Abstract
In this paper we implement the 7-point checklist, a set of dermoscopic criteria widely used by clinicians for melanoma detection, on smart handheld devices, such as the Apple iPhone and iPad. The application developed is using sophisticated image processing and pattern recognition algorithms, yet it is light enough to run on a handheld device with limited memory and computational speed. When combined with a commercially available handheld dermoscope that provides proper lesion illumination, this application provides a truly self-contained handheld system for melanoma detection. Such a device can be used in a clinical setting for routine skin screening, or as an assistive diagnostic device in underserved areas and in developing countries with limited healthcare infrastructure.
I. INTRODUCTION
The new generation of smart handheld devices with sophisticated hardware and operating systems has provided a portable platform for running medical diagnostic software, such as the heart rate monitoring [1], diabetes monitoring [2], and experience sampling [3] applications, which combine the usefulness of medical diagnosis with the convenience of a handheld device. Their light operating systems, such as the Apple iOS and Google Android, the support for user friendly touch gestures, the availability of an SDK for fast application development, the rapid and regular improvements in hardware, and the availability of fast wireless networking over Wi-Fi and 3G make these devices ideal for medical applications. In this paper we present an innovative application for melanoma detection running on Apple iOS based smart devices, like the iPhone and iPad. Our long term objective is to develop an automated system for melanoma detection deployed on handheld devices, based on well known diagnostic criteria used by dermatologists to analyze dermoscopic images. Dermatologist use pattern analysis to detect the presence of certain features or patterns of texture and color in a skin lesion. Some widely used criteria are the ABCD rule [4], Menzies rule [5], and the 7-point checklist [6]. The latter includes seven dermoscopic features that can be detected with high sensitivity and decent specificity by even less experienced clinicians [6]. The seven points of the list are subdivided into three major and four minor criteria, reflecting their importance in defining a melanoma. To score a lesion, the presence of a major criterion is given two points and that of a minor one point. If the total score is greater than or equal to 3, the lesion is classified as melanoma.
We have recently developed a library of image processing and texture analysis algorithms for low level features of a lesion that runs on embedded devices. In this paper we present an application for automatic detection of melanoma, called SkinScan. It uses high level features, such as those on the 7-point checklist, and is implemented on Apple iOS4, as shown in Figure 1. This application can be easily configured for the diagnosis of other skin conditions, such as wounds and ulcers.
Fig. 1.
The SkinScan application running on the Apple iPhone.
II. METHODS
The 7-point checklist includes the following criteria:
-
Major criteria
Atypical pigment network: Black, brown, or gray network with irregular meshes and thick lines.
Blue-whitish veil: Confluent gray-blue to whitish-blue diffuse pigmentation associated with pigment network alterations, dots/globules and/or streaks.
Atypical vascular pattern: Linear irregular or dotted vessels not clearly combined with regression structures and associated with pigment network alterations, dots/globules and/or streaks.
-
Minor criteria
Irregular streaks: Irregular more or less confluent, linear structures not clearly combined with pigment network lines.
Irregular pigmentation: Black, brown, and/or gray pigment areas with irregular shape and/or distribution.
Irregular dots/globules: Black, brown, and/or gray round to oval, variously sized structures irregularly distributed.
Regression structures: Associated white scar-like and gray blue, peppering, multiple blue gray dots.
A. Dataset
In this study, we used images from a large commercial library of skin cancer images annotated by expert dermatologists [8] that were uploaded to the phone. A total of 347 images were selected: 110 were classified by the 7-point list as melanoma and the remaining 237 were classified as benign. Intra-observer and inter-observer agreement could be low for certain criteria [9]. To demonstrate the feasibility of our automated system, we only chose images considered as low difficulty by the experts [8]. There were 385 low difficulty images in the database [8] and our segmentation methods could provide a satisfactory boundary for 347 (90.13%) of them. All images were segmented manually by one of the authors to provide a ground truth against which the automated techniques employed by our application were compared.
B. Feature Extraction
A skin lesion image can be acquired using the smart phone camera (with or without an external attachment which can provide illumination and magnification) or can be loaded from the photo library to provide the diagnosis in real time. To identify a region of interest (ROI), an image is first converted to greyscale, and then fast median filtering [10] for noise removal is performed, and followed by ISODATA segmentation [11], and several morphological operations. From the ROI, we extract color and texture features relating to each criterion on the 7-point checklist, as follows.
1) Texture Features
They provide information on the various structural patterns [8] of 7-point checklist, such as pigmentation networks, vascular structures, and dots and globules present in a skin lesion. Elbaum et al. [12] used wavelet coefficients as texture descriptors in their skin cancer screening system MelaFind®. Our previous work [13] has demonstrated the effectiveness of Haar wavelet coefficients and local binary patterns [14] for melanoma detection.
Haar Wavelet
From the ROI we select non-overlapping K × K blocks of pixels, where K is a user defined variable. Computation time for feature extraction in directly proportional to the block size K. The block of pixels is decomposed using a three-level Haar wavelet transform [15] to get 10 sub-band images. We extract texture features by computing statistical measures, like the mean and standard deviation, on each sub-band image, which are then combined to form a vector Wi = {m1, sd1, m2, sd2,......}. Haar wavelet extraction for texture feature is as follows:
Convert the color image to greyscale; select a set of points in the ROI using a rectangular grid of size M pixels.
Select patches of size K×K pixels centered on the selected points.
Apply a 3-level Haar wavelet transform on the patches.
For each sub-band image compute statistical measures, namely mean and standard deviation, to form a feature vector Wi = {m1, sd1, m2, sd2, …}.
For all feature vectors Wi extracted, normalize each dimension to zero-mean and unit-variance.
Apply K-means clustering [16] to all feature vectors Wi from all training images to obtain L clusters with centers C = {C1, C2, …, CL}.
For each image build an L-bin histogram Hi. For feature vector Wi, increment the jth bin of the histogram such that min j||Cj − Wi||.
The value of parameter M is a trade-off between accuracy and computation speed. When the algorithm runs on a handheld device we want to reduce computation time, so we choose M = 10 for the grid size, K = 24 for patch size, and L = 200 as the number of clusters in the feature space. By exhaustive parameter exploration in our previous studies [13], we found that these parameters are reasonable settings.
Local Binary Pattern (LBP)
LBP is a robust texture operator [14] defined on a greyscale input image. It is invariant to monotonic transformation of intensity and invariant to rotation. It is derived using a circularly symmetric neighbor set of P members on a circle of radius R denoted by . The parameter P represents the quantization of angular space in the circular neighborhood, and R represents the spatial resolution. A limited number of transitions or discontinuities (0/1 changes in LBP) are allowed to reduce the noise and for better discrimination of features. We restrict the number of transitions in LBP to P, and transitions greater than that are considered equal. An occurrence histogram of LBP with useful statistical and structural information is computed as follows:
Convert the color image to greyscale.
Select pixels belonging to ROI and compute local binary pattern .
Build an occurrence histogram, where we increment the jth bin of the histogram if number of transitions in LBP is j.
Repeat steps 2 and 3 for all pixels in ROI.
We build the occurrence histograms for LBP16,2 and LBP24,3 and concatenate them to form a feature vector Li.
2) Color Features
Detection of the 7-point checklist criteria, such as blue-whitish veil and regression, which consist of mixtures of certain colors, can be achieved by analyzing the color intensity of pixels in the lesion. Stoeker et al. [17] investigated the discriminatory capability of color histograms in classification of malignant lesions from benign lesions. To reduce the variance due to the lighting conditions in which dermoscopic images were taken, we considered also the HSV and LAB color spaces, which are invariant to illumination changes [18].
Color Histograms
To extract the color information of a lesion, we compute a color histogram from the intensity values of pixels belonging to the ROI. Additional images in the HSV and LAB color spaces are obtained from the original RGB image. The intensity range of each channel is divided into P fixed-length intervals. For each channel we build a histogram to keep count of the number of pixels belonging to each interval, resulting in a total of nine histograms from three color spaces. Statistical features, such as standard deviation and entropy (Eq. 1), of the nine histograms are also extracted as features for classification. More specifically, entropy is defined as
| (1) |
where
The color histogram feature extraction steps are as follows:
Obtain skin lesion image in HSV and LAB color space from input RGB image
For each channel in all three color spaces build a separate P bin histogram
-
For all pixels belonging to ROI, increment the jth bin of histogram where . Ic and Mc are pixel intensity and maximum intensity in the specified color space.
Compute the standard deviation and entropy of the histogram.
Repeat steps 3 and 4 for all the channels in RGB, HSV, and LAB color space.
Color histogram and statistical features are combined to form a feature vector Ci.
C. Classification
The features from color histogram Ci, Haar wavelet Hi, and LBP Li are combined to form Fi = {Ci, Hi, Li}. For each criterion in the 7-point checklist we perform a filtered feature selection to obtain a subset with the highest classification accuracy. Correlation coefficient values are used as the ranking criterion in the filters. The size of the subset and the parameters of the linear support vector machine (SVM) are obtained by grid search. Each criterion requires both training and testing.
Training
The training algorithm is as follows:
Segment the input image to obtain region of interest.
Extract Color histogram, Haar wavelet, and Local binary pattern. Concatenate them to form feature vector Fi.
Repeat steps 1 and 2 for all training images.
Perform filter feature selection to choose a subset of features Si from Fi.
Input Si to linear SVM to obtain maximum margin hyperplane.
Testing
Image classification is performed as follows:
Read the input image and perform steps 1 and 2 from the training algorithm.
For each criterion use the SVM coefficients obtained from training to make a prediction.
Scores from all major and minor criteria are summed up. If the score is greater than or equal to 3 then lesion is classified as melanoma, otherwise as benign.
III. RESULTS
In this section we present an instantiation of the 7-point checklist on the Apple iPhone. We compare the classification accuracy of each criterion separately against the overall final classification by expert physicians [8]. We have developed a menu based application that implements the automated procedure for classification, as shown in Figure 2. The end user can take an image of the skin lesion using the 5 megapixel built in camera with LED flash, or load the image from media library. The image is analyzed in quasi real time and the final result displayed on the screen.
Fig. 2.
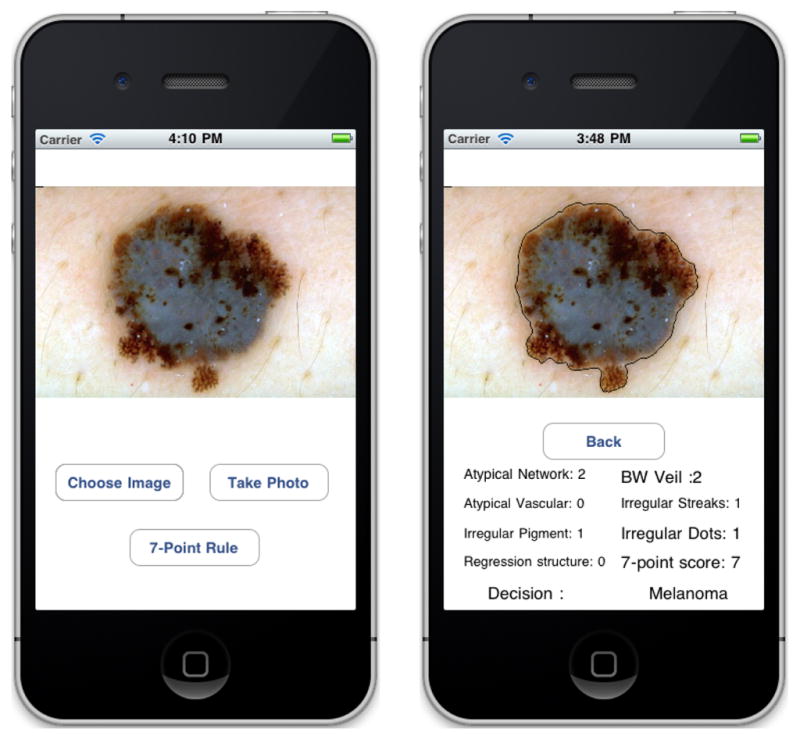
SkinScan application on Apple iPhone device
A. Classification Results
We performed 10-fold cross validation on the set of 347 images. We divided the dataset into 10 folds, nine folds with 11 melanoma and 23 benign lesions and the remaining fold with 11 melanoma and 30 benign lesions. Of the 10 folds, we used nine for training and one for test. We performed 10 rounds of validation where we chose each fold for testing and the rest for training, to get 10 experiments. In the following sub-sections we compare the classification accuracy of each criterion and the overall decision of the 7-point checklist with dermatology and histology.
Classification Results for each Criterion
In Table I we present the sensitivity and specificity of our algorithm in classification of each of the 7-point checklist criterion. We have lower accuracy for the regression structures, because they are usually indistinguishable from the blue-whitish veil via dermoscopy [8]. However, this is not an issue, as is it only necessary to obtain a minimum score of 3 to correctly detect a melanoma.
TABLE I.
Classification Results for all Criteria.
| Sensitivity | Specificity | |
|---|---|---|
| Atypical Pigment Network | 72.86% | 70.40% |
| Blue-Whitish Veil | 79.49% | 79.18% |
| Atypical Vascular Pattern | 75.00% | 69.66% |
| Irregular Streaks | 76.74% | 79.31% |
| Irregular Pigmentation | 69.47% | 74.21% |
| Irregular Dots and Globules | 74.05% | 74.54% |
| Regression Structures | 64.18% | 67.86% |
Classification and Overall Decision
In Table III we compare the sensitivity and specificity of our algorithms with the decision made by expert clinicians via dermoscopy. In Table II we present the confusion matrix computed using the sum of ten confusion matrices from the ten test sets, given from the 10-fold cross validation.
TABLE III.
Classification accuracy
| Sensitivity | Specificity | |
|---|---|---|
| 7-Point Checklist | 87.27% | 71.31% |
| Ignoring 7-Point Checklist | 74.78% | 70.69% |
TABLE II.
Confusion Matrix of the automated Decision.
| Confusion Matrix | Predicted | ||
|---|---|---|---|
| Melanoma | benign | ||
| Dermoscopy | Melanoma | 96 | 14 |
| Benign | 68 | 169 | |
We also performed another classification experiment using SVM, where we ignore the 7-point checklist and directly classify each skin lesion as melanoma or benign. The feature vectors, feature selection scheme, and final ground truth (melanoma/benign) are the same as the classification using the automated 7-point checklist. Table III shows that classification accuracy is much lower when we ignore the 7-point checklist.
Execution Time
We compare the time needed for classification using the ISODATA segmentation algorithm [11] on the Apple iPhone 3G with a typical desktop computer (2.26 GHz Intel Core 2 Duo with 2GB RAM). The classification time includes time taken for feature extraction. Table IV shows computation time in seconds for both platforms. It can be seen that the whole procedure takes under 10 sec to complete. This proves that the application is light enough to run on a smartphone which has limited computation power.
TABLE IV.
Mean computation time on the iPhone 4 and desktop computer
| Mean time (sec) | Apple iPhone 3G | Desktop computer |
|---|---|---|
| Segmentation | 2.3910 | 0.1028 |
| Classification | 7.4710 | 0.2415 |
IV. CONCLUSIONS AND DISCUSSION
In this paper, we showed an implementation of 7-Point checklist on the Apple iPhone. The application, written in C/C++, is light enough to run on devices with limited memory and computation speed. We implemented sophisticated image processing and pattern recognition algorithms that run with limited resources and provide classification results in quasi real time. These finding demonstrate the feasibility of developing medical diagnostic applications on off-the-shelf smart handheld devices, which have the advantages of low cost and portability.
Acknowledgments
This work was supported in part by NIH grant no. 1R21AR057921 and by grants from UH-GEAR and the Texas Learning and Computation Center at the University of Houston.
Contributor Information
Tarun Wadhawan, Department of Computer Science.
Ning Situ, Department of Computer Science.
Hu Rui, Department of Electrical and Computer Engineering.
Keith Lancaster, Department of Electrical and Computer Engineering.
Xiaojing Yuan, Email: xyuan@uh.edu, Department of Engineering Technology.
George Zouridakis, Email: zouridakis@uh.edu, Departments of Engineering Technology, Computer Science, and Electrical and Computer Engineering, University of Houston, Houston, TX 77204-3058 USA; phone: +1-713-743-8656; fax: +1-713-743-1250.
References
- 1.Morris M, Guilak F. Mobile heart health: Project highlight. Pervasive Computing, IEEE. 2009 April–june;8(2):57–61. [Google Scholar]
- 2.Logan A, McIsaac W, Tisler A, et al. Mobile phone-based remote patient monitoring system for management of hypertension in diabetic patients. American journal of hypertension. 2007;20(9):942–8. doi: 10.1016/j.amjhyper.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 3.Hicks J, Ramanathan N, Kim D, Monibi M, Selsky J. Wireless health 2010, WH ’10. New York, NY, USA: ACM; 2010. And wellness: an open mobile system for activity and experience sampling; pp. 34–43. [Google Scholar]
- 4.Stolz W, Riemann A, Cognetta AB, Pillet L. Abcd rule of dermoscopy: a new practical method for early recognition of malignant melanoma. Eur J Dermatol. 1994;4:521–527. [Google Scholar]
- 5.Menzies SW, Ingvar C, Crotty KA, McCarthy WH. Frequency and morphologic characteristics of invasive melanomas lacking specific surface microscopic features. Archives of Dermatology. 1996;132(10):1178–1182. [PubMed] [Google Scholar]
- 6.Argenziano G, Fabbrocini G, Carli P, De Giorgi V, Sammarco E, Delfino M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions. Arch Dermatol. 1998;134:1563–70. doi: 10.1001/archderm.134.12.1563. [DOI] [PubMed] [Google Scholar]
- 7.Wadhawan T, Situ N, Lancaster K, Yuan X, Zouridakis G. Proceeding. ISBI; 2011. Skinscan: A portable library for melanoma detection on handheld devices. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Argenziano G, Soyer HP, Giorgi VD, Piccolo D, Carli P, Delfino M, Ferrari A, Wellenhof R, Massi D, Mazzocchetti G, Scalvenzi M, Wolf I. EDRA: medical publishing and new media. 2000. Feburary, 2000. Dermoscopy: a tutorial. [Google Scholar]
- 9.Argenziano G, Soyer HP, Chimenti S, et al. Dermoscopy of pigmented skin lesions: Results of a consensus meeting via the internet. J Am Acad Dermatol. 2003;48:679–93. doi: 10.1067/mjd.2003.281. [DOI] [PubMed] [Google Scholar]
- 10.Huang T, Yang G, Tang G. A fast two-dimensional median filtering algorithm. Acoustics, Speech and Signal Processing, IEEE Transactions; 13–18, Feb 1979. [Google Scholar]
- 11.Dunn JC. A fuzzy relative of the isodata process and its use in detecting compact well-separated clusters. Journal of Cybernetics. 1973;3:32–57. [Google Scholar]
- 12.Elbaum M, Kopf AW, Rabinovitz HS, Langley RGB, Kamino H, Mihm MC, et al. Automatic differentiation of melanoma from melanocytic nevi with multispectral digital dermoscopy: A feasibility study. Journal of the American Academy of Dermatology. 2001;44(2):207–218. doi: 10.1067/mjd.2001.110395. [DOI] [PubMed] [Google Scholar]
- 13.Situ N, Yuan X, Chen J, Zouridakis G. Malignant melanoma detection by bag-of-features classification. Engineering in Medicine and Biology Society, 2008. EMBS 2008; 30th Annual International Conference of the IEEE; 2008. [DOI] [PubMed] [Google Scholar]
- 14.Ojala T, Pietikinen M, Topi M. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2002;24:971–987. [Google Scholar]
- 15.Stollnitz EJ, DeRose TD, Salesin DH. Wavelets for computer graphics: A primer. IEEE Computer Graphics and Applications. 1995;15:76–84. [Google Scholar]
- 16.Hartigan JA, Wong MA. A k-means clustering algorithm. Applied Statistics. 1979;28:100–108. [Google Scholar]
- 17.Stanley RJ, Stoecker WV, Moss RH. A relative color approach to color discrimination for malignant melanoma detection in dermoscopy images. Skin Research and Technology. 2007;13(1):62–72. doi: 10.1111/j.1600-0846.2007.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Sande K, Gevers T, Snoek C. Evaluating color descriptors for object and scene recognition. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2010;32(9):1582–96. doi: 10.1109/TPAMI.2009.154. [DOI] [PubMed] [Google Scholar]


