 3(c)).
Figure 3.
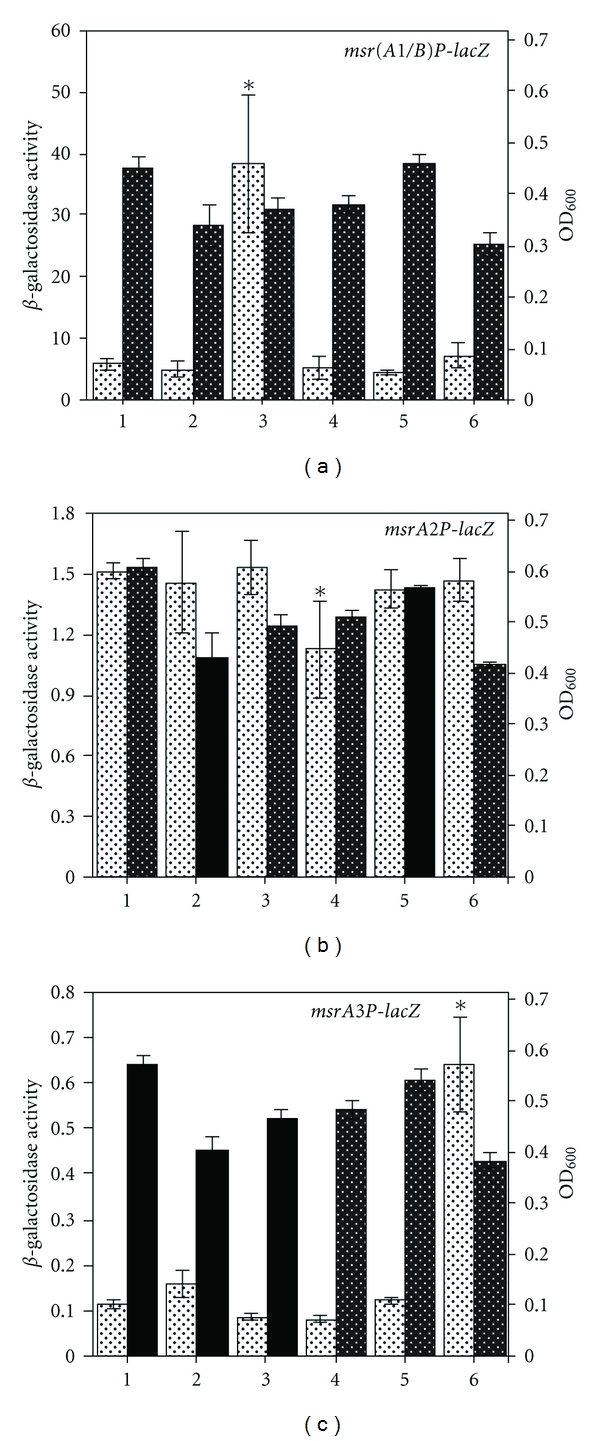
Expression of the msr(A1/B), msrA2, and msrA3 loci in S. aureus SH1000 under different environmental stress conditions. Cultures of S. aureus SH1000 msr(A1/B)P-lacZ, msrA2P-lacZ, and msrA3P-lacZ reporter strains were grown to OD600 of 0.3 at 37°C and treated separately with H2O2 (15 mM) (2), oxacillin (1.2 μg/mL) (3), pH 5.0 (4), pH 9.0 (5), and TSB with 1.5 M added NaCl (6) for 1 h. β-galactosidase activity (lighter bar) and growth (OD600) (darker bar) were subsequently determined. β-galactosidase activity and growth of cells in TSB control are represented in bars 1. Values indicate averages of data from three independent experiments ± standard deviation (SD).
3.4. Expression of S. aureus msr Genes under Chemically Induced Oxidative Stress Conditions
Expression of msr genes was determined in actively growing S. aureus cells that were exposed for 1 h to various chemicals to induce oxidative stress. The concentration utilized was selected for their ability to show relatively slower growth of treated cultures compared to untreated culture. In these experiments, contrary to the expectations, chemically induced oxidative stress did not induce the expression of any of the msr genes (Figures 4(a), 4(b), and 4(c)). Many of these chemicals, most notably, cumene hydroperoxide, methyl viologen, diamide, and NEM, repressed the expression of the msr genes to a significant level (Figures 4(a), 4(b), and 4(c)).
Figure 4.
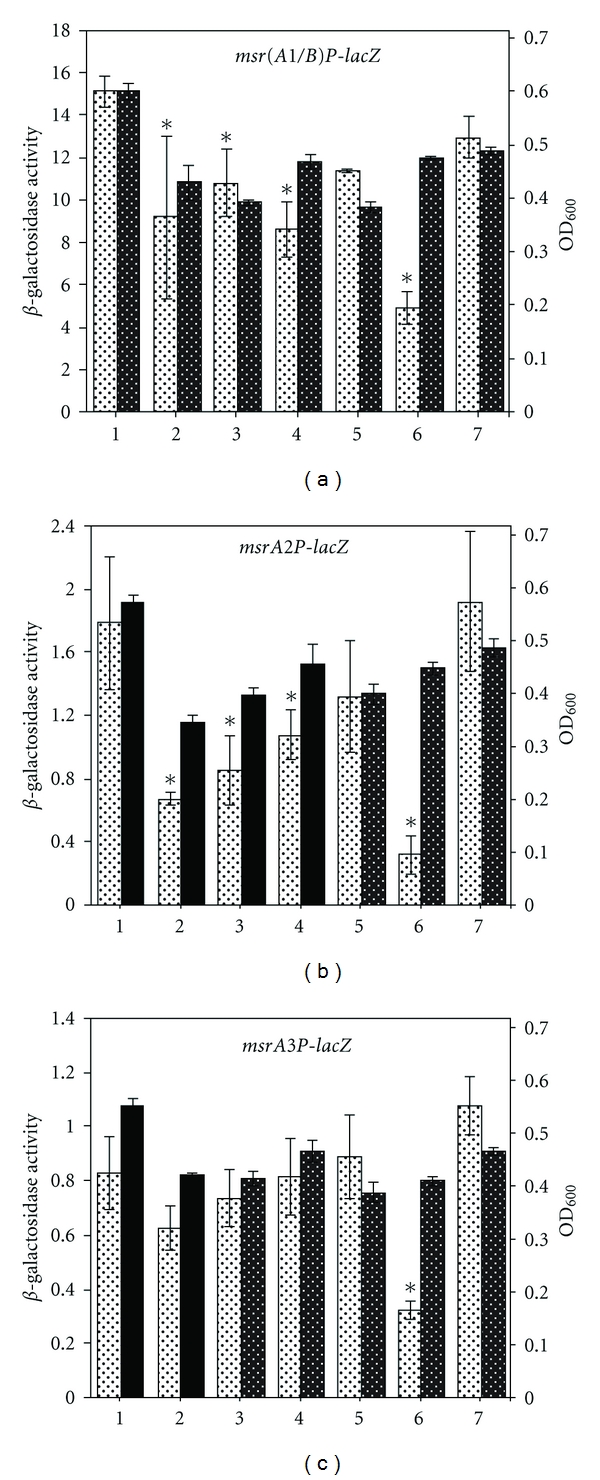
Expression of the msr(A1/B), msrA2, and msrA3 loci in S. aureus SH1000 in the presence of different oxidizing chemical agents. Cultures grown to OD600 of 0.3 at 37°C were treated separately with the following stresses for 1 h: diamide (5 mM) (2), N-ethylmaleimide (NEM) (0.05 mM) (3), methyl viologen (MV) (20 mM) (4), menadione (MD) (0.05 mM) (5), cumene peroxide (CuOOH) (0.0125%) (6), and sodium nitroprusside (SNP) (5 mM) (7). β-galactosidase activity (lighter bar) and growth (OD600) (darker bar) were subsequently determined. β-galactosidase activity and growth of cells in TSB control are represented in bars 1. Values indicate averages of data from three independent experiments ± standard deviation (SD).
4. Discussion
Survival of S. aureus under various environmental stresses is a key determinant of its pathogenicity. During colonization and invasion of a host, staphylococci are continuously exposed to toxic conditions. Following S. aureus invasion, the host responds by recruitment of polymorphonuclear leukocytes and macrophages to the site of infection so that they can ingest the staphylococci. Uptake of bacteria triggers oxygen-dependent microbicidal pathways in the phagocytic cells that generate reactive oxygen species (ROS) such as hydrogen peroxide, hydroxyl radicals, singlet oxygen, and hypochlorous acid [4]. Degradation of phagocytosed bacterial cells in lysosomes is also facilitated by its acidic environment [28]. To defend itself against the oxidative stress of ROS from neutrophils, S. aureus has several strategies in place that enables it to successfully colonize and survive in the host. S. aureus produces antioxidant enzymes such as superoxide dismutases that convert superoxide anion to hydrogen peroxide, catalase that converts hydrogen peroxide to water and oxygen, alkyl hydroperoxide reductases that detoxify hydrogen peroxide, peroxynitrites and hydroperoxides, and the carotenoid pigment staphyloxanthin that is also involved in the detoxification of ROS [29, 30]. In addition, S. aureus also contains methionine sulfoxide reductase enzyme system, which has been shown to be protective from oxidative stress [28].
S. aureus produces four Msr enzymes. In this study, we examined the strengths of the three msr promoters: msrA1/B promoter that drives the transcription of msrA1 and msrB genes, msrA2 promoter that drives the transcription of msrA2 gene, and msrA3 promoter that drives the transcription of msrA3 gene. β-galactosidase activity analysis of msr reporter strains revealed that the expression of msr in S. aureus is growth phase dependent. The expression of msrA1/B locus is highest during the stationary phase of growth, whereas the expression of msrA2 and msrA3 was higher during the early-to-mid exponential growth phase. Similar stationary-phase-induced expression of msr genes has been documented in E. coli [20], Helicobacter pylori [19], and Xanthomonas campestris pv. phaseoli [31]. Overall, the expression of the msrA1/B locus in S. aureus was observed to be much higher compared to the msrA2 and msrA3 genes. During exponential growth phase, high levels of antioxidant enzymes minimize the intracellular accumulation of oxidants [31], thus offering a likely explanation for lower expression of the more active msrA1/B locus during this stage of growth in S. aureus. During the stationary phase, nutrient limitation, accumulation of toxic metabolic byproducts, such as ROS, and decreased activity of antioxidant enzymes, such as catalase and superoxide dismutase, increase the likelihood of oxidative damage to cells [31, 32]. The increased expression of msrA1/B would serve to alleviate the oxidant-induced damage during the stationary phase. This phenomenon is similar to the induction of MsrA in stationary phase observed in E. coli [20].
The expression of msr genes was also studied under a variety of stress conditions. This study showed that oxacillin and osmotic stress induced the expression of msrA1/B gene locus and msrA3 expression, respectively. Besides oxacillin (msrA1/B) and salt (msrA3), other stress conditions tested in this study had no impact on the expression of msr genes. The increased expression of msrA1/B in response to oxacillin has previously been demonstrated in S. aureus [23–25]. S. aureus is one of the most osmotolerant pathogens capable of growing in medium containing upto 3.5 M NaCl [33]. Osmotic stress results in the shrinkage and decreased turgor pressure in bacterial cells. In response, bacteria restore turgor pressure by accumulating osmoprotective solutes, such as glycine betaine, choline, proline, and taurine. It has been previously shown that an exposure of exponentially growing S. aureus cells to 2.5 M NaCl significantly increased cell size, and the normal cell size was subsequently restored by the addition of glycine betaine. In addition, muropeptide analysis revealed significant alteration in the morphological structure of cell wall in the presence of NaCl [34]. Under normal conditions, peptidoglycan layers of S. aureus cell wall are cross-linked via pentaglycine bridges to provide strong structural framework [3]. However, the cells exposed to NaCl exhibited altered glycine content in the pentaglycine and reduced cross-linking. These structural abnormalities were corrected by glycine betaine [34]. It is, therefore, plausible that the increased expression of msrA3 under osmotic stress may be related to maintaining cell wall integrity in S. aureus. This response would seem to be analogous to the expression of msrA1/B in the presence of cell wall-active antibiotics. However, more work needs to be done to fully understand the significance of msrA3 and its induced expression under osmotic stress in S. aureus.
Induced expression of msr genes has been observed in many bacteria under various stress conditions. In E. coli, depletion of glucose or nitrogen in the growth media led to a three- to four-fold increase in MsrA activity [20]. Cells exposed to peroxide, peroxynitrite, or dipyridyl (iron-chelator) stress showed a 3-fold msr induction in H. pylori [19]. Various oxidizing chemicals such as menadione (10-fold), tert-butyl hydroperoxide (6-fold), H2O2 (3-fold), and N-ethylmaleimide (2-fold) induced msrA expression in Xanthomonas campestris pv. phaseoli [31]. In Streptococcus gordonii, an increase in pH (6.2 to 7.3) induced msrA expression [35]. Chemical stress of phenol or chlorophenol induced msrA expression by 4-fold and 5-fold, respectively, in the soil bacterium Ochrobactrum anthropic [22]. In Bacillis subtilis, paraquat, a superoxide generating chemical, induced msrA expression by 3.5-fold [36].
The lack of overall msr induction in S. aureus under oxidative stress was surprising considering that such conditions have been shown to induce the expression of msr genes in other organisms. However, it has been speculated that even if msr genes are not induced in response to oxidative stress in some species, these gene products are still required to ensure appropriate survival under stress [12]. This was illustrated in E. coli, where the oxidizing agents, H2O2 and paraquat, failed to induce msrA expression. However, disk-inhibition studies on solid medium revealed significantly increased growth inhibition of msrA mutants in response to H2O2 [20]. Similarly, a mutation in msrA1 rendered S. aureus more susceptible to H2O2 stress, but no induction of msrA1/B was noticed on exposure to H2O2 [13, 16].
In S. aureus, the basal level of msr expression is probably sufficient to protect the cells from oxidative damage. Alternatively, other stress responsive genes may be able to respond more efficiently to the stress conditions tested in this study, thus bypassing the need for an induction of the msr genes. In microarray experiments, at least 25 stress-related genes were upregulated in S. aureus upon exposure to ROS. Some of these genes encode enzymes such as catalase, thioredoxin, thioredoxin reductase, superoxide dismutase, alkyl hydroperoxide reductase, and glutathione peroxidase [37]. Nitric oxide produced from sodium nitroprusside reacts with oxygen or superoxide to generate reactive nitrogen species that attack thiols, metal centers, and macromolecules. Proteomic analysis showed that in response to nitric oxide stress in S. aureus, a total of 35 proteins were synthesized in elevated amounts [38]. Another study showed a differential regulation of 638 staphylococcal genes in response to nitrosative stress caused by sodium nitrite [39]. Transcriptomic analysis of S. aureus in response to hydrogen peroxide-induced oxidative stress revealed differential expression of 343 genes after 10 min and 20 min exposure [40]. Altogether, these results suggest that the induction of additional oxidative stress response genes prevents ROS-induced damage in S. aureus.
In summary, the findings of this study suggest that the expression of the msrA1/B locus is highest during the stationary growth phase while the expression of msrA2 and msrA3 is highest during the early- to mid-exponential phases of growth. The msrA1/B locus is under the control of a more powerful promoter compared to msrA2 and msrA3 gene promoters. The expression of msrA1/B locus is significantly induced by oxacillin, while the expression of msrA3 is significantly increased in response to osmotic stress. As the oxidative stress conditions did not affect msr gene expression, it would be of interest in the future to see if S. aureus msr mutants (msrA1, msrB, msrA1:msrB, msrA2, msrA3, a complete msrA, or a complete msr mutant) show any differential sensitivity to oxidative stress or other stress conditions, which is currently under investigation.
Acknowledgments
The authors thank Jane C. Johnson for her valuable assistance with statistical analysis. This work was supported in part by a Warner/Fermaturo & ATSU Board of Trustees Research Funds and grant 1R15AI090680-01 from the National Institutes of Health to Vineet K. Singh and a Grant from KCOM Biomedical Sciences Graduate Program to Kuldeep Singh.
References
- 1.Bamberger DM, Boyd SE. Management of Staphylococcus aureus infections. American Family Physician. 2005;72(12):2474–2481. [PubMed] [Google Scholar]
- 2.Lowy FD. Medical progress: staphylococcus aureus infections. New England Journal of Medicine. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Murray P, Rosenthal KS, Pfaller M. Medical Microbiology. 5th edition. Philadelphia, Pa, USA: Elsevier Mosby; 2002. [Google Scholar]
- 4.Coico R, Sunshine G. Immunology: A Short Course. 6th edition. Hoboken, NJ, USA: Wiley-Blackwell; 2009. [Google Scholar]
- 5.Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. International Microbiology. 2000;3(1):3–8. [PubMed] [Google Scholar]
- 6.Shacter E. Quantification and significance of protein oxidation in biological samples. Drug Metabolism Reviews. 2000;32(3-4):307–326. doi: 10.1081/dmr-100102336. [DOI] [PubMed] [Google Scholar]
- 7.Grimaud R, Ezraty B, Mitchell JK, et al. Repair of oxidized proteins: identification of a new methionine sulfoxide reductase. Journal of Biological Chemistry. 2001;276(52):48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 8.Moskovitz J, Poston JM, Berlett BS, Nosworthy NJ, Szczepanowski R, Stadtman ER. Identification and characterization of a putative active site for peptide methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. Journal of Biological Chemistry. 2000;275(19):14167–14172. doi: 10.1074/jbc.275.19.14167. [DOI] [PubMed] [Google Scholar]
- 9.Moskovitz J, Singh VK, Requena J, Wilkinson BJ, Jayaswal RK, Stadtman ER. Purification and characterization of methionine sulfoxide reductases from mouse and Staphylococcus aureus and their substrate stereospecificity. Biochemical and Biophysical Research Communications. 2002;290(1):62–65. doi: 10.1006/bbrc.2001.6171. [DOI] [PubMed] [Google Scholar]
- 10.Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(7):4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodrigo MJ, Moskovitz J, Salamini F, Bartels D. Reverse genetic approaches in plants and yeast suggest a role for novel, evolutionarily conserved, selenoprotein-related genes in oxidative stress defense. Molecular Genetics and Genomics. 2002;267(5):613–621. doi: 10.1007/s00438-002-0692-3. [DOI] [PubMed] [Google Scholar]
- 12.Ezraty B, Aussel L, Barras F. Methionine sulfoxide reductases in prokaryotes. Biochimica et Biophysica Acta. 2005;1703(2):221–229. doi: 10.1016/j.bbapap.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Singh VK, Moskovitz J, Wilkinson BJ, Jayaswal RK. Molecular characterization of a chromosomal locus in Staphylococcus aureus that contributes to oxidative defence and is highly induced by the cell-wall-active antibiotic oxacillin. Microbiology. 2001;147(11):3037–3045. doi: 10.1099/00221287-147-11-3037. [DOI] [PubMed] [Google Scholar]
- 14.Hayes CS, Illades-Aguiar B, Casillas-Martinez L, Setlow P. In vitro and in vivo oxidation of methionine residues in small, acid- soluble spore proteins from Bacillus species. Journal of Bacteriology. 1998;180(10):2694–2700. doi: 10.1128/jb.180.10.2694-2700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olry A, Boschi-Muller S, Marraud M, Sanglier-Cianferani S, Van Dorsselear A, Branlant G. Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis. Journal of Biological Chemistry. 2002;277(14):12016–12022. doi: 10.1074/jbc.M112350200. [DOI] [PubMed] [Google Scholar]
- 16.Singh VK, Moskovitz J. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology. 2003;149(10):2739–2747. doi: 10.1099/mic.0.26442-0. [DOI] [PubMed] [Google Scholar]
- 17.Wizemann TM, Moskovitz J, Pearce BJ, et al. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(15):7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moskovitz J, Weissbach H, Brot N. Cloning and expression of a mammalian gene involved in the reduction of methionine sulfoxide residues in proteins. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(5):2095–2099. doi: 10.1073/pnas.93.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alamuri P, Maier RJ. Methionine sulfoxide reductase in Helicobacter pylori: interaction with methionine-rich proteins and stress-induced expression. Journal of Bacteriology. 2006;188(16):5839–5850. doi: 10.1128/JB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moskovitz J, Rahman MA, Strassman J, et al. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. Journal of Bacteriology. 1995;177(3):502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasindran SJ, Saikolappan S, Dhandayuthapani S. Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiology. 2007;2(6):619–630. doi: 10.2217/17460913.2.6.619. [DOI] [PubMed] [Google Scholar]
- 22.Tamburro A, Allocati N, Masulli M, Rotilio D, Di Ilio C, Favaloro B. Bacterial peptide methionine sulphoxide reductase: co-induction with glutathione S-transferase during chemical stress conditions. Biochemical Journal. 2001;360(3):675–681. doi: 10.1042/0264-6021:3600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh VK, Jayaswal RK, Wilkinson BJ. Cell wall-active antibiotic induced proteins of Staphylococcus aureus identified using a proteomic approach. FEMS Microbiology Letters. 2001;199(1):79–84. doi: 10.1111/j.1574-6968.2001.tb10654.x. [DOI] [PubMed] [Google Scholar]
- 24.Pechous R, Ledala N, Wilkinson BJ, Jayaswal RK. Regulation of the expression of cell wall stress stimulon member gene msrA1 in methicillin-susceptible or -resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2004;48(8):3057–3063. doi: 10.1128/AAC.48.8.3057-3063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utaida S, Dunman PM, Macapagal D, et al. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology. 2003;149(10):2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor, NY, USA: C.S.H. Laboratory; 1989. [Google Scholar]
- 27.Chan PF, Foster SJ, Ingham E, Clements MO. The Staphylococcus aureus alternative sigma factor σ(B) controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. Journal of Bacteriology. 1998;180(23):6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hampton MB, Kettle AJ, Winterbourn CC. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood. 1998;92(9):3007–3017. [PubMed] [Google Scholar]
- 29.DeLeo FR, Diep BA, Otto M. Host defense and pathogenesis in Staphylococcus aureus infections. Infectious Disease Clinics of North America. 2009;23(1):17–34. doi: 10.1016/j.idc.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu GY. Molecular pathogenesis of Staphylococcus aureus infection. Pediatric Research. 2009;65(5):71R–77R. doi: 10.1203/PDR.0b013e31819dc44d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vattanaviboon P, Seeanukun C, Whangsuk W, Utamapongchai S, Mongkolsuk S. Important role for methionine sulfoxide reductase in the oxidative stress response of Xanthomonas campestris pv. phaseoli. Journal of Bacteriology. 2005;187(16):5831–5836. doi: 10.1128/JB.187.16.5831-5836.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatterjee I, Becker P, Grundmeier M, et al. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. Journal of Bacteriology. 2005;187(13):4488–4496. doi: 10.1128/JB.187.13.4488-4496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott WJ. Water relations of Staphylococcus aureus at 30°C. Australian Journal of Biological Sciences. 1953;6(4):549–564. [PubMed] [Google Scholar]
- 34.Vijaranakul U, Nadakavukaren MJ, De Jonge BLM, Wilkinson BJ, Jayaswal RK. Increased cell size and shortened peptidoglycan interpeptide bridge of NaCl-stressed Staphylococcus aureus and their reversal by glycine betaine. Journal of Bacteriology. 1995;177(17):5116–5121. doi: 10.1128/jb.177.17.5116-5121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vriesema AJM, Dankert J, Zaat SAJ. A shift from oral to blood pH is a stimulus for adaptive gene expression of Streptococcus gordonii CH1 and induces protection against oxidative stress and enhanced bacterial growth by expression of msrA. Infection and Immunity. 2000;68(3):1061–1068. doi: 10.1128/iai.68.3.1061-1068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You C, Sekowska A, Francetic O, Martin-Verstraete I, Wang Y, Danchin A. Spx mediates oxidative stress regulation of the methionine sulfoxide reductases operon in Bacillus subtilis. BMC Microbiology. 2008;8, article no. 128 doi: 10.1186/1471-2180-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voyich JM, Braughton KR, Sturdevant DE, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. Journal of Immunology. 2005;175(6):3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 38.Hochgräfe F, Wolf C, Fuchs S, et al. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. Journal of Bacteriology. 2008;190(14):4997–5008. doi: 10.1128/JB.01846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlag S, Nerz C, Birkenstock TA, Altenberend F, Götz F. Inhibition of staphylococcal biofilm formation by nitrite. Journal of Bacteriology. 2007;189(21):7911–7919. doi: 10.1128/JB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang W, Small DA, Toghrol F, Bentley WE. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. Journal of Bacteriology. 2006;188(4):1648–1659. doi: 10.1128/JB.188.4.1648-1659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. δb modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. Journal of Bacteriology. 2002;184(19):5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreiswirth BN, Lofdahl S, Betley MJ. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305(5936):709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 43.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


