Abstract
This paper introduces a new method to quantify and characterize shape changes during early facial development without the use of landmarks. Landmarks are traditionally used in morphometric analysis, but very few can be identified reliably across all stages of embryonic development. This method uses deformable registration to produce a dense vector field describing the point correspondences between two images. Low and mid-level features are extracted from the deformable vector field to find regions of organized differences that are biologically relevant. These methods are shown to detect regions of difference when evaluated on chick embryo images warped with small magnitude deformations in regions critical to midfacial development.
I. INTRODUCTION
Facial development involves the coordinated growth of multiple three-dimensional structures, which grow towards each other in pairs and fuse into a single craniofacial structure [5]. Any disruption of these structures or the timing of their growth can result in deformations of the midface. These disruptions can be caused by a complex combination of genetic and environmental factors. A detailed characterization of developmental shape change is needed to provide information on which factors play a major role.
Study of the developing face is complicated by the difficulty of measuring its three-dimensional growth. Traditional craniofacial shape analysis methods rely on landmarks that are manually placed on an image by an expert [8] [9]. Landmarks are chosen at locations that are easy to identify across individuals, so the relative difference in position of the landmarks can be compared. However, for embryos, very few landmarks are available that can be reliably identified across samples from different developmental stages.
This work introduces a new method for comparing shape differences without the use of landmarks, developed for the analysis of optical projection tomography (OPT) images of chick embryos at different developmental stages. OPT imaging is highly suited for imaging small specimens up to 2cm in diameter with isotropic resolutions up to 3 microns. In this method, deformable registration is used to provide a dense vector field describing the point correspondences between two images at each point. This is equivalent to using continuous landmark data. The use of this vector field to describe shape differences poses a challenge in terms of dimensionality. This challenge is addressed by novel measures to identify regions where the shape changes are likely to be biologically relevant.
II. RELATED WORK
A. Geometric Morphology
Geometric morphometrics (GM) is the most commonly used method for measuring the difference in shape between two biological samples. This method relies on the use of landmarks, which can be precisely identified on each individual and are placed manually by an expert. Landmarks are typically located at the junctures of identifiable structures. The gold standard for aligning landmarks across samples and obtaining their relative shape information is the Procrustes method [2], [6]. Comparisons between aligned landmarks can be made across groups using statistical analysis [8], [9].
B. Deformation-Based Morphology
Ashburner et al. introduced the term deformation-based morphology to describe a method for analyzing group differences in brain shape using deformation vector fields, which arose as a spin-off from the problem of brain registration [1]. The deformation vector field of each individual describes the spatial normalization of that image to a standard anatomical space. In [1] multivariate statistics similar to those used in traditional GM analysis are applied to the vector field parameters of a group. These statistics are used to discriminate between groups with different gross morphometry.
Deformation morphology has also been applied to more standard morphometric measurement applications. Olafsdottir et al. present a computational mouse atlas in [4] that represents the average of a group of normal, wild-type mice. This atlas is warped to each subject from a normal or mutated strain. Principal Component Analysis (PCA), Independent Component Analysis (ICA) and Sparse Principal Component Analysis (SPCA) are all applied to reduce the data dimensionality and find the modes of the deformation that can discriminate between groups.
III. DATA AND PREPROCESSING
To investigate facial shape differences of chick embryos, the external contour of a sample needs to be extracted from the OPT scan image. Noise in the image can cause bright pixels that are reflected outside the object, gaps or holes, or indistinct borders between the object and the background. In addition, the internal structure of the sample, such as the brain cavity and eye lenses, can result in a complex segmentation rather than the external contour that is desired. For this application, a method based on geodesic active contours is used to address these issues and locate the surface of the sample [3]. Once the contours have been extracted, an affine transformation is applied to align the images and remove differences in pose.
IV. METHODOLOGY
The goal of the method presented in this work is to find and describe significant differences between a pair of chick embryo images. A deformable registration is used to assess local differences at every point between two images, or an image and a mirrored copy. Regions of interest are identified and features are extracted that will be used to characterize the regions.
The deformable registration determines the spatial transform mapping points from a source to homologous points on an object in a target image. The output is a dense deformation vector field in which the vector at each point describes the spatial transformation of that point. When applied to three-dimensional images of two objects, these vectors reflect the structural differences between the source and target objects. For this application, a B-spline deformable transform using a mutual information metric was chosen, since it is widely applicable and computationally efficient.
To interpret the deformation vector field in a meaningful way, it is necessary to define which differences between two images are significant. For this application, differences in a region showing organization are assumed to be significant. This may indicate areas where an underlying process is directing the difference in shape, in contrast to random fluctuations. While all vectors in such a region may not have similar values for properties such as magnitude or angle, variations should occur smoothly over the surface.
A. Low-Level Vector Field Properties
To locate regions in the deformable vector field with some form of organization, three low-level vector properties were chosen. The properties selected are:
deformation vector magnitude,
cosine distance between the deformation vector and the surface normal vector,
cosine distance between the deformation vector and a predefined reference vector.
Each property can be extracted from the vector field and used as low-level features or to calculate mid-level features.
B. Mid-Level Vector Field Properties
Once the low-level features have been extracted from the vector field, they can be used to calculate mid-level features which identify areas of organization.
1) Average Neighborhood Similarity Measure
The average neighborhood similarity is defined as the average difference between a point and its neighbors within a radius r. When the local similarity is calculated for a vector field property, the value at each point represents the difference between that point and its neighborhood average. A low value of average neighborhood difference indicates that the voxels surrounding the center point are similar in value. Groups of points that are spatially connected and have a low level of neighborhood difference indicate a region of interest.
2) Local Entropy Measure
Entropy can be interpreted as a measure of disorder or unpredictability, so it is a natural choice for a metric when looking for organization in a data set. Given N observations {x1, x2, …, xN}, which occur with probabilities {p1, p2, …, pN}, the Shannon entropy is inversely proportional to the log of the probability of observation i, which indicates that the less likely the observation, the higher its entropy will be [7]. A set of observations where all values are equally likely will have low predictability, since there are no dominant values, which results in high entropy. Conversely, a group of observations with higher predictability, which are clustered around a few values, will have lower entropy.
The local entropy measure is used at each point to calculate the entropy of a neighborhood with radius r centered on that point. Points with low entropy are centered on a neighborhood where the values contain a high degree of predictability or order. Spatially connected regions that have similar, low levels of entropy indicate areas of interest.
C. Clustering and Merging Similar Regions
Once low-level or mid-level features have been identified they are clustered to form regions. For low-level features, these are regions of similarity. For mid-level features, the regions represent areas with feature organization. K-means clustering was chosen for this task because it is computationally efficient and effective. One of the main challenges in using the conventional K-means algorithm is that the value of K needs to be estimated or known in advance. This problem was avoided by choosing a value of K higher than the expected value, since similar regions will be merged in a later step. Spatial constraints can be enforced so that spatially disconnected clusters are split apart and clusters with a very small number of voxels can be eliminated.
The last step of the algorithm merges clusters with different means that are part of the same spatially varying pattern. The goal is to identify neighboring clusters where the voxels on the cluster borders have a high level of similarity. To accomplish this, the edge set of voxels from each cluster border is identified. For each pair of neighboring clusters, the vector property values for neighboring voxels on each side of the border are compared to get a value expressing the similarity at the border of the two clusters. Lastly, a similarity threshold is applied to the border similarity values and this determines whether the clusters should be merged.
V. EXPERIMENTAL RESULTS
In this section, results are presented which motivate the use of the methods described in the previous section. Examples of results from the K-means clustering method of low-level vector characteristics, average neighborhood similarity measure, and local entropy measure are shown.
A. Synthetic Images
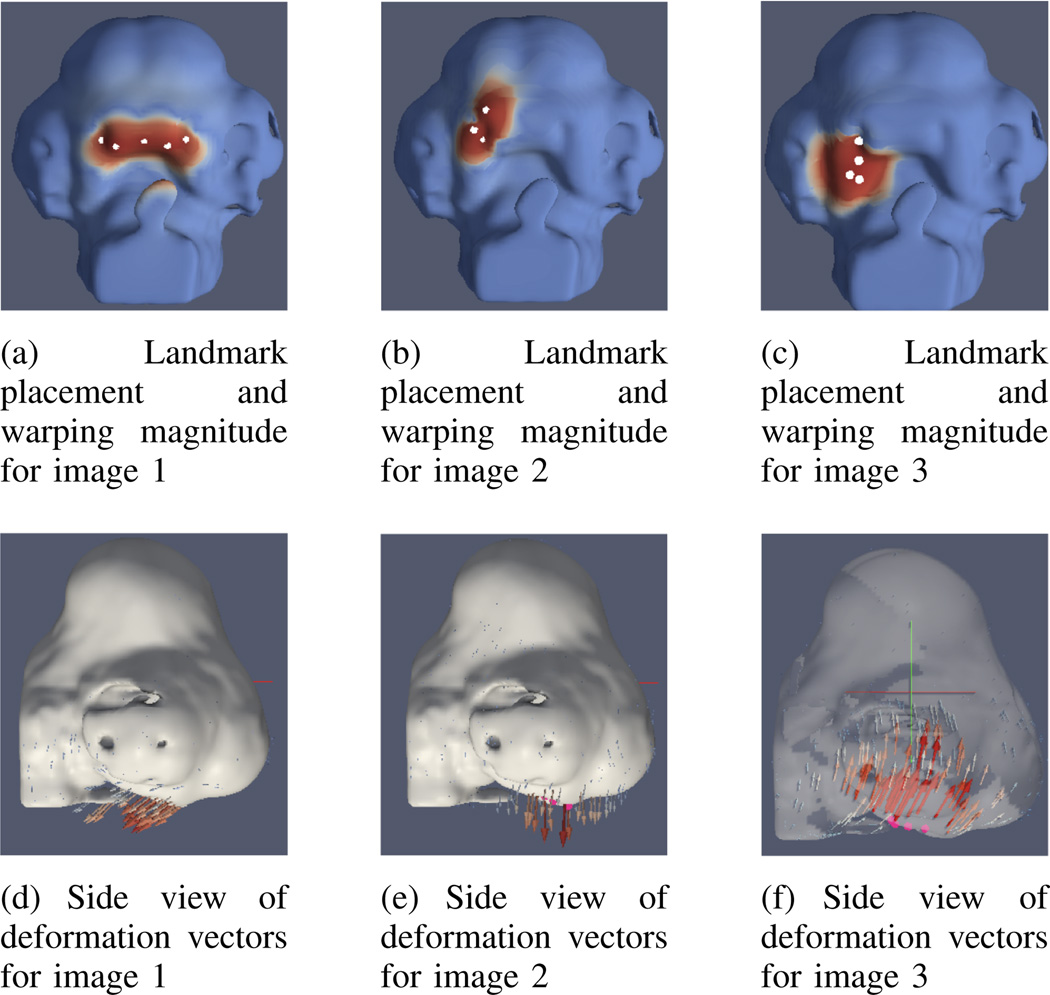
To test the methods presented in this work, a synthetic data set was created. One image of a real chick embryo head was chosen as a reference and deformed to create images with predetermined differences. The synthetic images were generated using landmark warping to create small magnitude deformations in regions critical to the development of the midface. In landmark warping, a set of landmarks are identified in an image and their desired location in the deformed output image is specified. The transformed landmark points are interpolated to define a transformation that is applied to the whole image. Figure 1 shows initial landmark placement, interpolated warping magnitude, and deformation vectors for three landmark-warped images used for evaluation.
Fig. 1.
Figures 1(a) through 1(c) show landmark points marked in white. Image color indicates the interpolated transform magnitude, with highest values shown in red and lowest in blue. The deformation vectors for these three images are shown in figures 1(d) through 1(f).
B. K-means Clustering of Low-Level Vector Characteristics
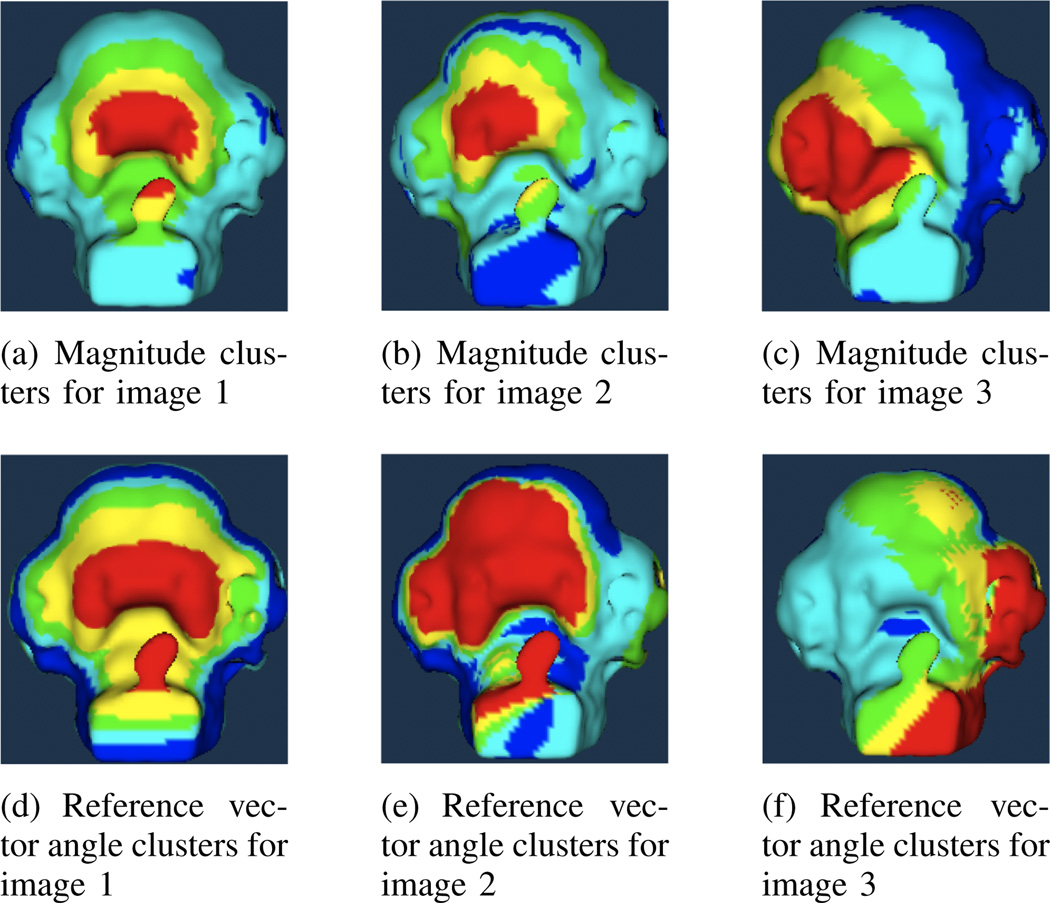
For these experiments, each landmark-warped image was registered to the original image, resulting in a vector field describing their differences. Vector characteristics were extracted from the fields and the K-means clustering method was applied with K = 5. The merging step was omitted so initial cluster mean values could be compared across images and related back to ground truth values. Figure 2 shows examples of the results for clustering of the vector magnitude and the reference vector angle. In all figures, clusters with the highest mean value are shown in red and those with the lowest mean value are shown in blue. In figures 2(a) through 2(c) it can be seen that the vector magnitude clusters with the highest mean are located in areas of high magnitude of the original warping transforms, shown in figures 1(a) through 1(c). In figures 2(d) through 2(f), the reference vector angle values find clusters around similar regions. The area incorporated is larger, since it includes regions where the transform magnitude is lower but the angle is similar as it is interpolated across the image. In these images the cluster means represent direction rather than magnitude. The first two images show the cluster around the warped region in red. The third image shows this region in cyan, indicating the different direction of its landmark warping. Normal vector angle results are not shown, since they are not suited to landmark-warped images, which are deformed without respect to surface curvature.
Fig. 2.
Magnitude and reference vector clusters for three landmark-warped images.
To validate these findings, the clusters can be compared to the transformation used to create the data set. The vector magnitude cluster with the highest mean should occur where the magnitude of the landmark transformation used to deform the original image is the highest. To show this, a second synthetic data set of 25 images was developed by placing a 5x5 grid over the midface region. A warped image was generated for each grid square using 4 landmarks placed in that square. Table I shows the ranked magnitude clusters, mean value of the landmark transformation magnitude for the cluster regions averaged across the 25 regions, and the standard deviation of the mean values. It also indicates the total number of deformation landmark locations present in each cluster. It can be seen that the cluster rank corresponds to increasing average landmark transformation magnitude. Furthermore, the cluster with the highest rank identifies the location of all the landmarks used to define the transformations.
TABLE I.
Relationship between vector magnitude clusters and landmark warping transforms, averaged over 25 images
| Cluster Rank |
Mean Warping Transform Average |
Mean Warping Transform STD |
Landmarks in cluster (100 total) |
|---|---|---|---|
| 1 | 2.026 | 0.577 | 100 |
| 2 | 0.474 | 0.217 | 0 |
| 3 | 0.206 | 0.069 | 0 |
| 4 | 0.087 | 0.025 | 0 |
| 5 | 0.022 | 0.004 | 0 |
C. Neighborhood Similarity Measure
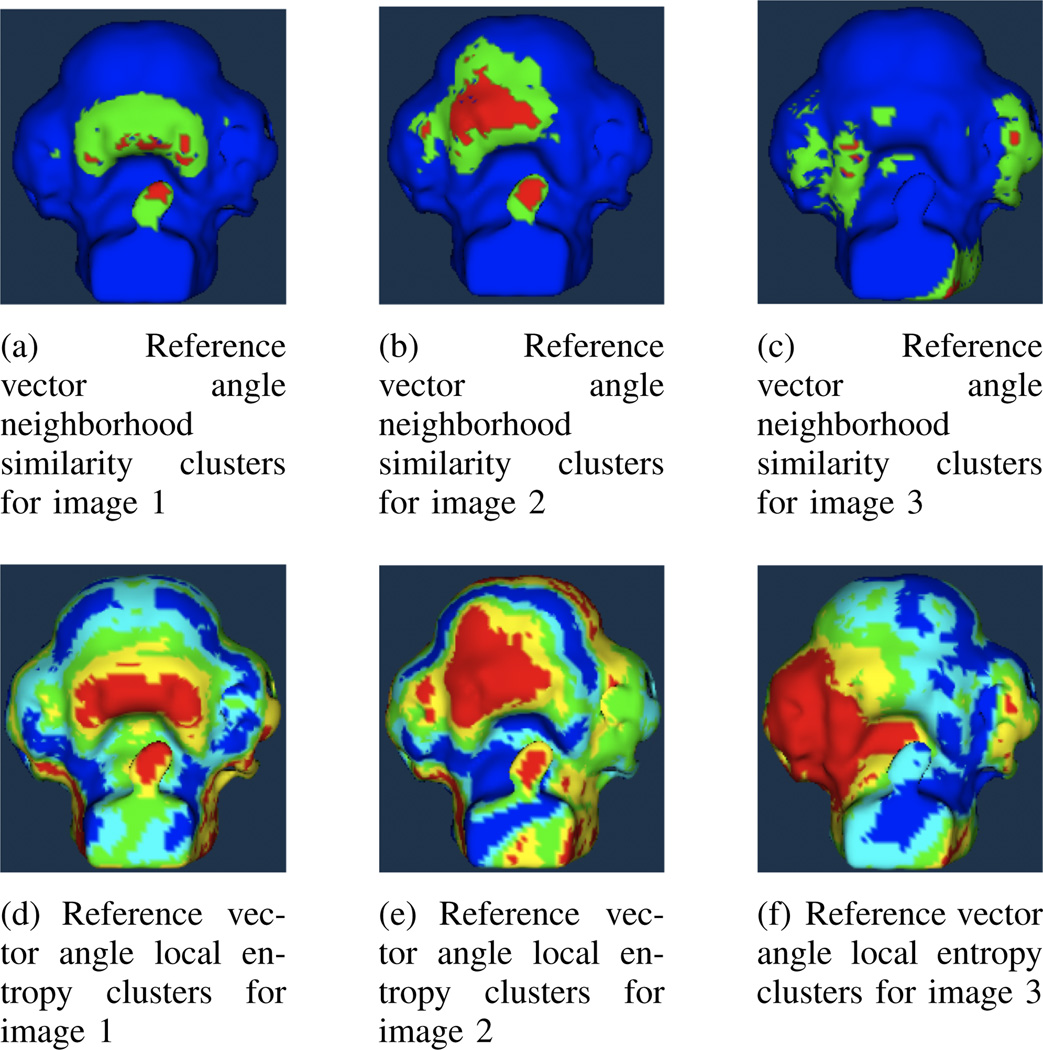
Figures 3(a) through 3(c) show example results from the neighborhood similarity measure. This measure is shown applied to the reference vector values for each of the three warped images. High values of neighborhood similarity indicating regions of interest are shown in red. Clusters with low values are shown in blue. The landmark-warped regions have high levels of neighborhood similarity in each case, with the highest values located near the landmark points.
Fig. 3.
Neighborhood similarity and local entropy of reference vector angles for three landmark-warped images.
D. Local Entropy Measure
Figures 3(d) through 3(f) show example results from the local entropy measure applied to the reference vector angle values for the three warped images. For local entropy, low values indicate regions of interest, so clusters with the lowest mean values are highlighted in red and clusters with the highest mean values are shown in blue. It can be seen that the landmark-warped regions are surrounded by clusters with the lowest entropy in each case.
VI. CONCLUSION AND FUTURE WORK
In this paper, a new method is introduced that is shown to be capable of detecting differences between two images without the use of landmarks. The future work planned will extend this method towards the goal of developing a framework to produce quantitative characterizations of normal embryonic growth patterns.
The experimental results, summarized in table II, show that the vector field features, K-means clustering method, average neighborhood difference measure, and local entropy measure are capable of detecting regions of significant difference between two images. Future work will explore how the clusters detected by each technique can be combined to identify regions of organized differences between images. Features will be extracted to describe these regions and their spatial relationships and will be used to calculate an average distance between two images.
TABLE II.
Summary of Feature Clustering Results
| Feature | Results |
|---|---|
| Magnitude | Landmark-warped regions identified by clusters with highest mean |
| Reference Vector Angle | Regions similar to magnitude identified, mean represents direction |
| Normal Vector Angle | Not suited to landmark warping |
| Local Similarity | Highest mean clusters located in landmark-warped region |
| Local Entropy | Landmark-warped region identified by clusters with highest mean |
The ability to describe and differentiate between developmental stages and between normal and abnormal growth patterns will provide an important tool to further the understanding of the complex and interrelated genetic and environmental factors that lead to common defects of the midface. This method will also be used in the future to analyze tooth and bone shape variation. It can be used to assess shape change in many applications and is especially relevant when the specimens are highly curved or have few definite landmarks.
Acknowledgments
This research was supported by NIH/NIDCR under grant numbers 1U01DE020050-01 (PI: L. Shapiro) and DE018456 (PI: T. Cox).
References
- 1.Ashburner J, Hutton C, Frackowiak R, Johnsrude I, Price C, Friston K. Identifying global anatomical differences: deformation-based morphometry. Human Brain Mapping. 1998;6(5–6):348–357. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<348::AID-HBM4>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gower JC. Generalized procrustes analysis. Psychometrika. 1975;40(1):33–51. [Google Scholar]
- 3.Kass M, Witkin A, Terzopoulos D. Snakes: Active contour models. International journal of computer vision. 1988;1(4):321–331. [Google Scholar]
- 4.Olafsdottir H, Darvann TA, Hermann NV, Oubel q, Ersboll BK, Frangi AF, Larsen P, Perlyn CA, Morriss-Kay GM, Kreiborg S. Computational mouse atlases and their application to automatic assessment of craniofacial dysmorphology caused by the crouzon mutation fgfr2c342y. Journal of Anatomy. 2007;211(1):37–52. doi: 10.1111/j.1469-7580.2007.00751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons TE, Kristensen E, Hornung L, Diewert VM, Boyd SK, German RZ, Hallgrímsson B. Phenotypic variability and craniofacial dysmorphology: increased shape variance in a mouse model for cleft lip. Journal of Anatomy. 2008;212(2):135–143. doi: 10.1111/j.1469-7580.2007.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohlf FJ, Slice D. Extensions of the procrustes method for the optimal superimposition of landmarks. Systematic Biology. 1990;39(1):40. [Google Scholar]
- 7.Shannon CE. A mathematical theory of communication. ACM SIGMOBILE Mobile Computing and Communications Review. 2001;5(1):3–55. [Google Scholar]
- 8.Zelditch ML, Lundrigan BL, Garland T., Jr Developmental regulation of skull morphology i: Ontogenetic dynamics of variance. Evolution & development. 2004;6(3):194–206. doi: 10.1111/j.1525-142X.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 9.Zelditch ML, Mezey J, Sheets HD, Lundrigan BL, Garland T. Developmental regulation of skull morphology ii: Ontogenetic dynamics of covariance. Evolution & Development. 2006;8(1):46–60. doi: 10.1111/j.1525-142X.2006.05074.x. [DOI] [PubMed] [Google Scholar]