Abstract
The transcription factors that control lineage specification of haematopoietic stem cells (HSCs) have been well described for the myeloid and lymphoid lineages, whereas transcriptional control of erythroid (E) and megakaryocytic (Mk) fate is less understood. We here use conditional removal of the GATA-1 and FOG-1 transcription factors to identify FOG-1 as required for the formation of all committed Mk- and E-lineage progenitors, whereas GATA-1 was observed to be specifically required for E-lineage commitment. FOG-1-deficient HSCs and preMegEs, the latter normally bipotent for the Mk and E lineages, underwent myeloid transcriptional reprogramming, and formed myeloid, but not erythroid and megakaryocytic cells in vitro. These results identify FOG-1 and GATA-1 as required for formation of bipotent Mk/E progenitors and their E-lineage commitment, respectively, and show that FOG-1 mediates transcriptional Mk/E programming of HSCs as well as their subsequent Mk/E-lineage commitment. Finally, C/EBPs and FOG-1 exhibited transcriptional cross-regulation in early myelo-erythroid progenitors making their functional antagonism a potential mechanism for separation of the myeloid and Mk/E lineages.
Keywords: haematopoiesis, lineage commitment, stem cells, transcription
Introduction
Lineage specification in the haematopoietic system is highly regulated at the transcriptional level, and for many lineages specific transcription factors have been identified as critical for their specification. Thus, formation of granulocyte-macrophage progenitors (GMPs) is regulated by C/EBPα during steady-state haematopoiesis (Zhang et al, 2004) and by C/EBPβ during stress granulopoiesis (Hirai et al, 2006), whereas Pax5 and GATA-3 control B and T lymphopoiesis, respectively (Urbanek et al, 1994; Ting et al, 1996). One roadblock to understanding how lineage commitment occurs in the myeloid branch of the definitive haematopoietic system has been our limited ability to precisely identify bipotent and monopotent progenitors with defined lineage potentials. In particular, progenitors for megakaryocytic (Mk) and erythroid (E) cells have remained elusive, complicating the identification of their specifying factors. Recently, the identification and prospective isolation of bipotent Mk/E progenitors (preMegEs), unipotent Mk and E progenitors (MkP and preCFU-E/CFU-E, respectively), as well as an early myeloid-committed progenitor (preGM) (Pronk et al, 2007) has opened up the possibility of studying the separation of the GM, Mk and E lineages in greater detail.
Using distinct cell sorting strategies cell populations containing progenitors with both Mk and E potential, but devoid of myeloid or lymphoid potential, have been identified (Akashi et al, 2000; Pronk et al, 2007). Therefore, the divergence of the Mk and E lineages is the last decision made before fully committed Mk and E progenitors arise. Candidate lineage specifying factors are Klf1 (also known as EKLF) and Fli-1. Klf1 is preferentially expressed in Mk/E bipotent and E-committed progenitors, but is downregulated in committed Mk progenitors (Frontelo et al, 2007). Conversely, Fli-1 expression is downregulated upon E-lineage commitment and upregulated in Mk progenitors. In murine embryoid bodies and human CD34+ cord blood cells ectopic Klf1 expression suppressed both Fli-1 expression and Mk progenitor formation, whereas mutation or knockdown of Klf1 (or its human homologue) was found to increase Mk differentiation (Frontelo et al, 2007; Bouilloux et al, 2008). Conversely, Fli-1 loss-of-function mutation, in both mouse and human, caused Mk differentiation defects (Hart et al, 2000; Starck et al, 2010), whereas its overexpression promoted megakaryopoiesis and suppressed erythropoiesis (Athanasiou, 1996, 2000). Finally, Fli-1 antagonizes Klf1 activity through protein–protein interaction (Starck et al, 2003), indicating Fli-1–Klf1 cross-antagonism as a potential mechanism for Mk–E-lineage separation. However, since Klf1 and Fli-1 deficiency causes defective erythroid (Nuez et al, 1995; Perkins et al, 1995) and megakaryocyte differentiation (Hart et al, 2000; Spyropoulos et al, 2000; Starck et al, 2010), respectively, rather than lineage commitment defects, it is possible that the function of their antagonism is lineage ‘lockdown’, with other transcriptional regulators involved in actual lineage specification.
The current paradigm for separation of the GM and Mk/E lineages states that PU.1 and GATA-1 maintain the multipotent state through a combination of mutual functional inhibition and autoregulation (Burda et al, 2010). This model is supported by gain-of-function studies where PU.1 expression induces myeloid-lineage commitment, and GATA-1 induces erythroid/megakaryocytic commitment or potential, in heterologous or multipotent cell types (Burda et al, 2010). Conditional PU.1 deletion in the adult haematopoietic system prevents the formation of both the common lymphoid progenitor (CLP) population and common myeloid progenitor (CMP) population (which includes Mk/E-committed cells; Pronk et al, 2007) and leads to the generation of self-renewing myeloid cell populations (Dakic et al, 2005; Iwasaki et al, 2005), consistent with a role in early myeloid-lineage specification. The results of GATA-1 loss-of-function studies are more ambiguous, since GATA-1 deletion has not been reported to enhance myelopoiesis or impair Mk/E-lineage commitment (Gutierrez et al, 2008). The requirements for Mk/E-lineage commitment has therefore remained elusive.
In addition to PU.1 also C/EBPα and C/EBPβ have been shown to mediate myeloid-lineage commitment. In the absence of C/EBPα, GMPs are not formed during steady-state haematopoiesis (Zhang et al, 2004), whereas C/EBPβ promotes GMP formation during stress haematopoiesis (Hirai et al, 2006). Based on the hypothesis that mediators of Mk/E-lineage commitment would exhibit an antagonistic relationship to C/EBPs similar to that of PU.1–GATA-1 and Klf1–Fli-1 we examined C/EBP-deficient haematopoiesis, and found that preGMs blocked from proceeding to the GMP stage upregulate Gata1, Gata2, Zfpm1 (encoding FOG-1), Fli-1 and Sfpi1 (encoding PU.1). As it was previously observed that FOG-1 was able to confer Mk/E potential on myeloid cells in a GATA-1-dependent manner we initially focused on these two factors. By conditionally deleting Zfpm1 and Gata1, we observed that loss of the former prevented specification of all Mk/E-committed progenitors, whereas Gata1 was required for the commitment of bipotent preMegEs towards an erythroid fate. Therefore, FOG-1 and GATA-1 act sequentially to generate and specify Mk/E progenitors during definitive haematopoiesis.
Results
Depletion of C/EBPs alters lineage potential and gene expression of preGMs
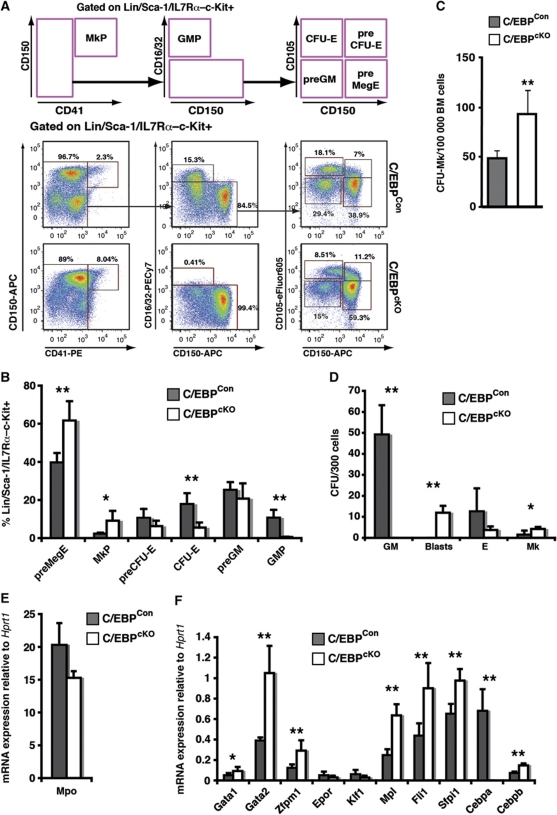
Based on the hypothesis that factors promoting Mk/E and myeloid commitment would behave antagonistically we conditionally depleted C/EBP transcription factors, known to promote myeloid progenitor formation, from the haematopoietic system. We generated mice carrying two floxed Cebpa alleles and one floxed Cebpb allele, along with the Mx1-Cre transgene (Cebpafl/fl; Cebpbfl/+; Mx1-Cretg/+: C/EBPcKO mice) and corresponding control mice lacking the Mx1-Cre transgene (Cebpafl/fl; Cebpbfl/+: C/EBPCon mice). One Cebpb allele was maintained as wild type since we observed that Mx1-Cre mediated deletion of all four Cebpa and Cebpb alleles led to rapid lethality (L Luciani and C Nerlov, unpublished). Recombination was initiated by injection of poly(I-C), which in the presence of Mx1-Cre leads to near-complete recombination in haematopoietic tissues (Kirstetter et al, 2006). After 12 days, the effect of C/EBP depletion was analysed by progenitor phenotyping (Pronk et al, 2007). We observed an overall expansion of the stem/progenitor compartment (data not shown), consistent with results previously obtained from Cebpa knock-in mice (Porse et al, 2005; Bereshchenko et al, 2009), which likely reflect loss of cell-cycle control in the stem cell compartment. C/EBPcKO bone marrow stem/progenitor cells were unable to generate GMPs, consistent with results previously obtained for conditional Cebpa knock-out mice (Zhang et al, 2004), while phenotypic myeloid-committed preGM cells were formed (Figure 1A and B). In addition, we saw a significant increase in MkPs as a proportion of the Lin/Sca-1/IL7Rα–c-Kit+ progenitor population, as well as an increase in the number of Mk colony forming cells (Figure 1C). The presence of preGM progenitors, which normally show predominantly myeloid potential (Pronk et al, 2007), in the absence of further differentiation along the myeloid lineage, led us to investigate the gene expression and lineage potential of these cells. Colony forming assays on sorted preGMs under GM, Mk and E conditions showed an overall decrease in clonogenic potential, but with the proportion of CFU-Mk being increased in both relative and absolute terms (Figure 1D). This was accompanied by upregulation of Mk lineage-associated genes, including Mpl, Fli1, Zfpm1, Gata1 and Gata2 (Figure 1E and F). Most of these encode transcriptional regulators with lineage regulatory potential. In particular, GATA-1 and FOG-1 have previously been implicated in extinction of myeloid-lineage programming and acquisition of Mk/E potential (Kulessa et al, 1995; Querfurth et al, 2000). We, therefore, selected these two factors for further study.
Figure 1.
C/EBP depletion expands stem/progenitor cells and activates Mk potential. (A) Representative FACS profiles of C/EBPCon and C/EBPcKO bone marrow 12 days after poly(I-C) treatment. The top panel shows the gating strategy and gates used to define the progenitor populations. (B) Progenitor population sizes determined as in (A) expressed as percentage of the Lin/Sca-1/IL7Rα–c-Kit+ fraction for C/EBPCon (n=4) and C/EBPcKO (n=5) mice. (C) Total C/EBPCon and C/EBPcKO bone marrow cells were plated in MegaCult assays, and the number of Mk colonies determined. Each measurement was performed in quadruplicate. Error bars indicate standard deviations (**P<0.01; Student's t-test). (D) Lineage potential of C/EBPCon and C/EBPcKO preGM cells was assayed separately under GM (M3534), E (M3436) and Mk (MegaCult) conditions. In the case of GM conditions, colonies with immature blast morphology (blasts) are shown separately from normal GM colonies. The average number of colony forming units (CFUs) per 300 cells plated is given. Each value represents the average of four individual mice, each measured in triplicate. (E, F) Gene expression analysis of C/EBPCon and C/EBPcKO preGM cells (C/EBPCon: n=4; C/EBPcKO: n=5). Data show average mRNA expression normalized to Hprt1, each mouse assayed in triplicate. Error bars indicate standard deviations and asterisks indicate statistical significance (*P<0.05; **P<0.01; Student's t-test).
GATA-1 is dispensable for preMegE formation, but required for their progression along the erythroid lineage
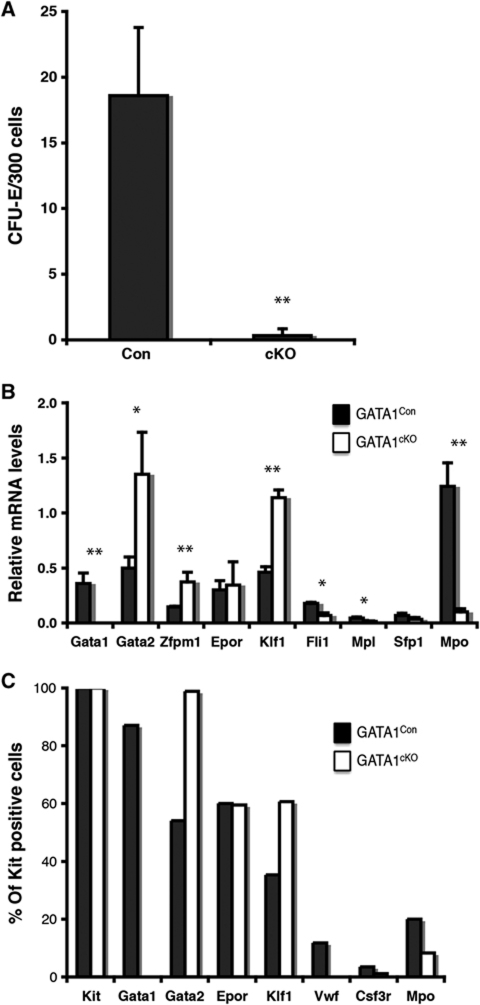
Loss of GATA-1 has previously been observed to block erythroid progenitors from terminally differentiating, but effects on lineage specification have not been addressed. We generated male mice containing a floxed Gata1 allele (in the hemizygous state, since the gene is X-linked) and the Mx1-Cre transgene (Gata1fl/Y; Mx1-Cretg/+: GATA-1cKO mice), as well as the corresponding control mice (Gata1+/Y; Mx1-Cretg/+: GATA-1Con mice). Twelve days after starting poly(I-C) treatment, we observed no effect on platelet or leukocyte numbers (Figure 2A and B), but a specific loss of peripheral red blood cells (Figure 2B). The lack of any significant effect of GATA-1 deficiency on platelets is consistent with a previous study demonstrating a mild megakaryocyte differentiation defect in the absence of GATA-1 (Gutierrez et al, 2008). In accordance with that study, we also observed accumulation in the bone marrow of early erythroid progenitors, apparently blocked at the pro-erythroblast stage (Figure 2C). However, a significant decrease in the total number of erythroid progenitors (Figure 2D) suggested that also their specification could be impaired. Flow cytometric phenotyping of the progenitor compartment showed a complete loss of the earliest stages of committed erythroid progenitors, preCFU-Es and CFU-Es (Figure 2E–G). This was accompanied by a significant increase in MkPs, whereas preGM/GMP numbers were unaffected. The GM versus Mk/E bifurcation, therefore, seems not to involve GATA-1, whereas the divergence of the E and Mk lineages does. However, the possibility existed that erythroid progenitors formed, which were undetectable by surface marker expression. To resolve this, we sorted preMegEs from GATA-1Con and GATA-1cKO mice, and tested their erythroid colony forming ability. This showed that GATA-1-deficient preMegEs were unable to form erythroid colonies (Figure 3A). Potentially, this could be caused by the inability of GATA-1-deficient preMegEs to respond to erythropoietin, the cytokine driving the CFU-E assay, rather than their inability to form erythroid cells. However, gene expression analysis of GATA-1Con and GATA-1cKO preMegEs showed that Gata1 expression was lost upon deletion, whereas Epor expression was normal. Gata2, Klf1 and Zfpm1 were upregulated (Figure 3B), indicating that the encoded factors, all of which are associated with erythroid differentiation, are insufficient to mediate lineage commitment in the absence of GATA-1. Analysis of gene expression at the single cell level gave a similar result: the proportion of Epor expressing cells was not altered, whereas Klf1 and Gata2 expressing cells were increased (Figure 3C). Overall, these results indicate that GATA-1 is required for erythroid development at the level of lineage commitment, whereas GATA-1 deficiency did not have any detectable effect on divergence of the GM lineages from the Mk/E lineages.
Figure 2.
Loss of GATA-1 prevents formation of committed erythroid progenitors. (A) Differential count of peripheral blood cells 12 days after poly(I-C) treatment (GATA-1Con: n=10; GATA-1cKO: n=8). WBCs, total white blood cells; NEs, neutrophils; LYs, lymphocytes; MOs, monocytes; PLT, platelets. (B) Red cell parameters measured as in (A). HCT, haematocrit; Hb, haemoglobin level; RBC, red blood cell count. Error bars show standard deviations. Asterisks indicate statistical significance (*P<0.05; **P<0.0001; ***P<0.00001). (C) Total bone marrow cells from GATA-1Con (left panel) and GATA-1cKO (right panel) mice day 12 after poly(I-C) injection were stained for CD71 and Ter119 to identify erythroid progenitor populations. Values shown are the size of gated populations as percentage of the total number of cells. I, pro-erythroblasts; II, basophilic erythroblasts; III, polychromatophilic erythroblasts; IV, orthochromatophilic erythroblasts. (D) Bar graphs represent percentage of the total number of cells in BM for each population gated as in (C). (E) Representative flow cytometric analysis of the experimental progenitor population from GATA-1Con and GATA-1cKO mice 12 days after poly(I-C) treatment. The schematic representation (top) indicates the gating strategy used for separating the different myelo-erythroid progenitor sub-populations from GATA-1Con (middle panels) and GATA-1cKO (bottom panels). The size of each population as percentage of the parental population is shown next to each gate. (F) Bar graphs represent the average size of each myelo-erythroid progenitor population as percentage of the Lin/Sca-1/IL7Rα–c-Kit+ fraction in GATA-1Con (n=6) and GATA-1cKO (n=6) poly(I-C)-treated mice. (G) Absolute number of myelo-erythroid progenitor from each of the populations analysed in (F). Numbers represent cells retrieved from tibias and femurs of each mouse.
Figure 3.
Loss of erythroid colony forming potential by GATA-1-deficient preMegEs. (A) CFU-E forming capacity of GATA-1Con and GATA-1cKO preMegEs. preMegE cells were sorted from GATA-1Con and GATA-1cKO mice 12 days after poly(I-C) injection and assayed for CFU-E activity (M3436). The values represent the average of three individually sorted mice, each assayed in duplicate. (B) Gene expression analysis of GATA-1Con and GATA-1cKO preMegEs obtained as in (A) was performed using real-time PCR. Values represent the average obtained from three individually sorted mice, each assayed in triplicate. Error bars indicate standard deviations and asterisks indicate statistical significance (*P<0.05; **P<0.01; Student's t-test). (C) Single cell level gene expression in GATA-1Con and GATA-1cKO preMegEs analysed by nested multiplex PCR. Values represent the percentage of cells scored as positive for the indicated transcripts. More than 85 single cells were assayed per genotype, and those scoring positive for Kit mRNA were included in the analysis (<3% were negative).
FOG-1 is required for megakaryocyte and erythroid-lineage commitment
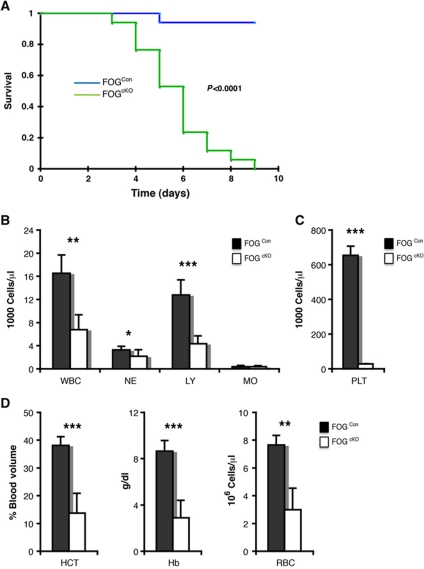
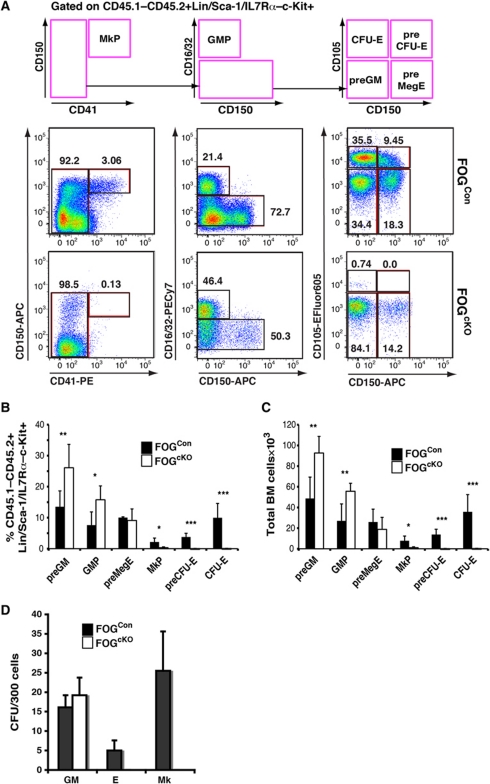
GATA-1 is co-expressed with GATA-2 in most Mk/E progenitors and mature cells, and redundancy between these factors could explain why Gata1 deletion did not affect GM progenitor formation. Both GATA-1 and GATA-2 use FOG-1 as a co-factor during Mk/E differentiation (Chang et al, 2002). We, therefore, conditionally inactivated the Zfpm1 gene as a way to access the combined function of the two GATA factors, since FOG-1 is not known to have GATA-independent activities. Initial experiments, using Zfpm1fl/fl; Mx1-Cretg/+ (FOGcKO genotype) and Zfpm1+/+; Mx1-Cretg/+ (FOGCon genotype) mice resulted in very rapid lethality of the FOGcKO mice upon poly(I-C) injection. Since FOG-1 is expressed in several non-haematopoietic tissues, we transplanted total bone marrow from FOGcKO and FOGCon mice (CD45.2 allotype) into wild-type recipients (CD45.1/2 allotype). One month after transplantation the exclusive reconstitution of the recipient haematopoietic system by the donor cells was verified by analysis of the peripheral blood (PB; Supplementary Figure S1), and recombination induced by poly(I-C) injection. This led to lethality within 8 days of the FOGcKO mice, with no loss of viability of FOGCon mice (Figure 4A), accompanied by severe anaemia and thrombocytopenia (Figure 4B–D). However, the transient effects of interferon-β induced by poly(I-C) on the haematopoietic stem/progenitor compartment precluded detailed analysis of progenitor populations before the lethal effects of the deletion occurred. To circumvent this problem, we next included wild-type CD45.1 competitor cells in the transplantation experiment in a 1:1 ratio with FOGcKO and FOGCon cells. No lethality was observed after poly(I-C) treatment, and mice could be analysed at steady state 3 months after induction of recombination (Supplementary Figure S2A). Under these conditions, blood values were largely normal, although a decrease in platelet numbers was observed (Supplementary Figure S2B–G). In the donor-derived CD45.1–CD45.2+ BM fraction, we observed that FOGcKO cells displayed a strong bias towards myeloid differentiation: preGMs and GMPs were increased whereas preCFU-E/CFU-E and MkP populations were virtually absent (Figure 5A), both when measured as proportion of the Lin–c-Kit+ progenitor pool (Figure 5B) and as absolute cell number (Figure 5C). No effect was observed on the CD45.1+CD45.2– competitor cells (Supplementary Figure S3), indicating that the effects of FOG-1 depletion on progenitor formation were cell intrinsic. These results showed an inability of FOG-1-deficient haematopoietic stem cells (HSCs) to progress towards MkE differentiation. To confirm this, we performed colony forming assays on the CD45.1–CD45.2+LSKFlt3– fraction: here, FOGCon cells showed CFU-GM, CFU-Mk and CFU-E activity as expected, whereas elevated CFU-GM activity was found in FOGcKO cells, along with a complete loss of MkE potential (Figure 5D).
Figure 4.
Haematopoietic FOG-1 is essential for survival of adult mice. (A) Kaplan–Meier survival curve of wild-type mice transplanted with FOG1Con (n=17) and FOG1cKO bone marrow cells (n=17) and injected with poly(I-C) (150 μg/mouse) 4 weeks after transplantation. The graph shows the fraction of live mice for each genotype. Statistical significance between groups was calculated using the log-rank test. (B) Differential count of peripheral blood cells 5 days after poly(I-C) treatment (FOG1Con: n=7; FOG1cKO: n=8). WBCs, total white blood cells; NEs, neutrophils; LYs, lymphocytes; MOs, monocytes. (C) Platelet (PLT) count measured as in (B). (D) Red cell parameters measured as in (B). HCT, haematocrit; Hb, haemoglobin level; RBC, red blood cell count. Error bars show standard deviations. Asterisks indicate statistical significance (*P<0.05; **P<0.0001; ***P<0.00001).
Figure 5.
FOG-1-deficient haematopoietic stem cells are unable to generate megakaryocyte-erythroid progenitors or colonies. (A) Representative flow cytometric analysis of the experimental progenitor population (CD45.1–CD45.2+Lin/Sca-1/IL7Rα–c-Kit+) from FOGCon (middle panels) and FOGcKO (bottom panels). The schematic representation (top) indicates the gating strategy used for separating the different myelo-erythroid progenitor sub-populations. The size of each population as percentage of the parental population is shown next to each gate. (B) Bar graph showing the average size of each myelo-erythroid progenitor population as percentage of the CD45.1–CD45.2+Lin/Sca-1/IL7Rα–c-Kit+ fraction in FOG1Con (n=10) and FOG1cKO (n=10) competitively transplanted mice. Data were pooled from two independent experiments. (C) Absolute number of myelo-erythroid progenitor from each of the populations analysed in (B). Numbers represent cells retrieved from tibias and femurs of each mouse. (D) Lineage potential in vitro of CD45.1–CD45.2+Lin–Sca-1+c-Kit+Flt3– stem/multipotent progenitor fraction sorted from FOG1Con and FOG1cKO competitively transplanted mice. Three hundred cells were plated in semisolid medium under Mk (MegaCult-C), E (M3436) and GM conditions (M3534), respectively. The lineage potential of each cell population is expressed as the percentage of the total number of colonies formed under all three conditions. Independently transplanted mice (FOG1Con: n=6; FOG1cKO: n=6) were analysed for each genotype in two separate experiments, each performed in triplicate (GM, E) or in quadruplicate (Mk). Error bars show standard deviations and asterisks indicate statistical significance (*P<0.05; **P<0.01; ***P<0.001; Student's t-test).
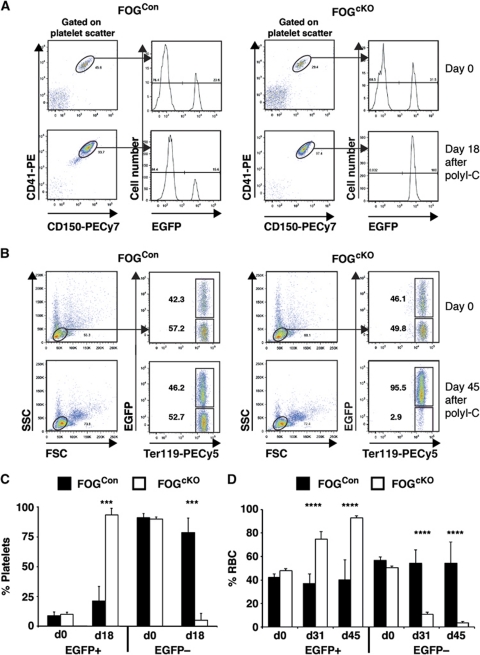
While the absence of FOG-1-deficient committed phenotypic and Mk/E colony forming cells provided strong evidence for a differentiation block along these lineages, it could not be formally excluded that the absence of phenotypic progenitors was due to a change in their surface phenotype, and that FOG-1-deficient HSCs still were able to generate platelets and erythrocytes. The CD45 allotype-specific antibodies cannot be used to address this issue, since neither platelets nor erythrocytes express CD45. In order to address this question, we therefore took advantage of the Vwf-EGFP (Sanjuan-Pla et al, in preparation) and the miR-144/451-EGFP (Rasmussen and O'Carroll, 2011) reporter mouse lines, where platelets and erythrocytes, respectively, are labelled by EGFP. By creating chimeras between unrecombined FOGCon/FOGcKO and Vwf-EGFP mice, followed by poly(I-C) injection, we observed that in the FOGcKO chimeras essentially all EGFP− platelets were lost after 18 days (Figure 6A, C). Similar experiments using the miR-144/451-EGFP knockin showed that EGFP− erythrocytes were lost, with somewhat slower kinetics (Figure 6B, D). Since the output of short-lived myeloid cells from FOGcKO HSCs was maintained in competitively repopulated recipients for several months after poly(I-C) induction (Figure 5A), the loss of EGFP− platelets and erythrocytes indicates that production of these cell types is indeed blocked in the absence of FOG-1.
Figure 6.
FOG-1-deficient haematopoiesis does not produce erythrocytes or platelets. (A) CD45.1/2 recipient mice were co-transplanted with 500 000 Vwf-EGFPtg/+ and 2 000 000 of either FOGCon or FOGcKO bone marrow cells. PB was analysed by flow cytometry 4 weeks after transplantation. Mice were then injected with poly(I-C). Eighteen days after the first poly(I-C) injection (two injections, 2 day intervals) PB analysis was performed. Platelets were identified by scatter and co-expression of CD41 and CD150, and the percentage of platelets expressing EGFP determined. Representative plots are shown. (B) CD45.1/2 recipient mice were co-transplanted with 1 000 000 mir144/451EGFP/+ and 2 000 000 of either FOGCon or FOGcKO bone marrow cells. PB was analysed by flow cytometry 4 weeks after transplantation. Mice were then injected with poly(I-C). In all, 31 and 45 days after the first poly(I-C) injection (three injections, 2 day intervals), PB analysis was performed (only 45 day time point is shown). Erythrocytes were identified by scatter and expression of Ter119, and the percentage of erythrocytes expressing EGFP determined. Representative plots are shown. (C) Summary of data obtained in (A), showing the percentage of EGFP+ and EGFP– platelets before and after poly(I-C) injection. FOGCon: N=7; FOGcKO: N=7. Error bars show standard deviations. Asterisks indicate statistical significance (***P<0.001; Student's t-test). (D) Summary of data obtained in (B), showing the percentage of EGFP+ and EGFP– erythrocytes before and after poly(I-C) injection. FOGCon: N=4; FOGcKO: N=5. Error bars show standard deviations. Asterisks indicate statistical significance (****P<0.0001; Student's t-test).
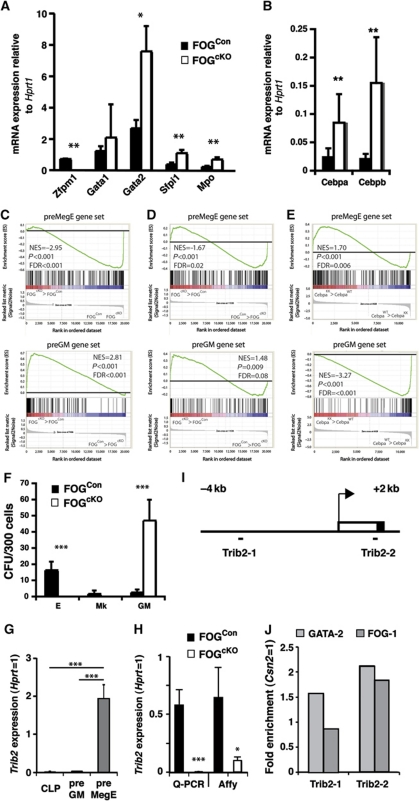
Some residual commitment towards the Mk and E lineages was suggested by the presence of FOGcKO cells with the surface phenotype of bipotent preMegEs. Cell sorting and qPCR gene profiling of this population confirmed the absence of Zfpm1 expression; expression of Gata2 was increased, whereas Gata1 mRNA levels were not significantly altered. Interestingly, myeloid genes (Sfpi1, Mpo) were upregulated in FOGcKO preMegEs (Figure 7A), as were both Cebpa and Cebpb (Figure 7B). Gene profiling at the global level showed that the gene expression pattern of FOGcKO preMegEs was highly significantly depleted of mRNAs associated with the normal preMegE phenotype, with an equally strong upregulation of preGM-associated genes (as defined by Pronk et al, 2007; Figure 7C). Since preMegEs and preGMs define the first committed progenitors derived from the HSC compartment, we used these gene sets to determine whether a lineage commitment bias was introduced into HSCs upon FOG-1 depletion. We observed similar reprogramming in the LSKFlt3− HSC fraction (Figure 7D), with significant depletion of preMegE genes and upregulation of preGM genes. The failure of FOG-1-deficient progenitors to upregulate Mk/E gene expression was, therefore, preceded by defective Mk/E programming of HSCs. Using gene sets defining mature myeloid, erythroid and megakaryocytic cells, we previously observed depletion of mature GM genes in HSCs homozygous for a Cebpa point mutation that disables C/EBPα DNA binding (K313KK or KK allele) (Bereshchenko et al, 2009). Using the early committed progenitor gene sets, we found that the Cebpa KK mutation had the converse effect on HSC lineage programming compared with FOG-1 conditional knockout, leading to preGM gene depletion and preMegE gene enrichment (Figure 7E). The inability of FOG-1-deficient HSCs to produce Mk/E colonies and give rise only to GM myeloid colonies, combined with the myeloid reprogramming of FOGcKO preMegEs raised the question whether GM potential was retained in this phenotypically Mk/E-committed cell population. Sorted FOGCon and FOGcKO preMegEs exhibited a different behaviour when plated under GM and E conditions: the control preMegEs showed high E and Mk potential, and very limited GM potential; FOGcKO preMegEs readily formed colonies only under GM conditions, but generated no E or Mk colonies (Figure 7F). These results are consistent with the loss of Mk/E-lineage commitment from preMegEs being accompanied, and possibly caused by, their inability to extinguish myeloid lineage-associated gene expression and myeloid-lineage potential. To identify putative regulators of FOG-1-dependent extinction of myeloid potential in preMegEs, we mined the gene profiling data to identify those genes that were (i) highly expressed in preMegEs, but not in preGMs and CLPs and (ii) significantly downregulated in FOGcKO preMegEs relative to FOGCon preMegEs (Supplementary Table S1). One of these genes was Trib2, which is known to induce the degradation of C/EBPα and C/EBPβ (Figure 7G). We confirmed the loss of Trib2 expression in FOGcKO preMegEs by Q–PCR (Figure 7H). Finally, we identified putative GATA binding sites in the Trib2 gene (Figure 7I), one of which was found to bind both GATA-2 and FOG-1 in chromatin immune-precipitation (ChIP) experiments performed on sorted preMegEs (Figure 7J).
Figure 7.
Loss of FOG-1 prevents extinction of the GM program in preMegE cells. (A, B) Quantitative RT–PCR on sorted bone marrow preMegE isolated from FOG1Con and FOG1cKO competitive-transplanted mice. Data show average mRNA expression normalized to Hprt1 from three independently transplanted mice, each mouse assayed in triplicate. (C) Gene set enrichment analysis comparing FOG1cKO and FOG1Con preMegEs and (D) LSKFlt3− cells. Panels show enrichment of preMegE (top)- and preGM (bottom)-specific gene sets in the FOG1cKO genotype relative to FOG1Con. The normalized enrichment score (NES), nominal P-value and false discovery rate (FDR) are indicated on each plot. (E) Gene set enrichment analysis comparing CebpaKK and wild-type (WT) control LSK cells. Panels show enrichment of preMegE (top)- and preGM (bottom)-specific gene sets in the CebpaKK genotype relative to wild type. The NES, nominal P-value and FDR are indicated on each plot. (F) Lineage potential of FOG1Con and FOG1cKO PreMegEs, assayed as in Figure 1D. Error bars show standard deviations. Asterisks indicate statistical significance (*P<0.05; **P<0.01; ***P<0.001; Student's t-test). (G) Expression of Trib2 in progenitor subsets based on Affymetrix array analysis of CLP (n=3), preGM (n=3) and preMegE populations (n=5). Expression values are normalized to Hprt expression. Error bars indicate standard deviations. Asterisks indicate statistical significance (***P<0.001; Student's t-test). (H) Expression of Trib2 in control and FOG-1-deficient preMegEs based on real-time Q–PCR (Con: N=3; cKO; N=3) and Affymetrix analysis (Con: N=3; cKO; N=3). Asterisks indicate statistical significance (*P<0.05; ***P<0.001; Student's t-test). (I) Trib2 promoter region, showing the position of the amplicons surrounding putative GATA/FOG binding sites, identified by sequence analysis. Arrow indicates the transcriptional start site; open box 5′ non-coding part of exon 1; closed box coding sequence. (J) ChIP was performed on sorted preMegEs (50 000 cells/IP), using anti-GATA-2, anti-FOG-1 and control IgG antibodies, followed by PCR amplification of Trib2 and control Csn2 amplicons. Values are expressed as enrichment (antibody/IgG ratio) relative to the enrichment observed for the Csn2 amplicon. Each value is the average of triplicate determinations.
Discussion
The results obtained in this manuscript identify FOG-1 and GATA-1 as regulators of Mk/E-lineage specification. Depleting the haematopoietic system of C/EBPs blocked myelopoiesis at the preGM to GMP transition. This was accompanied by increased expression of the genes encoding Mk/E-lineage regulators such as GATA-1, GATA-2 and FOG-1, as well as increased Mk potential within the phenotypic preGM compartment. Of these, FOG-1 was shown to be required for the coordinated loss of GM priming and acquisition of Mk/E priming associated with preMegE formation. This identifies FOG-1 as a transcription factor critical for the generation of all Mk/E-committed cells, whereas GATA-1 was specifically required for the specification of preCFU-E/CFU-Es from preMegEs. Therefore, FOG-1 and GATA-1 act sequentially during specification of committed Mk/E progenitors to separate these from GM progenitors, and subsequently to segregate the Mk and E lineages.
GATA-1 is required for erythroid-lineage commitment
The essential role of GATA-1 in erythropoiesis has long been established. Gata1 null embryos die around embryonic day 10.5 due to defective primitive erythropoiesis (Fujiwara et al, 1996). More recently, deletion of Gata1 during definitive haematopoiesis revealed a block of erythroid progenitor maturation at the pro-erythroblast stage (Gutierrez et al, 2008). However, our inability to prospectively identify committed unilineage E progenitors has so far precluded analysis of the role of GATA-1 in lineage commitment processes. Using the progenitor phenotyping scheme developed by Bryder and colleagues (Pronk et al, 2007) we here show that acute Gata1 deletion leads to a complete block of preCFU-Es and CFU-Es, the earliest erythroid-committed progenitors thus far identified. Loss of GATA-1 did not affect the numbers of preMegEs, preGMs or GMPs, but was accompanied by an increase of MkPs, indicating that in the absence of GATA-1 the allocation of progenitors at the Mk/E versus GM bifurcation is not significantly affected, whereas preMegEs showed only Mk-lineage commitment. Interestingly, other transcription factors involved in erythroid differentiation (Klf1, Gata2, Zfpm1, Epor) were not downregulated in preMegEs, showing that the impairment in E progenitor formation was not due to a general loss of erythroid regulators, but to a specific requirement for GATA-1 at the Mk- versus E-lineage bifurcation.
Other regulatory mechanisms have been identified that influence the relative Mk–E lineage output: miR-150 represses c-Myb expression, and miR-150 overexpression (Lu et al, 2008) or hypomorphic Myb alleles (Emambokus et al, 2003; Mukai et al, 2006) both led to overproduction of megakaryocytes, whereas miR-150 knockdown or c-Myb overexpression had the opposite effect. Activation of Notch signalling also has the potential to increase Mk differentiation: exposure to Notch ligands or transduction with the Notch1 intracellular domain increased megakaryocytic output in sorted murine stem and progenitor cell populations in vitro, and Notch inhibition reduced the number of megakaryocyte-erythroid progenitors (MEPs) and megakaryocytes in vivo (Mercher et al, 2008). These results are consistent with a role for these mechanisms in Mk versus E unilineage progenitor specification. It will now be important to determine whether this is the case, and to which extent Myb, miR-150 and Notch functionally interact with GATA-1 during this process.
FOG-1 is required for formation of all committed Mk and E progenitors
In the absence of FOG-1, we observed a complete loss of progenitors committed to the Mk and E lineages: MkPs, preCFU-Es, and CFU-Es were all absent, whereas myeloid-committed preGM and GMP progenitors were correspondingly increased. Loss of FOG-1 did not significantly alter the total number of myelo-erythroid progenitors, but simply altered their distribution. The current paradigm for separation of the GM and Mk–E lineages is thought to be PU.1–GATA-1 antagonism. PU.1 was observed to instruct myeloid differentiation in transformed avian multipotent progenitors while downregulating GATA-1 (Nerlov and Graf, 1998). Conversely, GATA-1 suppressed the myeloid phenotype of, and conferred MkE potential on, committed myeloid cells (Kulessa et al, 1995). In addition, PU.1 and GATA-1 (and GATA-2) showed functional antagonism through direct protein–protein interaction (Rekhtman et al, 1999; Zhang et al, 1999; Nerlov et al, 2000) as well as positive autoregulation through binding to their own promoters (Tsai et al, 1991; Okuno et al, 2005). Mathematical modelling has shown that a system with these properties is capable of maintaining an uncommitted metastable state that bifurcates into stable committed states when either of the two factors prevail (Huang et al, 2007).
The present results indicate that also FOG-1 plays a key role in this lineage bifurcation. FOG-1 null embryos die around embryonic day 11.5 and show arrested development of primitive erythrocytes and lack of megakaryocytes (Tsang et al, 1998). Furthermore, decreased erythropoiesis and thrombopoiesis, and increased myelopoiesis have been reported in morpholino-based FOG-1 knockdown in zebrafish (Amigo et al, 2009). It has, however, not been clear to which extent these abnormalities reflect lack of lineage commitment or subsequent differentiation defects. In addition, FOG-1 has been shown to repress eosinophil-specific gene expression through GATA-1/2 bound to eosinophil-specific promoters (Yamaguchi et al, 1999), and to block eosinophil specification (Querfurth et al, 2000). A similar role for FOG-1 in suppression of mast cell gene expression and differentiation has been demonstrated (Cantor et al, 2008). The repressor function of FOG-1 has been located to an N-terminal NuRD-recruitment domain (Hong et al, 2005). Recently, knock-in mutagenesis disabling NuRD interaction of FOG-1 resulted in ectopic expression of myeloid and mast cell genes in heterologous cell types (Gao et al, 2010; Gregory et al, 2010; Miccio et al, 2010). However, no major defects in lineage specification were observed. The present results show that complete loss of FOG-1 results in loss of Mk/E-lineage progenitor commitment. Competitive transplantation experiments using bone marrow genetically modified to allow tracking of platelets and erythrocytes showed that the lack of FOGcKO MkPs, preCFU-Es and CFU-Es was accompanied by a loss of mature cells of these lineages, arguing against the absence of these progenitors being due to an altered surface phenotype. The absence of Mk/E-lineage progenitors was also accompanied by loss of Mk/E-lineage programming and ectopic myeloid gene expression in the remaining phenotypic preMegEs. FOG-1-deficient preMegEs showed exclusively myeloid potential in vitro. This demonstrates a novel FOG-1 function in lineage specification, which is at least partially independent of NuRD complex interaction. Ectopic myeloid differentiation was observed from Mk/E progenitors in FOG-1–NuRD interaction mutant mice (Gregory et al, 2010), indicating that suppression of myeloid potential by FOG-1 at least partially requires NuRD recruitment, whereas the acquisition of Mk/E potential can proceed in its absence.
Transcriptional lineage priming of haematopoietic stem and progenitor cells has been shown to define their lineage potential (Hu et al, 1997; Mansson et al, 2007). We here observe that the inability of FOG-1-deficient HSCs to generate committed Mk/E progenitors is associated with their loss of preMegE-specific gene expression and upregulation of preGM-associated genes. The altered allocation of early myelo-erythroid progenitors from FOGcKO HSCs is, therefore, preceded by a corresponding imbalance in the gene expression programs defining the earliest progenitors in the two differentiation pathways, supporting the importance of HSC priming for subsequent lineage commitment. We have previously shown that Cebpa loss-of-function mutations decrease myeloid priming of HSCs while simultaneously impairing myeloid-lineage commitment (Bereshchenko et al, 2009). Using the preGM and preMegE gene sets, we here extend this analysis by showing that disruption of C/EBP function in HSCs perturbs early Mk/E and GM progenitor gene expression in a manner converse to that of FOG-1 depletion, consistent with the negative cross-regulation between FOG-1 and C/EBPs translating into opposing roles in global lineage programming and lineage commitment.
Suppression of myeloid progenitor specification by FOG-1
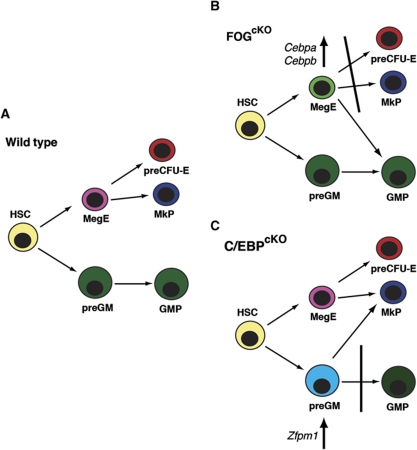
While loss of GATA-1 by itself did not disturb the allocation of Mk/E versus GM progenitors in vivo this could be explained by functional redundancy with GATA-2, which is also able to antagonize PU.1 activity. A larger issue is how FOG-1 interacts with the PU.1–GATA-1/2 antagonism. This potentially involves multiple mechanisms. First, since interaction of GATA-1 with PU.1 and FOG-1 has been observed to be mutually exclusive (Sugiyama et al, 2008), FOG-1 may act as a co-factor for GATA-1 on the GATA-1 promoter and impair the ability of PU.1 to repress Gata1 expression. Second, we observe cross-regulation of Zfpm1 and Cebpa/Cebpb during the GM versus Mk/E segregation: loss of FOG-1 led to upregulation of Cebpa and Cebpb in preMegEs, whereas C/EBP-depleted preGM cells showed increased Zfpm1 expression. In both cases, this correlated with altered lineage potential of the blocked progenitor population (summarized in Figure 8).
Figure 8.
Lineage perturbations in FOG-1- and C/EBP-deficient haematopoiesis. (A) The myelo-erythroid lineage bifurcation in normal haematopoiesis. (B) In FOG-1-deficient haematopoiesis, phenotypic preMegEs upregulate Cebpa and Cebpb, assume preGM-like gene expression and give rise to myeloid cells, but no committed Mk or E progenitors or colonies. (C) Conversely, C/EBP-deficient preGM cells are blocked in the progression to the GMP stage, upregulate Zfpm1 (encoding FOG-1) and Mk-specific genes, and show ectopic Mk potential.
Mining of microarray data identified the C/EBP antagonist Trib2 as a potential effector of FOG-1-mediated extinction of myeloid potential. Trib2 is selectively expressed in preMegEs, but not in CLPs and preGMs; its promoter region binds GATA-2/FOG-1 in sorted preMegEs; and its expression is strongly reduced in the absence of FOG-1. Trib2 is able to selectively degrade the long isoforms of C/EBPα and C/EBPβ (Keeshan et al, 2006; Naiki et al, 2007), which are the transcriptionally active isoforms (Nerlov, 2007). While it has been shown that viral transduction of Trib2 into the haematopoietic system is leukaemogenic (Keeshan et al, 2006) in the same manner as loss of the long C/EBPα p42 isoform (Kirstetter et al, 2008); however, it has so far not been addressed whether Trib2 expression alters myeloid-lineage allocation. Now, it will be of interest to directly examine the role of Trib2 in Mk/E-lineage formation. In addition, C/EBPs are known to positively regulate Sfpi1 expression (Kummalue and Friedman, 2003; Yeamans et al, 2007); this provides a potential interface between PU.1–GATA-1 and FOG-1–C/EBP cross-regulation. Finally, C/EBPs have been described to functionally antagonize GATA factors (Tong et al, 2005) through protein–protein interaction, suggesting a proteomic contribution of C/EBPs in addition to direct gene regulation. The simple model of PU.1–GATA-1 antagonism controlling the segregation of the Mk/E and GM lineages, therefore, likely needs to be replaced by a more complex model where PU.1 and C/EBPs collectively antagonize GATA-1/2 and FOG-1 through several independent mechanisms.
Materials and methods
Mouse lines
Zfmp1 (Katz et al, 2003), Gata1 (Lindeboom et al, 2003) Cebpa (Lee et al, 1997) and Cebpb (Lopez et al, 2009) conditional alleles and Mx1-Cre transgenic mice (Kuhn et al, 1995) were previously generated. miR144/451EGFP mice have been described (Rasmussen and O'Carroll, 2011). Details of the Vwf-EGFP line will be published elsewhere (A Sanjuan-Pla, C Nerlov and SE Jacobsen, in preparation). Poly(I-C) induced recombination, lethal irradiation and bone marrow transplantation were carried out as described (Kirstetter et al, 2008). Animal experiments were carried out with the approval of the EMBL Monterotondo Ethical Committee and the University of Edinburgh Ethical Committee.
Flow cytometry and colony assays
Measurement and analysis of haematological parameters, flow cytometric analysis and sorting of progenitors were performed as described (Kirstetter et al, 2006; Bereshchenko et al, 2009). A list of antibodies and dilutions used is provided in Supplementary Experimental Procedures. Media for colony forming assays for GM (M3436), E (M3534) and Mk lineages (MegaCult-C) were from Stem Cell Technologies (Vancouver, Canada) and used according to the manufacturer's instructions. Colonies generated in methylcellulose were scored as blasts, GM, BFU-E, and Mk colonies by morphologic criteria, and for megakaryocytic progenitors by acetylcholinesterase staining. Cells were plated in triplicate, or quadruplicate, and colonies were scored after 5 days (GM colonies) and 10 days (BFU-E and Mk colonies).
Gene expression analysis
RNA purification and Q–PCR analysis of sorted cells were as previously described (Ruffell et al, 2009), except that Hprt expression was used for normalization. Statistical significance was determined using Student's t-test. A list of TaqMan probe sets (Applied Biosystems, Foster City, CA, USA) used is provided in Supplementary Experimental Procedures.
Microarray analysis was performed at the Copenhagen University Hospital Microarray Unit. Total RNA from 5000 sorted cells was amplified using the WT-Ovation Exon Module, biotin labelled with FL-Ovation Biotin Module V2 kit (Nugen, San Carlos, CA, USA) and hybridized to Mouse Gene 1.0 ST GeneChip arrays (Affymetrix, Santa Clara, CA, USA) according to the manufacturers' instructions. Normalization and GSEA analysis (Subramanian et al, 2005) were performed as described (Bereshchenko et al, 2009) using CLP, preGM and preMegE gene sets (Pronk et al, 2007). Cebpa mutant HSC microarray data have previously been published (Bereshchenko et al, 2009). Microarray data have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/microarray-as/ae/) with the accession number E-MTAB-342.
Multiplex single-cell RT–PCR analysis was performed as previously described (Hu et al, 1997; Adolfsson et al, 2005; Mansson et al, 2007). The genes investigated have previously been validated for their lineage specificity in early committed progenitors of different lineages (Mansson et al, 2007). The primers used for nested multiplex single-cell PCR are listed in Supplementary Experimental Procedures.
For analysis of Trib2 expression, 200 preMegE cells were sorted into 10 μl CellsDirect (Invitrogen) mix containing 1 × CellsDirect Reaction Mix, 1 μl RT-Taq mix, and 200 nM primers against genes including Trib2 and Hprt. Reverse transcription was performed at 50°C for 15 min, followed by a 2-min incubation at 95°C and pre-amplification for 22 cycles of 95°C for 15 s and 60°C for 4 min. Pre-amplified samples were diluted to 50 μl with TE, and 2.5 μl of each sample was assayed in triplicate against UPL assays (Roche) for the target genes, also loaded in triplicate, on a Fluidigm 48.48 Dynamic Array. PCRs were performed on a BioMark HD instrument using the following conditions: 50°C for 2 min; 95°C for 10 min; 40 cycles of 95°C for 15 s, 58°C for 5 s, 60°C for 1 min. Thresholds were set manually for each assay prior to calling CTs. Reactions with no amplification were set to an arbitrary CT of 45, and relative expression levels were calculated by the ΔΔCt method. UPL (Roche)-based Q–PCR primers were Trib2: 5′-TGGAGGGAGACCACGTTTT-3′ and 5′-AGCAGCTGATCTCAAACACCT-3′, UPL probe 17; Hprt: 5′-TCCTCCTCAGACCGCTTTT-3′ and 5′-CCTGGTTCATCATCGCTAATC-3′, UPL probe 95. Microarray-based expression was calculated on RMA-processed data (FOGcKO versus FOGCon), and using GCRMA-normalized published data (Pronk et al, 2007) (comparison of CLP, preGM and preMegE populations).
Chromatin IP
In all, 50 000 PreMegEs were sorted into DPBS (Gibco) supplemented with 5% FBS, and crosslinked in DMEM/1% formaldehyde for 10 min at room temperature. The reaction was stopped by adding glycine to a final concentration of 0.125 M. Cells were washed twice in ice-cold HBSS, and lysed in 200 μl of lysis buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 1% SDS) on ice for 20 min. Chromatin was fragmented using Bioruptor (Diagenode) to obtain genomic fragments ranging from 200 to 500 bp. Samples were diluted with one volume of RIPA buffer containing protease inhibitor cocktail. ChIP assays were performed using 0.25 μg anti-GATA-2 (H-116) antibody, 0.5 μg anti-FOG-1 (M-20) antibody or an equal amount of control IgG (Santa Cruz). Following overnight incubation at 4°C, protein/DNA complexes were captured with 20 μl protein G Dynabeads (Invitrogen). After 2 h at 4°C, beads were washed 2 × with RIPA plus PIC buffer, 4 × with RIPA containing 500 mM NaCl plus PIC, 1 × LiCl buffer without protease inhibitor, 2 × TE buffer (10 mM Tris–HCl, pH 8.0, 10 mM EDTA) and finally resuspended in 100 μl cold TE buffer containing RNAse A (50 μg/ml) and incubated at 37°C for 30 min. Protease inhibitor cocktail (Roche) was added to all buffers. DNA was eluted twice with 200 μl elution buffer (20 mM Tris–HCl, pH 7.5, 5 mM EDTA, 50 mM NaCl) containing 1% SDS and Proteinase K. Crosslinks were reversed at 68°C for 2 h with shaking at 1300 r.p.m. Genomic DNA was recovered using phenol chloroform extraction and ethanol precipitation. Pellets were washed in 70% ethanol, briefly air dried, and resuspended in TE buffer (pH 8.0). The DNA was pre-amplified with specific primer using Taqman pre-amp master mix (Applied Biosystems). Putative GATA sites were located using TRANSFAC v2010.4 (Matys et al, 2006). Enrichment of the ChIP sample over input was confirmed by UPL-based Q–PCR. Enrichment was determined as the ratio between immune and non-immune IgG, and normalized to the values obtained for the Csn2 negative control. The following amplicons were used: Trib2-1: 5′-TTGTGACTAGAAATTGGGTACATGA-3′ and 5′-CAGCACATGTAGGGAAGCTG-3′, UPL probe #32; Trib2-2: 5′-CGAAGAGCTGTCGTCTATAAGGT-3′ and 5′-AACCTTGGCTCTCCGAGC-3′, UPL probe#20; Csn2: 5′-CGTCTTACTGTGCCTCTCCA-3′ and 5′-CATGGCCACAATTCTTGGTT-3′, UPL probe #82.
Supplementary Material
Acknowledgments
We thank Dr Stuart Orkin (Harvard University, Cambridge, MA, USA), Dr Sjaak Philipsen (Erasmus University, Rotterdam, The Netherlands) and Dr Ying-Hue Lee (Academia Sinica, Nankang, Taipei, Taiwan) for generously sharing mouse lines. This work was funded by the European Commission (EuroCSC STREP), the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Swedish Research Council (HematoLinne), as well as by a Strategic Grant and Programme Grant to CN from the Medical Research Council.
Author contributions: EM, SEJ and CN planned the experiments. AZ, AG, SM, EM, LL, AS-P and SL performed the experiments. SM carried out bioinformatics. DB assisted with flow cytometry. KR and DO provided mouse line. EM, SEJ and CN wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE (2005) Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121: 295–306 [DOI] [PubMed] [Google Scholar]
- Akashi K, Traver D, Miyamoto T, Weissman IL (2000) A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193–197 [DOI] [PubMed] [Google Scholar]
- Amigo JD, Ackermann GE, Cope JJ, Yu M, Cooney JD, Ma D, Langer NB, Shafizadeh E, Shaw GC, Horsely W, Trede NS, Davidson AJ, Barut BA, Zhou Y, Wojiski SA, Traver D, Moran TB, Kourkoulis G, Hsu K, Kanki JP et al. (2009) The role and regulation of friend of GATA-1 (FOG-1) during blood development in the zebrafish. Blood 114: 4654–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou M, Clausen PA, Mavrothalassitis GJ, Zhang XK, Watson DK, Blair DG (1996) Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell growth Differ 7: 1525–1534 [PubMed] [Google Scholar]
- Athanasiou M, Mavrothalassitis G, Sun-Hoffman L, Blair DG (2000) FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia 14: 439–445 [DOI] [PubMed] [Google Scholar]
- Bereshchenko O, Mancini E, Moore S, Bilbao D, Mansson R, Luc S, Grover A, Jacobsen SE, Bryder D, Nerlov C (2009) Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell 16: 390–400 [DOI] [PubMed] [Google Scholar]
- Bouilloux F, Juban G, Cohet N, Buet D, Guyot B, Vainchenker W, Louache F, Morle F (2008) EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood 112: 576–584 [DOI] [PubMed] [Google Scholar]
- Burda P, Laslo P, Stopka T (2010) The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia 24: 1249–1257 [DOI] [PubMed] [Google Scholar]
- Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH (2008) Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med 205: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AN, Cantor AB, Fujiwara Y, Lodish MB, Droho S, Crispino JD, Orkin SH (2002) GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc Natl Acad Sci USA 99: 9237–9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL (2005) PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med 201: 1487–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J (2003) Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J 22: 4478–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontelo P, Manwani D, Galdass M, Karsunky H, Lohmann F, Gallagher PG, Bieker JJ (2007) Novel role for EKLF in megakaryocyte lineage commitment. Blood 110: 3871–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH (1996) Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA 93: 12355–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Huang Z, Olivey HE, Gurbuxani S, Crispino JD, Svensson EC (2010) FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis. EMBO J 29: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory GD, Miccio A, Bersenev A, Wang Y, Hong W, Zhang Z, Poncz M, Tong W, Blobel GA (2010) FOG1 requires NuRD to promote hematopoiesis and maintain lineage fidelity within the megakaryocytic-erythroid compartment. Blood 115: 2156–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Tsukamoto S, Suzuki M, Yamamoto-Mukai H, Yamamoto M, Philipsen S, Ohneda K (2008) Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood 111: 4375–4385 [DOI] [PubMed] [Google Scholar]
- Hart A, Melet F, Grossfeld P, Chien K, Jones C, Tunnacliffe A, Favier R, Bernstein A (2000) Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity 13: 167–177 [DOI] [PubMed] [Google Scholar]
- Hirai H, Zhang P, Dayaram T, Hetherington CJ, Mizuno S, Imanishi J, Akashi K, Tenen DG (2006) C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol 7: 732–739 [DOI] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA (2005) FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J 24: 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T (1997) Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev 11: 774–785 [DOI] [PubMed] [Google Scholar]
- Huang S, Guo YP, May G, Enver T (2007) Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol 305: 695–713 [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, Fenyus ML, Elf S, Chan S, Kastner P, Huettner CS, Murray R, Tenen DG, Akashi K (2005) Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106: 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz SG, Williams A, Yang J, Fujiwara Y, Tsang AP, Epstein JA, Orkin SH (2003) Endothelial lineage-mediated loss of the GATA cofactor Friend of GATA 1 impairs cardiac development. Proc Natl Acad Sci USA 100: 14030–14035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, Rodriguez CG, Maillard I, Tobias JW, Valk P, Carroll M, Aster JC, Delwel R, Pear WS (2006) Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell 10: 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C (2006) Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol 7: 1048–1056 [DOI] [PubMed] [Google Scholar]
- Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E, Theilgaard-Monch K, Mansson R, Pedersen TA, Pabst T, Schrock E, Porse BT, Jacobsen SE, Bertone P, Tenen DG, Nerlov C (2008) Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell 13: 299–310 [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K (1995) Inducible gene targeting in mice. Science 269: 1427–1429 [DOI] [PubMed] [Google Scholar]
- Kulessa H, Frampton J, Graf T (1995) GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev 9: 1250–1262 [DOI] [PubMed] [Google Scholar]
- Kummalue T, Friedman AD (2003) Cross-talk between regulators of myeloid development: C/EBPalpha binds and activates the promoter of the PU.1 gene. J Leukoc Biol 74: 464–470 [DOI] [PubMed] [Google Scholar]
- Lee YH, Sauer B, Johnson PF, Gonzalez FJ (1997) Disruption of the c/ebp alpha gene in adult mouse liver. Mol Cell Biol 17: 6014–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom F, Gillemans N, Karis A, Jaegle M, Meijer D, Grosveld F, Philipsen S (2003) A tissue-specific knockout reveals that Gata1 is not essential for Sertoli cell function in the mouse. Nucleic Acids Res 31: 5405–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez RG, Garcia-Silva S, Moore SJ, Bereshchenko O, Martinez-Cruz AB, Ermakova O, Kurz E, Paramio JM, Nerlov C (2009) C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat Cell Biol 11: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, Pretz J, Schlanger R, Wang JY, Mak RH, Dombkowski DM, Preffer FI, Scadden DT, Golub TR (2008) MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell 14: 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, Buza-Vidas N, Qian H, Soneji S, Enver T, Sigvardsson M, Jacobsen SE (2007) Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity 26: 407–419 [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercher T, Cornejo MG, Sears C, Kindler T, Moore SA, Maillard I, Pear WS, Aster JC, Gilliland DG (2008) Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell 3: 314–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miccio A, Wang Y, Hong W, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, Blobel GA (2010) NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J 29: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai HY, Motohashi H, Ohneda O, Suzuki N, Nagano M, Yamamoto M (2006) Transgene insertion in proximity to the c-myb gene disrupts erythroid-megakaryocytic lineage bifurcation. Mol Cell Biol 26: 7953–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki T, Saijou E, Miyaoka Y, Sekine K, Miyajima A (2007) TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPbeta. J Biol Chem 282: 24075–24082 [DOI] [PubMed] [Google Scholar]
- Nerlov C (2007) The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol 17: 318–324 [DOI] [PubMed] [Google Scholar]
- Nerlov C, Graf T (1998) PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev 12: 2403–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, Querfurth E, Kulessa H, Graf T (2000) GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95: 2543–2551 [PubMed] [Google Scholar]
- Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F (1995) Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375: 316–318 [DOI] [PubMed] [Google Scholar]
- Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H, Akashi K, Moreau-Gachelin F, Li Y, Zhang P, Gottgens B, Tenen DG (2005) Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol Cell Biol 25: 2832–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH (1995) Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375: 318–322 [DOI] [PubMed] [Google Scholar]
- Porse BT, Bryder D, Theilgaard-Monch K, Hasemann MS, Anderson K, Damgaard I, Jacobsen SE, Nerlov C (2005) Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J Exp Med 202: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D (2007) Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1: 428–442 [DOI] [PubMed] [Google Scholar]
- Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, Graf T, Nerlov C (2000) Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev 14: 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, O'Carroll D (2011) The miR-144/451eGFP allele, a novel tool for resolving the erythroid potential of hematopoietic precursors. Blood 118: 2988–2992 [DOI] [PubMed] [Google Scholar]
- Rekhtman N, Radparvar F, Evans T, Skoultchi AI (1999) Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev 13: 1398–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, Nerlov C (2009) A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci USA 106: 17475–17480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, Watson DK (2000) Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol 20: 5643–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J, Cohet N, Gonnet C, Sarrazin S, Doubeikovskaia Z, Doubeikovski A, Verger A, Duterque-Coquillaud M, Morle F (2003) Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol Cell Biol 23: 1390–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J, Weiss-Gayet M, Gonnet C, Guyot B, Vicat JM, Morle F (2010) Inducible Fli-1 gene deletion in adult mice modifies several myeloid lineage commitment decisions and accelerates proliferation arrest and terminal erythrocytic differentiation. Blood 116: 4795–4805 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama D, Tanaka M, Kitajima K, Zheng J, Yen H, Murotani T, Yamatodani A, Nakano T (2008) Differential context-dependent effects of friend of GATA-1 (FOG-1) on mast-cell development and differentiation. Blood 111: 1924–1932 [DOI] [PubMed] [Google Scholar]
- Ting CN, Olson MC, Barton KP, Leiden JM (1996) Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 384: 474–478 [DOI] [PubMed] [Google Scholar]
- Tong Q, Tsai J, Tan G, Dalgin G, Hotamisligil GS (2005) Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol Cell Biol 25: 706–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SF, Strauss E, Orkin SH (1991) Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev 5: 919–931 [DOI] [PubMed] [Google Scholar]
- Tsang AP, Fujiwara Y, Hom DB, Orkin SH (1998) Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev 12: 1176–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M (1994) Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79: 901–912 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Nishio H, Kishi K, Ackerman SJ, Suda T (1999) C/EBPbeta and GATA-1 synergistically regulate activity of the eosinophil granule major basic protein promoter: implication for C/EBPbeta activity in eosinophil gene expression. Blood 94: 1429–1439 [PubMed] [Google Scholar]
- Yeamans C, Wang D, Paz-Priel I, Torbett BE, Tenen DG, Friedman AD (2007) C/EBPalpha binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood 110: 3136–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z (1999) Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA 96: 8705–8710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Himes JA, Akashi K, Tenen DG (2004) Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity 21: 853–863 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.