Abstract
Cancer cells frequently exhibit deregulation of coregulatory molecules to drive the process of growth and metastasis. One such group of ubiquitously expressed coregulators is the metastasis-associated protein (MTA) family, a critical component of nucleosome remodeling and histone deacetylase (NuRD) complex. MTA1 occupies a special place in cancer biology due to its dual corepressor or coactivator nature and widespread overexpression in human cancers. Here, we highlight recent advances in our understanding of the vital roles of MTA1 on transformation, epithelial-mesenchymal transition, and on the functions of key cancer-relevant molecules as a nexus of multiple oncogenes and tumor suppressors. In addition to its paramount role in oncogenesis, we also reveal several new physiological functions of MTA1, related to DNA-damage, inflammatory responses and infection, in which MTA1 functions as a permissive “gatekeeper” for cancer-causing parasites. Further, these discoveries unraveled the versatile multidimensional modes of action of MTA1, which are independent of the NuRD complex and/or transcription. Given the emerging roles of MTA1 in DNA repair, inflammation, and parasitism, we discuss the possibility of MTA1 targeted therapy for use in not only combating cancer but also other inflammation and pathogen-driven pathological conditions.
Keywords: Master Coregulator, Cancer, DNA-damage Response, Inflammation, Pathogen-driven Cancers
Introduction
Cancer is a collection of cumulative phenotypic changes that are caused by dysregulated genetic and epigenetic processes including, aberrant transcription of genes that drive cell survival, proliferation, immunological functions, invasion, and metastasis (1). Gene transcription is a highly ordered but dynamic process that is tightly regulated by sequence-specific transcription factors (TFs) and associated coregulatory proteins (2). Coregulators control TF-dependent gene expression through direct binding to TFs as well as indirectly by interacting with histones and thereby regulating the accessibility of TFs to DNA, resulting in the stimulation or repression of the transcription of specific genes (3). Accordingly, coregulators that activate gene transcription are referred to as co-activators while those that repress it are known as co-repressors, and those with both functions, in a context-dependent manner, are dual coregulators.
The critical role of coregulators in cancer is evident from the fact that cancer cells often overexpress “growth coactivators” and hijack these molecules to drive proliferation and other processes that are characteristics of a cancer cell. One group of ubiquitously expressed coregulators is the metastasis-associated protein (MTA) family, comprising six different gene products (MTA1, MTA1s, MTA1-ZG29p, MTA2, MTA3, and MTA3L) that originate from three different genes (4–8). MTA1, the founding member of the MTA family (9), holds a special place among the family and among coregulators due to several reasons such as its ability to repress or stimulate transcription in a context-dependent manner, controlling the steady-state of proteins via affecting protein ubiquitination, contributing to the DNA damage response, and maintaining an overexpressed state in a wide variety of human epithelial and hematological malignancies (5–7). Here, we highlight recent advances in our understanding of the key roles of MTA1 in the NuRD-complex that happen in a transcription-dependent or -independent manner, in processes that govern oncogenesis, epithelial-mesenchymal transition, DNA damage response, inflammation, and pathogen-driven cancers (Fig. 1).
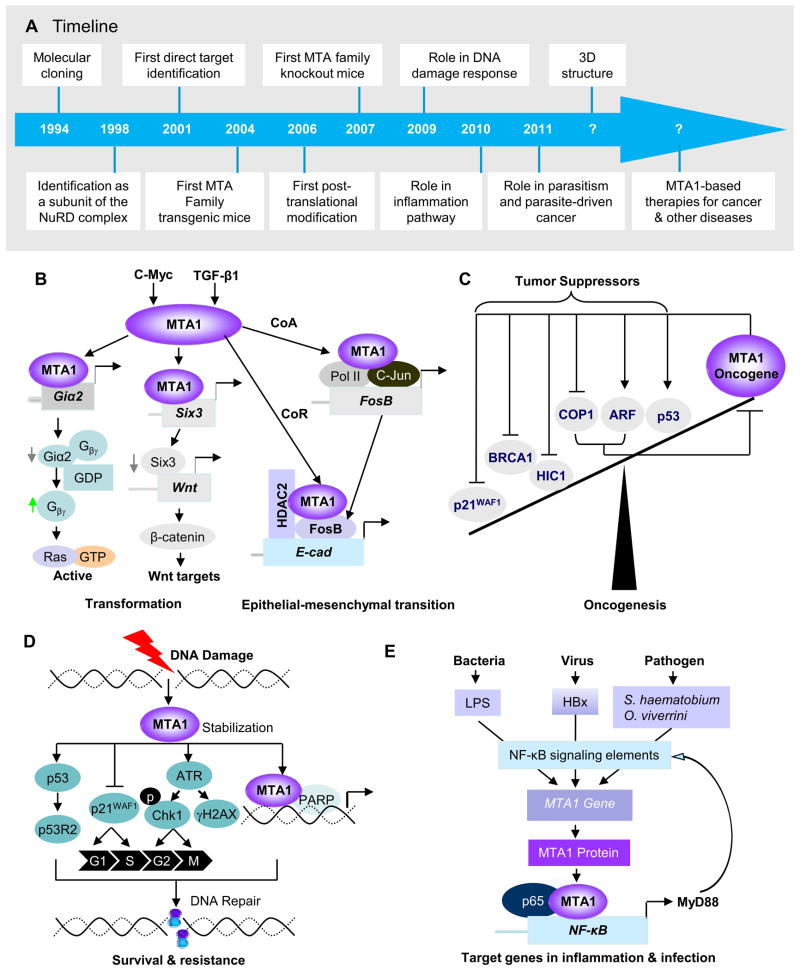
Figure 1.
Diverse functions of the MTA1/NuRD complex in cancer. (A) Timeline of major advance in the MTA1 research. (B) Role of MTA1 in the cell transformation and epithelial-mesenchymal transition through multiple signaling pathways. (C) Interaction of MTA1 with tumor suppressors. (D) Emerging role of MTA1 in DNA damage response through p53-depdnent and – independent mechanism. (E) MTA1 is a key molecular link between inflammation and cancer, and plays a critical role in infectious agent-driven cancer. CoA, co-activator; CoR, co-repressor.
Tracing the MTA1 Path to Cancer Progression
Despite the paramount importance of MTA1, its molecular functions remained a mystery until 1998 (Fig. 1A) when a proteomic analysis identified MTA1 as an integral component of the nucleosome remodeling and histone deacetylase (NuRD) complex (10), providing clues about possible functions of MTA1 in chromatin remodeling. However, its direct role in chromatin remodeling remained unknown until 2001 (Fig. 1A) when estrogen receptor-α (ERα) became the first direct target of MTA1 (11), establishing a direct link between MTA1 and the NuRD complex in the transcriptional repression of a specific gene. In addition, these findings also provided the first chromatin remodeling-based mechanism by which MTA1 overexpression may contribute to tamoxifen-resistant and aggressive phenotypes of human breast cancer by blocking the transactivation activity of ERα. Those early discoveries opened the door for a new era in elucidating the mechanistic role of MTA1 in cancer progression as well as its physiological functions. However, growing bodies of evidence have now firmly established that MTA1 functions are not limited to the MTA1/NuRD complex and/or its transcriptional effects as originally presumed.
Several studies over the past two decades have identified MTA1 as one of the most commonly overexpressed gene products in human cancers (Table S1). The mechanism involved has been proposed to be either transcriptional regulation or post-translational modifications, or both (see below). In addition, MTA1 is also detected in most normal mouse tissues, with significant levels present in brain, lung, ovary, mammary gland, and testis (11, 12). In general, MTA1 levels are upregulated in metastatic and/or aggressive cancer cell lines and tumors as compared to its levels in low metastatic cell lines and/or tumors (7–9), and are correlated with tumor progression and poor prognosis (Table S1). These observations are consistent with the original cloning of MTA1 as a differentially expressed gene from a rat metastatic mammary gland cell line (9). However, the hypothesis of a mechanistic role of MTA1 in cancer progression remained unsupported until 2001 when overexpression of MTA1 alone was able to confer an anchorage-independent phenotype to a non-invasive breast cancer cell line (11) while selective depletion of MTA1 by siRNA reduced the invasiveness (13).
The second set of observations that tied MTA1 overexpression a step closer to cancer, include the development of the first transgenic murine model expressing human MTA1 under the control of the mouse mammary tumor virus promoter (Fig. 1A) (14). It was observed that MTA1 overexpression could lead to the development of ductal hyperbranching and hyperplasic nodules in virgin glands, presumably due to the activation of anti-apoptotic B-cell lymphoma-extra large (Bcl-xl) and oncogenic cyclin D1 pathways, as well as the formation of mammary gland adeno-carcinomas in the older animals (14). This provided support to the notion of a potentially causative role of MTA1 in mimicking critical stages of tumor development in a physiologically relevant whole-animal model. Consistent with a hormone-independent mechanistic role of MTA1 (11), MTA1 overexpression also triggered hyperplasia in the mammary gland in both ovartazid female as well as male mice (14). Interestingly, most of the MTA1-MMTV-transgenic mice also developed widespread metastases to various organ systems over time (15). This provided the first experimental animal model that showed spontaneous metastasis driven by the MTA1-transgene. Surprisingly, most of these metastases were of lymphatic origin with diffuse large B-cell lymphomas (DLBCL), and accompanied by an upregulation of Pax5, a B-cell specific transcriptional factor (16) via MTA1/Pol II but not the NuRD complex, as was the case with the transcription of the BCAS3 coactivator (17). These findings were also relevant to human DLBCL because of a wide-spread co-overexpression of MTA1 and Pax5 in such specimens (16).
A definitive role of MTA1 in oncogenesis is also supported by the ability of MTA1 overexpression to transform the Rat1 fibroblasts by direct repression of Gi2alpha transcription owing to the corepressor activity of the MTA1/NuRD complex (Fig. 1B, left panel)(18). Since Gi2alpha negatively regulates the trimeric G-protein cycle, MTA1 repression of Gi2alpha resulted in a profound hyperstimulation of the Ras-Raf pathway(18). In fact, this study also found evidence of a close correlation between the levels of MTA1 and ERK1/2 activation, a downstream component of the Ras pathway, in a large cohort of breast tumors. Given the critical role of ERK in the Wnt/β-catenin pathway, which is aberrantly activated in human breast cancer (19), and the synergistically elevated levels of MTA1 and ERK activation, the hypothesis that MTA1 upregulation may contribute to canonical Wnt-signaling was formed. Furthermore, virgin mammary glands from MTA1 transgenic mice also show upregulation of two Wnt1 targets, β-catenin and cyclin D1. However, in spite of widely reported, increased Wnt1 expression in cancer, upstream modifiers that regulate the transcription of the Wnt1 gene in mammary epithelial cells are still not well-defined. In this context, two recent studies have revealed a definitive role of MTA1 in the stimulation of canonical Wnt1-signaling (Fig. 1B, middle panel)(20, 21). It was found that MTA1 stimulates the transcription of Wnt1 itself, and consequently, Wnt1 signaling, via repressing the Six3 homeodomain gene, which functions as a potent corepressor of Wnt1 transcription (20, 21). Consistent with a critical role of MTA1 in cancer progression, MTA1 is also essential for transformation by another well-studied oncogene, c-Myc- a target of a large number of signaling cascades (22). Experimental depletion of MTA1 severely compromises the ability of c-Myc to manifest its transforming activity (22). In brief, MTA1 represents and regulates nexuses of multiple oncogenes and qualifies to be labeled as a “regulator-of-regulators” of transformation (Fig. 1B).
Interaction of MTA1 with Tumor Suppressors
The process of oncogenesis is profoundly influenced by the status, activity, and upstream regulators of tumor suppressors which play a fundamental role in counteracting dysregulated cell proliferation. Since detectable levels of MTA1 are found in most normal cells, cancer progression and interaction with tumor suppressors, in general, occurs when the “fine-balance” and equilibrium of the level of MTA1 (i.e. overexpression beyond a threshold) is disrupted (Fig. 1C). Therefore, to understand the impact of MTA1 in cancer biology, it is essential to reveal its role in normal cells. In this context, emerging data suggest repression of the transcription of a tumor suppressor gene, breast and ovarian cancer susceptibility protein 1 (BRCA1) by MTA1 via physical recruitment of the MTA1/NuRD complex to an estrogen-responsive element in the BRCA1 promoter (23). It is noteworthy that the down-regulation or dysfunction of BRCA1 leads to accelerated tumor growth and metastasis (24) and therefore it remains possible that MTA1 inhibition of BRCA1 might contribute to the malignant transformation of normal cells.
The MTA1/NuRD complex was also identified as a transcriptional co-repressor of tumor suppressors, p21WAF1 (25) and hypermethylated in cancer 1 (HIC1)(26), both of which are direct targets of p53 (27, 28). The p21WAF1 protein promotes cell cycle arrest in response to many stimuli and functions as both a sensor and an effector of multiple anti-proliferative signals (29). HIC1 also play a role in the early stages of tumor progression, and HIC1 is shown to be hypermethylated and transcriptionally silent in several types of human cancer (30). MTA1 is reported to associate (31, 32) and deacetylate p53 (32, 33), and therefore it is also possible that MTA1 might indirectly modulate the levels of p53-regulated genes such as p21WAF1 and HIC1 by affecting the transcriptional activity of p53. However, several lines of experimental, genetic investigations from MTA1-knockout mice showed that p53 stabilization by MTA1 through competition with E3 ubiquitin ligase constitutive photomorphogenesis protein 1 (COP1) in p53 binding as well as destabilization of COP1 and murine double minute 2 (31) occurs in an MTA1/NuRD complex independent manner. In addition, the MTA1/NuRD complex is also known to directly repress the transcription of p21WAF1 in a p53-independent manner in the p53-null murine embryonic fibroblasts (MEFs) (25). Furthermore, a recent microarray-based analysis of p53-knockout MEFs overexpressing MTA1 uncovered a whole range of p53-independent MTA1 regulated genes(34), highlighting the significance of MTA1 in cancer biology in both a p53-independent and -dependent manner.
Apart from the aforementioned tumor suppressors, MTA1 is shown to regulate another p53-associated tumor suppressor gene, alternative reading frame (ARF) (also known as p14ARF in humans and p19ARF in mice), which plays an important protective role in oncogenic transformation and tumorigenicity (35). A recent study discovered MTA1 as a novel transcriptional coactivator of the ARF gene. This happens through the recruitment of the transcription factor c-Jun onto the ARF promoter in a p53-independent manner. Interestingly, ARF, in turn, negatively regulates MTA1 expression independent of p53 and c-Myc via both transcriptional and post-translational mechanisms (36). Thus, MTA1-mediated activation of ARF and ARF-mediated inhibition of MTA1 represent a p53-independent, bidirectional, autoregulatory mechanism in which these two opposites act in concert to regulate cell homeostasis and oncogenesis, depending on the cellular context and the environment (36). Given the putative growth suppressive roles of p53 and ARF, and the oncogenic activity of MTA1, it seems a paradox that MTA1 positively regulates both tumor suppressors (Fig. 1C). However, these findings indicate the “fail-safe” mechanism that is maintained in normal mammalian cells which safeguards the cells against oncogenic stimuli. Given the fact that the tumor suppressor p53 is mutated or deleted in more than 50% of human cancers (37), and the INK4a/ARF locus is found to be deleted or silenced in about 30% of all known types of malignancies (38), these genetic perturbations could help cancer cells override the normal mechanisms controlling cellular proliferation. Thus, it is a real possibility that the loss or silencing of p53 or ARF could contribute to MTA1-driven oncogenesis and tumor progression.
As discussed above, MTA1 has important functions in regulating gene transcription, but this might not be the only mechanism by which it contributes to oncogenesis. In this respect, MTA1 is shown to interact with several cancer-related proteins and these associations have also been implicated in oncogenesis. One such example is the tumor suppressor protein COP1, which functions as an E3 ubiquitin-protein ligase and targets protein substrates for ubiquitination and degradation. COP1 also acts as a tumor suppressor by negatively regulating the stability of the proto-oncogenes ETV1 (39) and c-Jun (40). A recent study found that MTA1 promotes auto-ubiquitination and degradation of COP1, revealing an additional level of MTA1 participation in oncogenesis by blocking the tumor suppressor activity of COP1 (41). Interestingly, COP1, in turn, targets MTA1 for ubiquitination and destruction by acting as an ubiquitin protein ligase (41). This finding partially explains why MTA1 is dominantly overexpressed in human cancer. Thus, both MTA1 and COP1 proteins form double vicious cycles, contributing to uncontrolled growth regulation. Similarly, the interaction between MTA1 and BRCA1 tumor suppressor which possesses an intrinsic E3 ubiquitin-protein ligase activity could also have parallel bidirectional auto regulation that might enable oncogenesis.
Dysregulated MTA1 Promotes Epithelial-Mesenchymal Transition
Since the overexpression but not normal levels of MTA1, is responsible for oncogenesis, an immediate compelling question about the upstream signals or factors that trigger MTA1 upregulation in human cancer is raised. A recent study identified transforming growth factor-β1 (TGF-β1) as a potent inducer of MTA1 transcription and expression as well as MTA1 as a downstream effector of TGF-β1 which mediates repression of epithelial-cadherin (E-cadherin) expression (Fig. 1B, right panel) (42). E-cadherin plays an important role in epithelial cell-cell adhesion and in the maintenance of tissue architecture. Therefore reduction or loss of E-cadherin expression promotes epithelial-to-mesenchymal transition (EMT) and tumor progression (43). The underlying basis of MTA1 mediated regulation of E-cadherin includes the dual coregulatory nature of MTA1 wherein the MTA1/Pol II coactivator complex stimulates the expression of FosB which, in-turn, interacts with MTA1/NuRD corepressor to inhibit the transcription of E-cadherin(42). Thus, activation of the TGF-β1-MTA1-E-cadherin axis is emerging as a central feature of EMT and metastasis in epithelial cells. In addition, MTA1 has also been shown to activate two related transcription factors SNAI1 and SLUG (also known as SNAI2) in ovarian cancer cells (44), although the details of this mechanism remain to be determined. Both SNAI1 and SLUG (45) proteins are direct transcriptional repressors of E-cadherin and their expression induces EMT. Thus, it is possible that oncogenic MTA1 exploits multiple signaling pathways to activate EMT programs, directly or indirectly, during cancer invasion and metastasis. In agreement with this notion, several recent findings show MTA1 to interject into the hypoxia-inducible factor 1α (46, 47) and Wnt1 pathways (20), which have been implicated in EMTs through multiple distinct mechanisms (48).
MTA1, a New Target and Component in the DNA Damage Response
Due to the paramount role of MTA1 in human cancer, it is reasonable to expect MTA1 to play a crucial role in the DNA repair process. In fact, a set of recent studies have discovered an essential role of MTA1 in the DNA damage response and more importantly, a cancer-relevant physiologic function of the normal levels of MTA1 in eukaryotic cells (Fig. 1D)(25, 31, 41, 49–51). Surprisingly, the involvement of MTA1 in DNA damage can be traced back to 1999, when Schmidt et al. found that DNA damage checkpoint protein ATR (ataxia telangiectasia mutated-and Rad3-related) associates with multiple components of the NuRD complex, which includes MTA1 (52). Ten years later in 2009, substantial molecular and genetic experimental evidence established a previously unrecognized functional role of MTA1 in ionizing radiation (IR)-induced double-strand break (DSB) repair (41). The genetic depletion of MTA1 renders cells to be hypersensitive to IR exposure, suggesting a defect in DSB repair in MTA1-deficient cells. Reassuringly, re-expressing MTA1 in such cells could rectify these defects, indicating the involvement of MTA1 in efficient DSB repair (41). The underlying mechanism by which MTA1 facilitates DNA repair is shown to occur in a p53-dependent and -independent manner (25, 31). Interestingly, this function of MTA1 might be conserved in MTA2 (53). However, this was not manifested in MTA1-knockout cells where the remaining members of the MTA family were unable to rescue the noted defects in DNA repair, highlighting the power of genetic supporting evidence. Further, the DNA damage role of MTA1 was emphasized by evidence that demonstrates the recruitment of MTA1 to sites of DNA damage in a poly(ADP-ribose) polymerase (PARP)-dependent manner, and depletion of MTA1 by siRNAs resulted in an increased cell sensitivity to IR in human cancer cells (50). These finding were strongly substantiated by several recent reports that highlight the role of CHD4 (50, 53–55) and HDAC1/2 (56, 57) in DNA damage repair. Together, it is becoming increasingly clear that multiple components of the NuRD complexes are indeed, implicated in DNA damage repair, emphasizing the evolutionally conserved functions of this family of chromatin-remodeling complexes in DNA repair.
In addition to DSB, MTA1 is equally essential for ATR-mediated DNA damage checkpoint function following ultraviolet (UV) radiation (49). In this context, MTA1 is required for the activation of the ATR-Claspin-checkpoint kinase 1 (Chk1) and ATR-H2AX pathways following UV treatment, and consequently, depletion of MTA1 results in the G2-M checkpoint defect and increases cellular sensitivity to UV-induced DNA damage (49). Together, these findings established the significant function of MTA1 in facilitating the repair of the damaged DNA, and hence survival of cancer cells, in the face of the severe, endogenous, oncogene-induced DNA damage as well as the damage induced by genotoxic therapy(58). Such a concept is plausible because the status of MTA1 might not only promote the fitness of cancer but also regulate the sensitivity of tumor cells to genotoxic therapy, as experimental depletion of MTA1 can enhance radio-sensitivity of the cultured cells to radiation. Based on these findings, MTA1-targeting approaches are likely to increase therapeutic benefits of radiation therapy in cancer.
MTA1, A Bridge between Inflammation and Cancer
Accumulating clinical, epidemiological, and molecular evidence has led to a generally accepted notion that inflammation and cancer are intimately linked and persistent inflammation over time, contributes towards the molecular processes involved in cancerous phenotypes (59, 60). Most of these pathways converge onto a central transcriptional player, nuclear factor-κB (NF-κB) (61), which in-turn, orchestrates a complex set of reactions involving multiple negative- and positive-feedback loops by virtue of transcriptional regulation. While searching for the upstream activator of MTA1, Pakala et al. recognized the presence of multiple NF-κB consensus motifs in the MTA1 promoter and for the first time showed MTA1 as an NF-κB-target gene and a primary target of inflammation (62). Accordingly, stimulation of macrophages with lipopolysaccharide (LPS), a prototypical endotoxin in response to various infections, stimulates MTA1 transcription via the NF-κB pathway as well as expression of NF-κB-target cytokines(62). Interestingly, MTA1 in-turn, became instrumental for the production of NF-κB-mediated pro-inflammatory cytokines, such as interleukin-1β, tumor necrosis factor α (TNFα), and macrophage inflammatory protein-2 in LPS-stimulated macrophages (Fig. 1E)(62). These findings also discovered MTA1’s corepressor or coactivator differential regulation of NF-κB-target genes under basal or stimulated conditions via MTA1/HDAC or MTA1/Pol II complexes, and thus function as one of the core referees that coordinate the details of the NF-κB signaling network and homeostasis maintenance during inflammatory responses(62). Since many of the protein components of NF-κB signaling are NF-κB-targets, subsequent studies found MTA1 protein to be involved in the regulation of the expression of myeloid-differentiation factor 88 (63) and transglutaminase 2(64), both key components of NF-κB signaling in a variety of inflammatory diseases and human cancers. These recent reports established MTA1 as a target and an essential component of the inflammatory response (Fig. 1E).
MTA1, A Host Master Router in Infectious Agent-driven Cancer
In keeping with its role as a primary target of inflammation (62–64), MTA1 is shown to be involved in the optimum stimulation of Hepatitis B virus (HBV) x-protein (HBx)-triggered NF-κB signaling as well as the expression of NF-κB targets genes, such as TNF-α, cyclooxygenase-2 (COX2), and inducible nitric oxide (Fig. 1E) (65, 66). Interestingly, MTA1, an established NF-κB-target gene, was also found to be upregulated by HBx, which fuels the amplification and duration of NF-κB signaling. Furthermore, both MTA1 and NF-κB are co-overexpressed in human liver cancer, which is positive for the HBx protein (65). Together, these observations suggest MTA1 status to be an important element that contributes to inflammation prolongation, and in-turn, HBV-driven liver tumorigenesis.
Schistosoma haematobium is classified as a group 1 carcinogen by the WHO and is established to cause bladder cancer and fibrosis in the liver, as a result of chronic inflammation. MTA1 is reported to be essential for the optimal growth of schistosomes to adulthood and successful parasitism in a murine MTA1-knockout experimental model (Fig. 1E) (67). The underlying mechanism included deregulation of cytokine interdependence and regulated Th1/Th2 cytokine responses, presumably largely due to chromatin remodeling of its immune targets in the MTA1-knocout mice. Similarly, the carcinogenic liver fluke Opisthorchis viverrini, which is responsible for inflammation-associated development of human cholangiocarcinomas also requires the MTA1 coregulator for survival in a murine model (Fig. 1E)(68). Relevance of these studies to humans was proved by IHC studies of ~300 liver tissue cores from confirmed cases of Opisthorchis viverrini, showing a widespread upregulation of MTA1 expression in the specimens(68). These findings suggest MTA1 to function as a host permissive factor for the optimal mediation of the infectious process of human helminth parasites which cause cancer. Collectively, these recent findings, for the first time, have revealed an inherent role of MTA1 in parasitism and cancer and thus, have opened new avenues of exciting research that would be beneficial for cancer patients globally, both in affluent and resource-limited settings.
The Next Horizon for MTA1 Research
Since the isolation of the MTA1 cDNA in 1994 (9), remarkable progress has been made in our understanding of the functions of MTA1, not only in human cancer but also in several other physiological processes such as the DNA-damage response, inflammation and immune modulation (Fig.1A). However, there are still many questions that remain to be answered. For instance, the triggers which disrupt the MTA1 equilibrium and allow it to manifest oncogenic activity as well as the normal cellular functions of MTA1 still remain to be defined. This equilibrium could be fine-tuned by post translational modifications of MTA1 but a comprehensive understanding about the cross-talks between these modifications needs to be characterized. We began to appreciate the physiological functions of MTA1 in various pathways and these emerging studies already suggest other significant physiological roles of MTA1. Therefore, redefining the nomenclature of MTA1 away from metastasis and reflecting on its complete characteristics should be considered. The snapshots of MTA1 functions show its co-repressor and co-activator abilities but how MTA1 deals with this conundrum and the determining elements of MTA1 that decide that this molecular switch will act as an activator or repressor is one of the central mechanistic questions to be resolved. This might be understood through studies that modulate MTA1 in a signal-dependent post-translational way, generating the code which might dictate its engagement in co-repressor vs. co-activator complexes. To understand the complete molecular mechanism of MTA1, high resolution crystal structures of MTA1 are needed. The authors hope to bring such on-going efforts to a meaningful conclusion in the near future. Such information will also pave the way for the rationale development of the target-specific cancer therapeutics. Given the emerging role of MTA1 in DNA repair, targeting MTA1 might prove efficacious when used in combination with DNA-damaging chemotherapeutic drugs and radiotherapy. More significantly, the host permissive nature of MTA1 allows it to be an effective drug target for pathogen-driven cancers which are prevalent in countries with limited economic resources. In summary, the past decade of research on MTA1 drastically transforms our understanding of the abilities and influences of coregulators and therefore we hope to realize the possibility of MTA1 targeted therapy against not only cancer but also several inflammation and pathogen-driven pathological conditions such as arthritis, atherosclerosis and respiratory disorders.
Supplementary Material
Acknowledgments
We are grateful to the past and present members of the Kumar laboratory for insightful discussions and to Ms. Amanda Lyon for editing the manuscript. We apologize to all of our colleagues whose work has not been cited here due to space limitations. This study was supported by National Institutes of Health Grants CA98823 and CA98823S1 to R.K.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 3.Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–52. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 4.Manavathi B, Kumar R. Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J Biol Chem. 2007;282:1529–33. doi: 10.1074/jbc.R600029200. [DOI] [PubMed] [Google Scholar]
- 5.Manavathi B, Singh K, Kumar R. MTA family of coregulators in nuclear receptor biology and pathology. Nucl Recept Signal. 2007;5:e010. doi: 10.1621/nrs.05010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar R, Wang RA, Bagheri-Yarmand R. Emerging roles of MTA family members in human cancers. Semin Oncol. 2003;30:30–7. doi: 10.1053/j.seminoncol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Toh Y, Nicolson GL. The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications. Clin Exp Metastasis. 2009;26:215–27. doi: 10.1007/s10585-008-9233-8. [DOI] [PubMed] [Google Scholar]
- 8.Kumar R, Wang RA, Mazumdar A, Talukder AH, Mandal M, Yang Z, et al. A naturally occurring MTA1 variant sequesters oestrogen receptor-alpha in the cytoplasm. Nature. 2002;418:654–7. doi: 10.1038/nature00889. [DOI] [PubMed] [Google Scholar]
- 9.Toh Y, Pencil SD, Nicolson GL. A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. J Biol Chem. 1994;269:22958–63. [PubMed] [Google Scholar]
- 10.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 11.Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, et al. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol. 2001;3:30–7. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Ma L, Zhao J, Liu X, Li Z, Zhang Y. Expression profile of MTA1 in adult mouse tissues. Tissue Cell. 2009;41:390–9. doi: 10.1016/j.tice.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Qian H, Lu N, Xue L, Liang X, Zhang X, Fu M, et al. Reduced MTA1 expression by RNAi inhibits in vitro invasion and migration of esophageal squamous cell carcinoma cell line. Clin Exp Metastasis. 2005;22:653–62. doi: 10.1007/s10585-006-9005-2. [DOI] [PubMed] [Google Scholar]
- 14.Bagheri-Yarmand R, Talukder AH, Wang RA, Vadlamudi RK, Kumar R. Metastasis-associated protein 1 deregulation causes inappropriate mammary gland development and tumorigenesis. Development. 2004;131:3469–79. doi: 10.1242/dev.01213. [DOI] [PubMed] [Google Scholar]
- 15.Bagheri-Yarmand R, Balasenthil S, Gururaj AE, Talukder AH, Wang YH, Lee JH, et al. Metastasis-associated protein 1 transgenic mice: a new model of spontaneous B-cell lymphomas. Cancer Res. 2007;67:7062–7. doi: 10.1158/0008-5472.CAN-07-0748. [DOI] [PubMed] [Google Scholar]
- 16.Balasenthil S, Gururaj AE, Talukder AH, Bagheri-Yarmand R, Arrington T, Haas BJ, et al. Identification of Pax5 as a target of MTA1 in B-cell lymphomas. Cancer Res. 2007;67:7132–8. doi: 10.1158/0008-5472.CAN-07-0750. [DOI] [PubMed] [Google Scholar]
- 17.Gururaj AE, Singh RR, Rayala SK, Holm C, den Hollander P, Zhang H, et al. MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc Natl Acad Sci U S A. 2006;103:6670–5. doi: 10.1073/pnas.0601989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohshiro K, Rayala SK, Wigerup C, Pakala SB, Natha RS, Gururaj AE, et al. Acetylation-dependent oncogenic activity of metastasis-associated protein 1 co-regulator. EMBO Rep. 2010;11:691–7. doi: 10.1038/embor.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 20.Kumar R, Balasenthil S, Manavathi B, Rayala SK, Pakala SB. Metastasis-associated protein 1 and its short form variant stimulates Wnt1 transcription through promoting its derepression from Six3 corepressor. Cancer Res. 2010;70:6649–58. doi: 10.1158/0008-5472.CAN-10-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar R, Balasenthil S, Pakala SB, Rayala SK, Sahin AA, Ohshiro K. Metastasis-associated protein 1 short form stimulates Wnt1 pathway in mammary epithelial and cancer cells. Cancer Res. 2010;70:6598–608. doi: 10.1158/0008-5472.CAN-10-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang XY, DeSalle LM, Patel JH, Capobianco AJ, Yu D, Thomas-Tikhonenko A, et al. Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein. Proc Natl Acad Sci U S A. 2005;102:13968–73. doi: 10.1073/pnas.0502330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molli PR, Singh RR, Lee SW, Kumar R. MTA1-mediated transcriptional repression of BRCA1 tumor suppressor gene. Oncogene. 2008;27:1971–80. doi: 10.1038/sj.onc.1210839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 25.Li DQ, Pakala SB, Reddy SD, Ohshiro K, Peng SH, Lian Y, et al. Revelation of p53-independent function of MTA1 in DNA damage response via modulation of the p21 WAF1-proliferating cell nuclear antigen pathway. J Biol Chem. 2010;285:10044–52. doi: 10.1074/jbc.M109.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Rechem C, Boulay G, Pinte S, Stankovic-Valentin N, Guerardel C, Leprince D. Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol Cell Biol. 2010;30:4045–59. doi: 10.1128/MCB.00582-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wales MM, Biel MA, el Deiry W, Nelkin BD, Issa JP, Cavenee WK, et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med. 1995;1:570–7. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- 28.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 29.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–14. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 31.Li DQ, Divijendra Natha Reddy S, Pakala SB, Wu X, Zhang Y, Rayala SK, et al. MTA1 coregulator regulates p53 stability and function. J Biol Chem. 2009;284:34545–52. doi: 10.1074/jbc.M109.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon HE, Cheon H, Lee MS. Metastasis-associated protein 1 inhibits p53-induced apoptosis. Oncol Rep. 2007;18:1311–4. [PubMed] [Google Scholar]
- 33.Kai L, Samuel SK, Levenson AS. Resveratrol enhances p53 acetylation and apoptosis in prostate cancer by inhibiting MTA1/NuRD complex. Int J Cancer. 2010;126:1538–48. doi: 10.1002/ijc.24928. [DOI] [PubMed] [Google Scholar]
- 34.Ghanta KS, Li DQ, Eswaran J, Kumar R. Gene profiling of MTA1 identifies novel gene targets and functions. PLoS One. 2011;6:e17135. doi: 10.1371/journal.pone.0017135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–59. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 36.Li DQ, Pakala SB, Reddy SD, Ohshiro K, Zhang JX, Wang L, et al. Bidirectional autoregulatory mechanism of metastasis-associated protein 1-alternative reading frame pathway in oncogenesis. Proc Natl Acad Sci U S A. 2011;108:8791–6. doi: 10.1073/pnas.1018389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 38.Haber DA. Splicing into senescence: the curious case of p16 and p19ARF. Cell. 1997;91:555–8. doi: 10.1016/s0092-8674(00)80441-9. [DOI] [PubMed] [Google Scholar]
- 39.Vitari AC, Leong KG, Newton K, Yee C, O’Rourke K, Liu J, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–6. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 40.Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E, et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest. 2011;121:1329–43. doi: 10.1172/JCI45784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li DQ, Ohshiro K, Reddy SD, Pakala SB, Lee MH, Zhang Y, et al. E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc Natl Acad Sci U S A. 2009;106:17493–8. doi: 10.1073/pnas.0908027106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pakala SB, Singh K, Reddy SD, Ohshiro K, Li DQ, Mishra L, et al. TGF-beta1 signaling targets metastasis-associated protein 1, a new effector in epithelial cells. Oncogene. 2011;30:2230–41. doi: 10.1038/onc.2010.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 44.Dannenmann C, Shabani N, Friese K, Jeschke U, Mylonas I, Bruning A. The metastasis-associated gene MTA1 is upregulated in advanced ovarian cancer, represses ERbeta, and enhances expression of oncogenic cytokine GRO. Cancer Biol Ther. 2008;7:1460–7. doi: 10.4161/cbt.7.9.6427. [DOI] [PubMed] [Google Scholar]
- 45.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–9. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 46.Moon HE, Cheon H, Chun KH, Lee SK, Kim YS, Jung BK, et al. Metastasis-associated protein 1 enhances angiogenesis by stabilization of HIF-1alpha. Oncol Rep. 2006;16:929–35. [PubMed] [Google Scholar]
- 47.Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. Embo J. 2006;25:1231–41. doi: 10.1038/sj.emboj.7601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 49.Li DQ, Ohshiro K, Khan MN, Kumar R. Requirement of MTA1 in ATR-mediated DNA damage checkpoint function. J Biol Chem. 2010;285:19802–12. doi: 10.1074/jbc.M109.085258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–80. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li DQ, Kumar R. Mi-2/NuRD complex making inroads into DNA-damage response pathway. Cell Cycle. 2010;9:2071–9. doi: 10.4161/cc.9.11.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt DR, Schreiber SL. Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry. 1999;38:14711–7. doi: 10.1021/bi991614n. [DOI] [PubMed] [Google Scholar]
- 53.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–9. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–9. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–40. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–51. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–9. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartek J, Lukas J. DNA repair: Cyclin D1 multitasks. Nature. 2011;474:171–2. doi: 10.1038/474171a. [DOI] [PubMed] [Google Scholar]
- 59.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 60.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–71. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 62.Pakala SB, Bui-Nguyen TM, Reddy SD, Li DQ, Peng S, Rayala SK, et al. Regulation of NF-kappaB circuitry by a component of the nucleosome remodeling and deacetylase complex controls inflammatory response homeostasis. J Biol Chem. 2010;285:23590–7. doi: 10.1074/jbc.M110.139469. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Pakala SB, Reddy SD, Bui-Nguyen TM, Rangparia SS, Bommana A, Kumar R. MTA1 coregulator regulates LPS response via MyD88-dependent signaling. J Biol Chem. 2010;285:32787–92. doi: 10.1074/jbc.M110.151340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghanta KS, Pakala SB, Reddy SD, Li DQ, Nair SS, Kumar R. MTA1 coregulation of transglutaminase 2 expression and function during inflammatory response. J Biol Chem. 2011;286:7132–8. doi: 10.1074/jbc.M110.199273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bui-Nguyen TM, Pakala SB, Sirigiri RD, Xia W, Hung MC, Sarin SK, et al. NF-kappaB signaling mediates the induction of MTA1 by hepatitis B virus transactivator protein HBx. Oncogene. 2010;29:1179–89. doi: 10.1038/onc.2009.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bui-Nguyen TM, Pakala SB, Sirigiri DR, Martin E, Murad F, Kumar R. Stimulation of inducible nitric oxide by hepatitis B virus transactivator protein HBx requires MTA1 coregulator. J Biol Chem. 2010;285:6980–6. doi: 10.1074/jbc.M109.065987. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Nair SS, Bommana A, Bethony JM, Lyon AJ, Ohshiro K, Pakala SB, et al. The metastasis-associated protein-1 gene encodes a host permissive factor for schistosomiasis, a leading global cause of inflammation and cancer. Hepatology. 2011;54:285–95. doi: 10.1002/hep.24354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nair SS, Bommana A, Pakala SB, Ohshiro K, Lyon AJ, Suttiprapa S, et al. Inflammatory response to liver fluke Opisthorchis viverrini depends on host master coregulator, MTA1, a marker for parasite induced cholangiocarcinoma. Hepatology. 2011 doi: 10.1002/hep.24518. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.