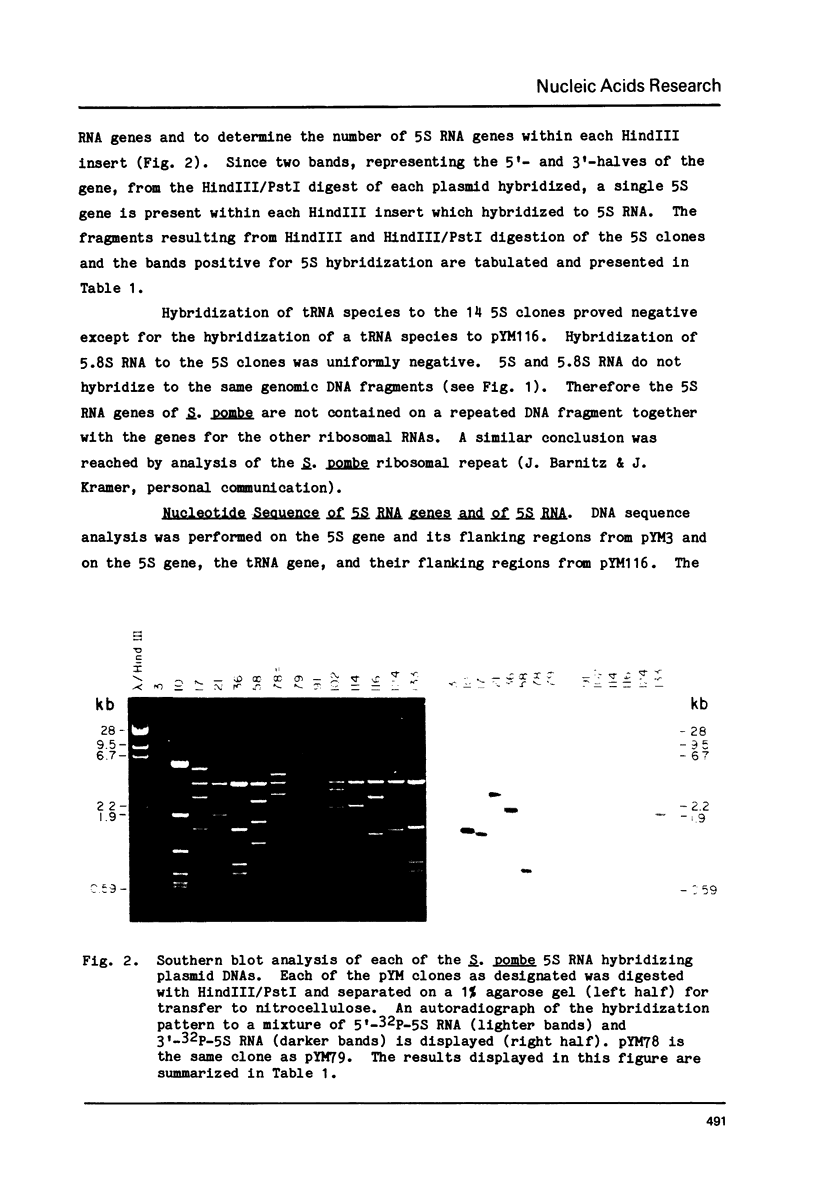
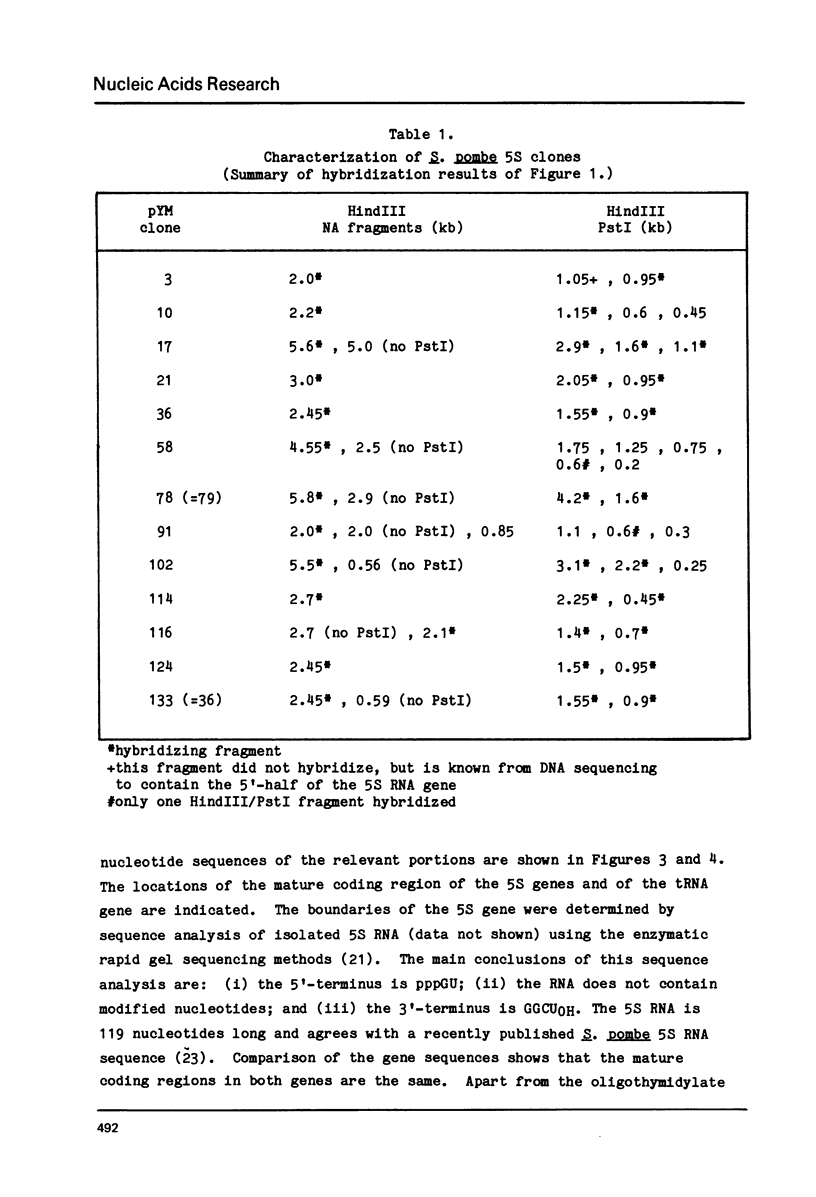
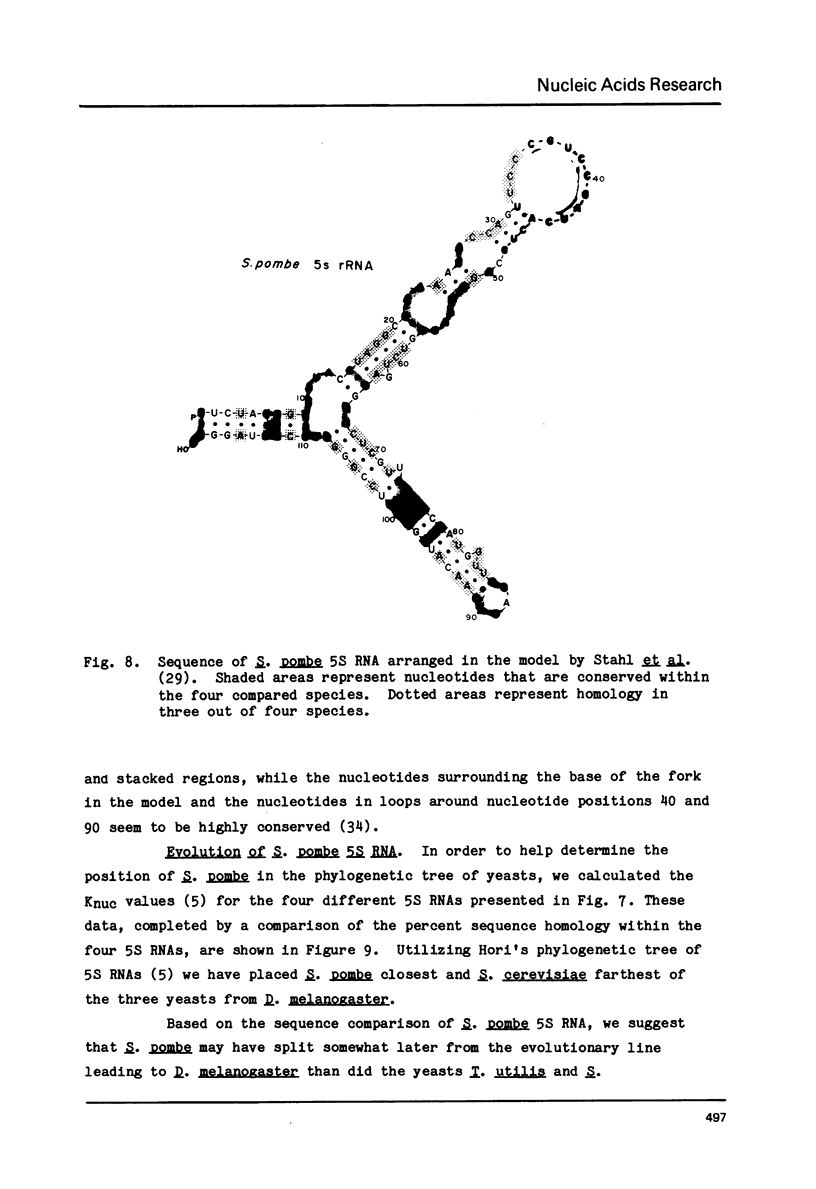
Abstract
The genomic arrangement and sequences of S. pombe 5S RNA genes are reported here. The 5S gene sequences appear to be dispersed within the genome, and are found independently of other rRNA genes. The sequences of two 5S genes examined show identical coding regions of 119 base pairs but have widely varying flanking sequences. A tRNAAsp gene is found in the 3' flanking region of one of the 5S genes. The tRNAAsp gene is faithfully transcribed in an X. laevis in vitro system, while the 5S genes are not transcribed in this system. The phylogenetic position of S. pombe is examined through comparison of 5S RNA sequences.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Cedergren R. J., Sankoff D., LaRue B., Grosjean H. The evolving tRNA molecule. CRC Crit Rev Biochem. 1981;11(1):35–104. doi: 10.3109/10409238109108699. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryer D. R., Eccleshall R., Marmur J. Isolation of yeast DNA. Methods Cell Biol. 1975;12:39–44. doi: 10.1016/s0091-679x(08)60950-4. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Vournakis J. N., Donis-Keller H., Chaconas G., Dougall D. K., Kafatos F. C. End labeling of enzymatically decapped mRNA. Nucleic Acids Res. 1977 Dec;4(12):4165–4174. doi: 10.1093/nar/4.12.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S rRNA sequences and their precursors. Nucleic Acids Res. 1980 Jan 11;8(1):r31–r47. doi: 10.1093/nar/8.1.197-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Gangloff J., Keith G., Ebel J. P., Dirheimer G. The primary structure of aspartate transfer ribonucleic acid from brewer's yeast. II. Partial digestions with pancreatic ribonuclease and T 1 ribonuclease and derivation of complete sequence. Biochim Biophys Acta. 1972 Jan 31;259(2):210–222. [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay B. K., Schmidt O., Gall J. G. In vitro transcription of cloned 5S RNA genes of the newt Notophthalmus. J Cell Biol. 1981 Aug;90(2):323–331. doi: 10.1083/jcb.90.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya H., Miyazaki M., Takemura S. The nucleotide sequence of 5S ribosomal RNA from Schizosaccharomyces pombe. J Biochem. 1981 May;89(5):1663–1666. doi: 10.1093/oxfordjournals.jbchem.a133365. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Lasar Raman evidence for a new cloverleaf secondary structure for eucaryotic 5 S RNA. J Mol Biol. 1978 Oct 15;125(1):95–105. doi: 10.1016/0022-2836(78)90256-5. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Petes T. D. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980 Mar;19(3):765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 1975;12:45–64. doi: 10.1016/s0091-679x(08)60951-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Schmidt O., Mao J. I., Silverman S., Hovemann B., Söll D. Specific transcription of eukaryotic tRNA genes in Xenopus germinal vesicle extracts. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4819–4823. doi: 10.1073/pnas.75.10.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber M. E., Dietrich R. Gene-enzyme relationships in the tryptophan pathway of Schizosaccharomyces pombe. Experientia. 1973 Sep 15;29(9):1152–1154. doi: 10.1007/BF01946778. [DOI] [PubMed] [Google Scholar]
- Sege R., Söll D., Ruddle F. H., Queen C. A conversational system for the computer analysis of nucleic acid sequences. Nucleic Acids Res. 1981 Jan 24;9(2):437–444. doi: 10.1093/nar/9.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Yanofsky C., Driftmier K., Metzenberg R. L., Alzner-DeWeerd B., RajBhandary U. L. Dispersed 5S RNA genes in N. crassa: structure, expression and evolution. Cell. 1981 Jun;24(3):819–828. doi: 10.1016/0092-8674(81)90107-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Silverman S., Gillam I. C., Tener G. M., Söll D. The nucleotide sequence of lysine tRNA2 from Drosophila. Nucleic Acids Res. 1979 Feb;6(2):435–442. doi: 10.1093/nar/6.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Becam A. M., Claisse M., Lederer F. Cytochrome c from Schizosaccharomyces pombe. 2. Amino-acid sequence. Eur J Biochem. 1978 May 16;86(2):407–416. doi: 10.1111/j.1432-1033.1978.tb12323.x. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stulz J., Ackermann T., Appel B., Erdmann V. A. Determination of base pairing in yeast 5S and 5.8S RNA infrared spectroscopy. Nucleic Acids Res. 1981 Aug 11;9(15):3851–3861. doi: 10.1093/nar/9.15.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980 Apr 3;284(5755):426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- Walseth T. F., Johnson R. A. The enzymatic preparation of [alpha-(32)P]nucleoside triphosphates, cyclic [32P] AMP, and cyclic [32P] GMP. Biochim Biophys Acta. 1979 Mar 28;562(1):11–31. doi: 10.1016/0005-2787(79)90122-9. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]