Abstract
Tissue-specific transcriptional activators initiate differentiation towards specialized cell types by inducing chromatin modifications permissive for transcription at target loci, through the recruitment of SWItch/Sucrose NonFermentable (SWI/SNF) chromatin-remodelling complex. However, the molecular mechanism that regulates SWI/SNF nuclear distribution in response to differentiation signals is unknown. We show that the muscle determination factor MyoD and the SWI/SNF subunit BAF60c interact on the regulatory elements of MyoD-target genes in myoblasts, prior to activation of transcription. BAF60c facilitates MyoD binding to target genes and marks the chromatin for signal-dependent recruitment of the SWI/SNF core to muscle genes. BAF60c phosphorylation on a conserved threonine by differentiation-activated p38α kinase is the signal that promotes incorporation of MyoD–BAF60c into a Brg1-based SWI/SNF complex, which remodels the chromatin and activates transcription of MyoD-target genes. Our data support an unprecedented two-step model by which pre-assembled BAF60c–MyoD complex directs recruitment of SWI/SNF to muscle loci in response to differentiation cues.
Keywords: chromatin, gene expression, muscle differentiation, myod, p38
Introduction
The muscle-specific basic helix-loop-helix (bHLH) transcription factor (TF) MyoD provides a general paradigm to elucidate the physical and functional interactions between tissue-specific TFs and the machinery that promotes chromatin modifications at specific loci (Sartorelli and Caretti, 2005; Tapscott, 2005). Among muscle-specific bHLH TFs, MyoD (and its functional paralogue Myf5) possesses the ability to activate transcription at previously silent muscle loci, via domains that mediate chromatin remodelling and nucleosome displacement at target genes (Gerber et al, 1997). Nucleosome disruption is frequently carried out by the SWItch/Sucrose NonFermentable (SWI/SNF) chromatin-remodelling complexes, which use energy from ATP hydrolysis for disruption of interactions between DNA and histone octamers. SWI/SNF-mediated chromatin remodelling permits the loading of the ‘transcriptosome’ on the regulatory elements of tissue-specific genes and the activation of gene transcription (de la Serna et al, 2006; Cairns, 2009). SWI/SNF complexes are composed of an ATPase subunit (either Brg1 or Brm) and a variable number of structural subunits assembled in different combinations (Clapier and Cairns, 2009; Wu et al, 2009). The involvement of the SWI/SNF complex in a variety of cell activities, including proliferation (Muchardt and Yaniv, 2001), transformation (Reisman et al, 2009), DNA damage signalling and repair (Gong et al, 2006; Park et al, 2006; Sinha et al, 2009), pluripotency and lineage determination (Lessard and Crabtree, 2010; Singhal et al, 2010) indicates a widespread function of SWI/SNF in the regulation of gene expression. Such a versatile activity appears to be conferred on SWI/SNF by a heterogeneous subunit composition and signal-dependent regulation (Wu et al, 2009). However, the mechanism that regulates SWI/SNF composition and chromatin distribution in response to specific cues, such as those that activate muscle-specific gene expression, remains unsolved (Albini and Puri, 2010).
MyoD is expressed in myoblasts well before the activation of muscle gene transcription (Weintraub, 1993), possibly in a conformation (homodimers) that precludes ‘productive’ binding to DNA-responsive elements (the E-Box sequence, CANNTG) (Puri and Sartorelli, 2000). On induction of differentiation, MyoD interactions with E2A gene products (e.g., E12 or E47) leads to the formation of heterodimers with an increased affinity for E-Box elements and ability to activate transcription (Tapscott, 2005). In myoblasts, MyoD interaction with chromatin has been the object of controversy (Bergstrom et al, 2002; Berkes et al, 2004; Simone et al, 2004; de la Serna et al, 2005; Mal, 2006). The recent advent of chromatin immunoprecipitation (ChIP)-based technologies for genome-wide analysis has indicated the existence of previously unappreciated interactions between MyoD and chromatin in undifferentiated myoblasts (Blais et al, 2005; Cao et al, 2006, 2010). Still, the function and regulation of MyoD in myoblasts remain obscure. In particular, it is unknown how MyoD recognizes and accesses target genes before induction of differentiation. In undifferentiated myoblasts, the expression of most of MyoD-target genes is prevented by the repressive chromatin conformation imposed by the nucleosomes, which typically preclude the access to transcriptional activators. Thus, a current gap of knowledge concerns the mechanism by which MyoD gains access to target genes in myoblasts, and how differentiation signals confer on MyoD the ability to recruit the machinery that reconfigures the chromatin into a conformation permissive for transcription from target genes.
Results
Physical interactions between MyoD and BAF60c
To elucidate the regulation of MyoD function in myoblasts, we carried out a two-hybrid screen, in which MyoD was used as bait. This system has the advantage of identifying potential proteins that interact with MyoD in the monomer/homodimer conformation predicted in myoblasts (Li et al, 1996), prior to the activation of muscle genes. Using the C-terminal domain of MyoD that contains a chromatin-remodelling domain (Gerber et al, 1997) (Supplementary Figure S1A), we identified a number of potential interacting proteins. From these we focused on BAF60b (SMARCD2) and BAF60c (SMARCD3)—two structural components of the SWI/SNF chromatin-remodelling complex, which is required for the activation of muscle gene transcription (de la Serna et al, 2001, 2005; Simone et al, 2004). Previous work established the essential role of BAF60c in cardiac and skeletal muscle development (Lickert et al, 2004; Takeuchi and Bruneau, 2009). However, the relative role of BAF60 variants a (SMARCD1), b and c in skeletal myogenesis has not been systematically addressed by previous studies. BAF60 variants show a tissue-specific distribution, with BAF60b and c being abundantly expressed in skeletal muscles (Supplementary Figure S1B), as also originally reported by Crabtree and colleagues (Wang et al, 1996). When we monitored the expression levels of BAF60 variants in mouse primary satellite cells, we noted that only BAF60c was upregulated during the differentiation process (Supplementary Figure S1C and F). The selective upregulation of BAF60c was also observed during differentiation of human primary myoblasts (Supplementary Figure S1D) and established skeletal muscle cell lines, such as mouse C2C12 myoblasts (Supplementary Figure S1E and F, right panel). This evidence indicates a relationship between BAF60c levels and myoblast differentiation. Among the two human BAF60c alternative variants—BAF60c1 and 2—BAF60c2 was the only form expressed in human skeletal myoblasts and upregulated in differentiated myotubes (Supplementary Figure S1G). We therefore decided to restrict our analysis to the BAF60c2 variant, which corresponds to mouse BAF60c and is hereafter referred to as BAF60c. In-vitro pull-down experiments using GST–MyoD full length or a mutant lacking the C-terminal domain confirmed the essential role of the C-terminal domain of MyoD for the interaction with BAF60c (Supplementary Figure S1H). The same interaction and requirement of MyoD C-terminus was also confirmed by co-immunoprecipitation of exogenously expressed Flag-tagged MyoD and Xp-tagged BAF60c (Supplementary Figure S1I).
Stage-specific interactions between MyoD, BAF60c and Brg1 during myogenic differentiation
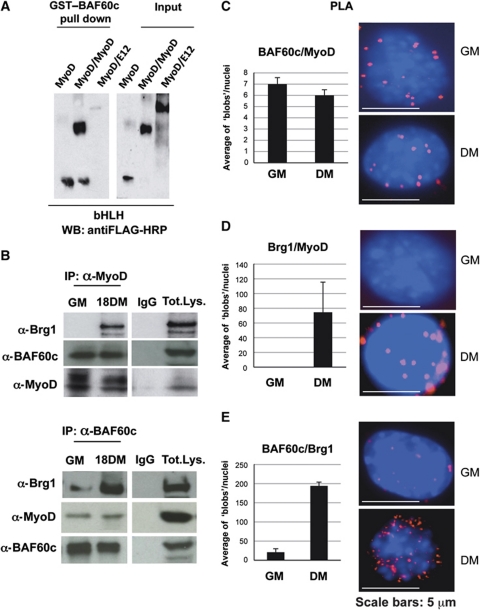
We further examined whether BAF60c could directly associate with MyoD in vitro. For this purpose, we performed in-vitro interaction studies with purified proteins, using GST–BAF60c in combination with either Flag-tagged purified MyoD, forced MyoD∼MyoD homodimer or MyoD∼E12 heterodimer. Pull-down assay showed that BAF60c interacted efficiently with MyoD and forced MyoD∼MyoD homodimers, while we observed a reduced ability of BAF60c to interact with the forced MyoD∼E12 heterodimers (Figure 1A). Because MyoD homodimers could only form in undifferentiated myoblasts (Li et al, 1996), these data suggest that an interaction between BAF60c and MyoD can take place in undifferentiated myoblasts, prior to the activation of the differentiation programme.
Figure 1.
BAF60c and MyoD interact in vitro and in vivo. (A) Pull-down assay was performed using GST–BAF60c and baculovirus-purified Flag-tagged MyoD, MyoD∼MyoD and MyoD∼E12 forced dimmers. (B) Co-immunoprecipitations from nuclear extracts of myoblasts and myotubes, with anti-MyoD and anti-BAF60c antibodies. (C–E) PLA was used to monitor nuclear ‘in situ’ interactions between MyoD, Brg1 and Flag–BAF60c during C2C12 differentiation. Each fluorescent dot, ‘blob’, represents the co-localization of the indicated proteins in myoblasts and myotubes. The quantification of the blobs is represented in the adjacent graphic. The BlobFinder software (Allalou and Wählby, 2009) was used to localize and quantify the blobs from images acquired with fluorescent microscopy. The average of blobs/nuclei in the graphic corresponds to the quantification of several images from three different experiments in myoblasts and myotubes.
Previous studies have shown both physical and functional interactions between the SWI/SNF enzymatic subunit Brg1 and the muscle regulatory factors MyoD and MEF2 only in differentiating muscle cells (Simone et al, 2004; de La Serna et al, 2005; Rampalli et al, 2007; Serra et al, 2007). Thus, we asked whether the interaction between BAF60c and MyoD detected in myoblasts could precede the association between MyoD and Brg1. Co-immunoprecipitation studies were used to monitor the combinatorial interactions between endogenous MyoD, Brg1 and BAF60c in undifferentiated myoblasts (cultured in growth medium—GM) and differentiating myoblasts (cultured for 18 h in differentiation medium—DM) (Figure 1B). For this purpose, we generated an antibody against BAF60c that specifically recognizes a sequence within the C-terminal region that is not conserved in BAF60a and b—see detailed description in Supplementary data. Immunoprecipitation with anti-MyoD antibodies revealed an interaction between MyoD and BAF60c in both undifferentiated myoblasts and differentiating myoblasts, while the interaction between MyoD and Brg1 was restricted to differentiating myoblasts (Figure 1B, upper panel). The other SWI/SNF catalytic subunit Brm did not interact with MyoD in any of the above conditions (data not shown). Immunoprecipitation with anti-BAF60c antibodies revealed an interaction between BAF60c and MyoD, and between BAF60c and Brg1 at both stages (Figure 1B, lower panel). Collectively, these results show physical interactions between MyoD and BAF60c in myoblasts in a complex distinct from the Brg1- and Brm-based SWI/SNF complexes. Moreover, these results indicate that in myoblasts, a fraction of BAF60c does not bind MyoD and is part of Brg1-based SWI/SNF complexes, which are involved in MyoD-independent regulation of gene expression. Importantly, the reciprocal interactions between Brg1, BAF60c and MyoD detected on induction of differentiation indicate the formation of an MyoD-associated BAF60c/Brg1-based SWI/SNF complex, which coincides with the activation of muscle gene expression. To further substantiate this finding, we used a proximity ligation assay (PLA) that detects the fluorescence signal generated by two antibodies when bound to proteins in such close proximity (at least 30 nm) that indicates a physical interaction (Fredriksson et al, 2002; Allalou and Wählby, 2009; see also Materials and methods). We monitored the interactions between endogenous MyoD, Brg1 and Flag–BAF60c at sequential stages of skeletal muscle differentiation. Flag–BAF60c was used in this assay because the Flag antibody generates an optimal signal for PLA, as compared with the endogenous BAF60c, and permits a direct comparison with mutated forms of BAF60c (see Supplementary Figure S5). The nuclear signal reflecting interactions between MyoD and BAF60c showed an intensity and number of foci (‘blobs’) that were comparable in undifferentiated and differentiating myoblasts (Figure 1C). In contrast, MyoD and Brg1 co-localization was restricted to the nuclei of differentiating C2C12 cells (Figure 1D). Likewise, co-localization of BAF60c and Brg1 dramatically increases in differentiating myoblasts, although interactions between Brg1 and BAF60c could also be detected (albeit to a much lesser extent) in undifferentiated myoblasts (Figure 1E). Interestingly, in differentiating cells, interactions between these proteins were predominantly detected in the nuclear periphery, which is enriched in transcriptionally active SWI/SNF complexes (Brickner, 2009). These results are consistent with the co-immunoprecipitation data shown in Figure 1B, and indicate that BAF60c can be present in distinct complexes during skeletal myogenesis. In undifferentiated myoblasts, BAF60c can be found in Brg1-based SWI/SNF complexes, which do not contain MyoD, and also associated with MyoD in a separate complex that does not contain the Brg1 and Brm ATPases. In differentiating myoblasts, the formation of a complex containing MyoD, BAF60c and Brg1 suggests that differentiation cues promote the incorporation of MyoD–BAF60c into a Brg1-based SWI/SNF complex. This evidence indicates a signal-dependent exchange in composition of MyoD-associated SWI/SNF complexes, and suggests that distinct BAF60c-containing complexes are involved in the regulation of gene expression at different stages of skeletal myogenesis. This notion is consistent with specialized functions of distinct SWI/SNF subcomplexes, as recently reported for the regulation of neurogenesis (Wu et al, 2007) and more generally for a versatile control of gene expression (Wu et al, 2009).
BAF60c and Brg1 co-regulate the expression of MyoD-target genes during myoblast differentiation
To gain functional insight into the specific role of BAF60c in skeletal myogenesis, we performed gene expression profiling on C2C12 myoblasts that were subjected to siRNA-mediated knockdown of either one of each BAF60 variants or the SWI/SNF catalytic subunits Brg1 and Brm. In an initial phenotypical screening, we noted that only downregulation of Brg1 and BAF60c prevented the formation of multinucleated myotubes (Supplementary Figure S2A and B). In contrast, in BAF60b- (Supplementary Figure S2A and B), BAF60a- or Brm-depleted C2C12 cells (data not shown), the formation of multinucleated myotubes proceeded normally. Interestingly, downregulation of BAF60b (but not BAF60a and Brm) could also reduce the expression of certain muscle genes, such as muscle creatine kinase (MCK) (Supplementary Figure S2C). Based on this evidence, as well as on our initial identification by two-hybrid system that both BAF60b and c are MyoD-interacting proteins (Supplementary Figure S1A, H and I), and the partial, functional redundancy between BAF60b and c (Takeuchi and Bruneau, 2009), we decided to restrict our analysis to C2C12 cells in which Brg1, BAF60b or c were individually knocked down.
We performed gene expression profile analysis in differentiating C2C12 cells that were depleted of Brg1, BAF60b or BAF60c, as compared with the expression profile of control C2C12 cells (scrambled RNAi). We first determined the genes that were induced during muscle differentiation, by comparing the gene expression profile of undifferentiated myoblasts (GM) and differentiating myocytes (DM, 18 h). The large majority of the genes induced in DM coincided in the populations of native C2C12 cells or scrRNAi C2C12 cells, and were enriched in genes implicated in muscle differentiation and other specialized functions of differentiated skeletal muscles, such as contraction, cytoskeleton and metabolism. Gene expression microarray analysis of C2C12 cells in which Brg1, BAF60b or c were downregulated by RNAi showed unique and overlapping roles of these SWI/SNF components in the activation of genes involved in muscle differentiation, contractile activity and metabolism (the complete list of genes downregulated by siRNA-mediated depletion of each of these SWI/SNF subunits is accessible through GEO Series accession number GSE24573; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24573). Depletion of Brg1 or BAF60c led to the downregulation of the large majority of muscle differentiation genes; however, some of the genes downregulated in BAF60b-depleted C2C12 were also included within the category of muscle genes.
Intersections of the genes downregulated by independent knockdown of Brg1, BAF60b or BAF60c identified categories of genes that can be co-regulated by combinations of these SWI/SNF components (Supplementary Figure S2D; Supplementary Table SI). Specifically, among the genes induced during C2C12 myoblast differentiation, we identified genes that were downregulated by Brg1, BAF60b and BAF60c (denominated common downregulated genes -Brg1/BAF60b/BAF60c- in Supplementary Figure S2D and listed in Supplementary Table SI). These genes required the expression of each of these SWI/SNF components and contained a significant number of muscle-specific genes previously identified as MyoD-target genes (Sartorelli et al, 1997; Bergstrom et al, 2002; Blais et al, 2005; Cao et al, 2006, 2010; Di Padova et al, 2007). Importantly, analysis of C2C12 cells depleted of either Brg1 or BAF60c identified a large number of downregulated genes that were not affected by BAF60b downregulation (unique downregulated genes—Brg1/BAF60c-, Supplementary Figure S2D; Supplementary Table SI). Those genes were also enriched with previously identified MyoD-target genes. By contrast, very few downregulated genes were identified in C2C12 cells depleted of either Brg1 or BAF60b, but not affected by BAF60c depletion (unique downregulated genes—Brg1/BAF60b-, in Supplementary Figure S2D and listed in Supplementary Table SI). From these genes, only two MyoD-target genes were annotated in this list (Supplementary Table SI). Gene ontology and process network analysis (Supplementary Figure S3A–C) confirmed that the vast majority of genes co-regulated by Brg1 and BAF60c were involved in muscle differentiation and related functions. When we considered all the genes downregulated by Brg1 and BAF60c (by merging the list of common downregulated genes Brg1/BAF60b/BAF60c and unique downregulated genes Brg1/BAF60c—see the ‘enriched list’ in Supplementary Table SI), we found that most of the muscle genes induced during C2C12 differentiation were included in this list. These data indicate a unique, essential role of BAF60c in the activation of muscle differentiation and suggests that Brg1 and BAF60c are essential components of the SWI/SNF complex that promotes muscle gene expression during skeletal myogenesis. These data also indicate an ‘ancillary’ role for BAF60b in the expression of certain muscle genes. Interestingly, a functional redundancy between BAF60b and c has previously been described during cardiomyogenesis (Lickert et al, 2004; Takeuchi and Bruneau, 2009), but the modality of regulation of common genes by BAF60b and c has to be determined. However, the present and previous studies (Lickert et al, 2004; Takeuchi and Bruneau, 2009) indicate a functional hierarchy between BAF60b and BAF60c, with BAF60c playing a more essential role in the activation of muscle genes. This critical role of BAF60c was further confirmed by independent experiments in which BAF60c knockdown inhibited morphological and biochemical differentiation in clones of mouse C2C12 cells in which BAF60c was stably downregulated by retroviral delivery of shRNA (Supplementary Figure S4A–C) and in rat L8 myoblasts (Supplementary Figure S4D and E) that were transiently transfected with BAF60c siRNA. Importantly, depletion of BAF60c in all these muscle cells led to an impaired formation of myotubes and reduced expression of differentiation markers, such as myogenin and myosin heavy chain (MyHC) (Supplementary Figure S4).
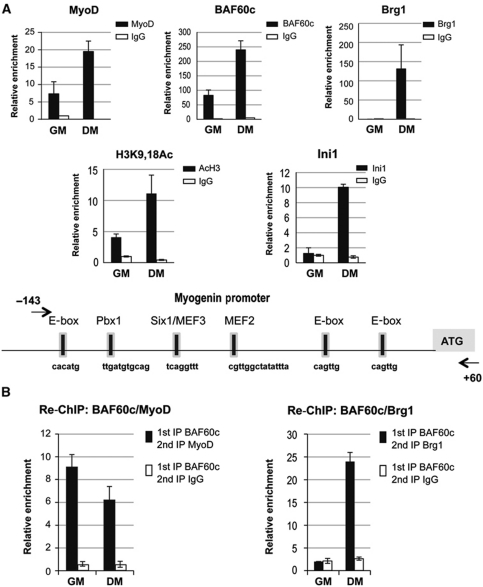
BAF60c/MyoD chromatin binding precedes Brg1-based SWI/SNF recruitment to MyoD-target genes
The enrichment in known MyoD-target genes within the category of genes co-regulated by Brg1 and BAF60c suggested that a Brg1/BAF60c-based SWI/SNF complex is recruited to the regulatory sequences of MyoD-target genes. From these genes, we selected myogenin for a detailed analysis of the chromatin recruitment of MyoD, Brg1 and BAF60c on the promoter elements. Myogenin promoter has been extensively analysed by previous studies, which have identified specific elements that mediate chromatin interactions of distinct TFs, including MyoD, the homodomain protein Pbx1, MEF2 and Six proteins (Edmondson et al, 1992; Knoepfler et al, 1999; Bergstrom et al, 2002; Berkes et al, 2004; Simone et al, 2004; de la Serna et al, 2005; Mal, 2006; Berghella et al, 2008; Palacios et al, 2010; Seenundun et al, 2010). ChIP analysis of the myogenin promoter revealed the presence of endogenous MyoD and BAF60c, but not Brg1, in proliferating myoblasts (Figure 2A), in which myogenin is not expressed. The structural SWI/SNF subunit Ini1 (BAF47), which is typically present in Brg1-based complexes, was also not detected on the myogenin promoter (Figure 2A). This evidence is consistent with the selective interaction of MyoD and BAF60c in undifferentiated myoblasts (Figure 1C) and their presence on myogenin promoter without Brg1-based SWI/SNF complex, prior to myogenin transcription. On induction of differentiation (differentiation medium—DM), which coincides with myogenin transcription, Brg1 and Ini1 were detected on the myogenin promoter, together with pre-existing MyoD and BAF60c (Figure 2A). This correlates with the enrichment in H3 acetylation (Figure 2A). To further substantiate the selective association of BAF60c with MyoD (but not Brg1) on myogenin promoter in undifferentiated myoblasts, we used re-ChIP analysis. To this purpose, we generated C2C12 cells stably expressing Flag-tagged BAF60c and used anti-Flag antibodies, which could immunoprecipitate larger amounts of BAF60c and therefore allowed efficient re-ChIP. Additionally, this strategy further discriminates between BAF60c and BAF60b. Re-ChIP analysis demonstrated co-occupancy of BAF60c and MyoD, but not Brg1, on the myogenin promoter in undifferentiated myoblasts (Figure 2B). However, in differentiating myoblasts, both MyoD and Brg1 were detected on BAF60c-bound chromatin at the myogenin promoter (Figure 2B). These results support the conclusion that in myoblasts, the myogenin promoter is occupied by an MyoD–BAF60c complex that is devoid of Brg1. On induction of differentiation, MyoD, BAF60c and the Brg1-containing SWI/SNF are recruited to the myogenin promoter, coinciding with the activation of myogenin transcription.
Figure 2.
BAF60c and MyoD are recruited to the myogenin promoter in myoblasts prior to the activation of transcription and the recruitment of Brg1-based SWI/SNF complex. (A) ChIP assays were performed in myoblasts and myotubes to monitor the recruitment of MyoD, BAF60c, Brg1, Ini1 and the relative enrichment in H3-K9-18 acetylation at myogenin promoter in proliferating, undifferentiated myoblasts (GM) and differentiating myoblasts (DM) (see Materials and methods for details). (B) Sequential ChIPs (re-ChIPs) were performed by immunoprecipitating the chromatin with antibodies against Flag–BAF60c followed by a second immunoprecipitation using MyoD (left panel) or Brg1 antibodies (right panel) in undifferentiated myoblasts (GM) and differentiating myoblasts (DM). ChIP values are normalized against the input and expressed as relative enrichment of the material precipitated by the indicated antibody on myogenin promoter (relative quantification using the comparative Ct method (2−(Ct sample−Ct input))). Error bars indicate mean±s.d. The graphic shown is representative of at least two independent experiments.
Previous studies showed that in undifferentiated myoblasts, the proximal element of the myogenin promoter is occupied by a nucleosome that precludes full access by the transcriptional activator MyoD (Gerber et al, 1997; de la Serna et al, 2001; Simone et al, 2004). However, an initial recruitment of MyoD on the distal region of the myogenin promoter has been described in myoblasts, via interaction with Pbx-Meis bound to a non-canonical E-box (Berkes et al, 2004). Our results showing that the MyoD–BAF60c complex could be first recruited to the myogenin promoter in the absence of the ATPase Brg1 are in agreement with these studies. Indeed, endonuclease assays show no remodelling of the myogenin promoter in undifferentiated myoblasts (Gerber et al, 1997; de La Serna et al, 2001; Simone et al, 2004; Serra et al, 2007). The recruitment of Brg1 and other SWI/SNF components to the myogenin promoter following the induction of differentiation and the consequent chromatin remodelling and gene transcription indicate that a differentiation-related event promotes the incorporation of MyoD–BAF60c into a Brg1-based SWI/SNF complex.
p38α-mediated phosphorylation of BAF60c promotes SWI/SNF recruitment to MyoD-target genes
The key role of p38 signalling in regulating the composition of the myogenic transcriptosome has been extensively established (Simone et al, 2004; Lluis et al, 2005; Rampalli et al, 2007). We have previously shown that p38-mediated phosphorylation of BAF60 correlates with the chromatin recruitment of the Brg1-based SWI/SNF complex to the myogenin promoter (Simone et al, 2004). However, these studies did not determine which specific BAF60 variant is phosphorylated by p38 kinases, nor did they address the functional consequences of this phosphorylation. Thus, we hypothesized that BAF60c phosphorylation by differentiation-activated p38 kinases could be the signal that promotes the recruitment of a Brg1-based SWI/SNF complex on the myogenin promoter in differentiating muscle cells.
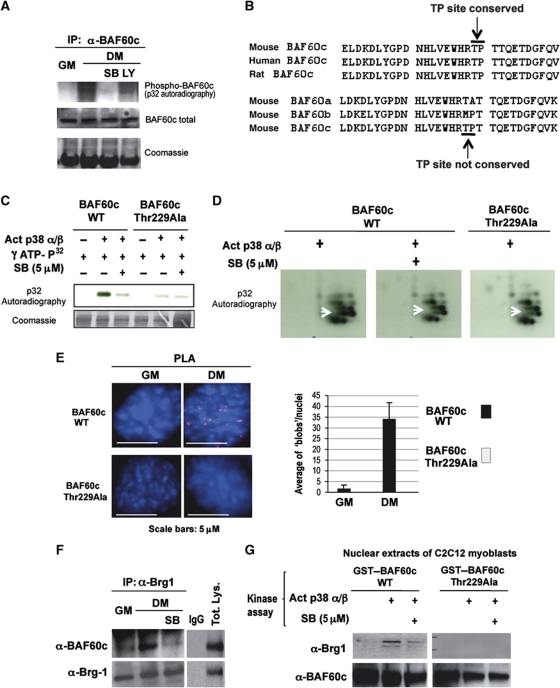
We detected a differentiation-dependent phosphorylation of endogenous BAF60c after 32P orthophosphate labelling of C2C12 (Figure 3A). The 32P incorporation was observed only upon induction of differentiation (DM) and was prevented by p38α/β blockade with SB203580 (SB), but not by the Pi3K inhibitor LY294002 (LY) (Figure 3A). Inspection of the amino-acid sequence of BAF60c revealed the presence of a consensus site for p38-mediated phosphorylation—the proline-directed threonine 229—that was conserved in BAF60c from mouse, human and rat, but not present in the other BAF60 variants (Figure 3B). In-vitro kinase assay (Figure 3C) and phospho-peptide analysis (Figure 3D) performed with purified, constitutively active p38α and β kinases and GST–BAF60c demonstrated that these kinases can directly phosphorylate BAF60c. Replacement of threonine 229 with alanine generated a p38 phosphorylation-resistant BAF60c mutant (BAF60c Thr229Ala) (Figure 3C and D).
Figure 3.
BAF60c phosphorylation by p38α/β kinases mediates Brg1 recruitment. (A) In-vivo 32P labelling of C2C12 cells induced to differentiate (DM) in the presence or not of p38α/β inhibitor (SB) or Pi3K inhibitor (LY) was followed by immunoprecipitation of endogenous BAF60c and detection of 32P incorporation after SDS gel-electrophoresis. (B) The p38 consensus site was identified by sequence analysis of BAF60c using BioEdit software (Hall, 1997). A proline-directed threonine 229, suitable for p38 phosphorylation and conserved in BAF60c of mouse, rat and human is indicated with arrow (TP site). Note that the p38 consensus site is not present in BAF60a or BAF60b. (C) In-vitro kinase assay using active p38α and β as kinases, GST–BAF60c as a substrate and radioactive γ-32P-ATP in the presence or absence of SB. (D) The bands from the gel displayed in (C) were cut, eluted, trypsin digested and run on a two-dimensional gel. Arrow indicates a spot that disappears in the BAF60c wt in the presence of SB and in the BAF60c Thr229Ala mutant. (E) PLA was performed in C2C12 cells expressing Flag-tagged BAF60c wt or the BAF60c Thr229Ala mutant, using an anti-proline directed phospho-threonine antibody (mouse) and an anti-Flag antibody (rabbit). (F) Co-immunoprecipitation of endogenous BAF60c and Brg1 from C2C12 undifferentiated (GM) or differentiating (DM) myoblasts, in the presence or absence of the p38α and β kinase inhibitor SB. (G) GST pull-down assay with GST–BAF60c wt or Thr229Ala mutant, first incubated with p38α and β kinases in a buffer permissive for phosphorylation, in the presence or not of SB, and then incubated with nuclear extracts from C2C12 cells. The precipitated material was blotted with anti-Brg1 and BAF60c antibodies.
We further analysed the p38-mediated threonine phosphorylation of BAF60c in vivo by transfecting Flag-tagged BAF60c wild-type (wt) and Thr229Ala mutant and monitoring their phosphorylation by PLA, using anti-phospho-threonine and anti-Flag antibodies. In this case, the signal (nuclear ‘blobs’) indicates threonine phosphorylation of Flag-tagged protein (BAF60c). Figure 3E shows a signal detected with Flag–BAF60c wt, but not with the Flag–BAF60c Thr229Ala mutant upon differentiation. We next determined the impact of p38-phosphorylation on BAF60c interaction with Brg1. We reasoned that if MyoD-associated BAF60c associates with Brg1 in differentiation conditions, but not in undifferentiated myoblasts, BAF60c phosphorylation on Threonine 229 by p38α/β kinases could be the signal that promotes the interaction between MyoD-associated BAF60c and Brg1. Indeed, p38α/β blockage by SB reduced the efficiency of co-immunoprecipitation of Brg1 with Baf60c from nuclear extracts of differentiating C2C12 cells (Figure 3F). A complementary assay was also used in which GST–BAF60c wt or Thr229Ala mutant were incubated with C2C12 nuclear extracts after previous phosphorylation in vitro by p38α/β. A GST pull-down assay showed that only p38-phosphorylated BAF60c interacted with the endogenous Brg1 present in the nuclear extracts, and this interaction was reduced by inhibition p38α/β kinases. In contrast, BAF60 Thr229Ala mutant did not interact with Brg1 in any of the experimental sample points (Figure 3G, lanes 4–6). This in-vitro assay presumably does not detect the in vivo interactions between endogenous BAF60c and Brg1, which occur in undifferentiated myoblasts independent of BAF60c phosphorylation (Figure 1E).
Indeed, PLA performed with BAF60c Thr229Ala mutant showed the same extent of co-localization with endogenous Brg1 and MyoD in undifferentiated (GM) and differentiating (DM) myoblasts (Supplementary Figure S5), whereas an increased signal was obtained with BAF60c wt and Brg1 in differentiating myoblasts (see Figure 1E). This evidence further demonstrates that in the absence of p38 activation (i.e., in undifferentiated myoblasts), unphosphorylated BAF60c forms two distinct complexes with Brg1 and MyoD, respectively. Conversely, p38-directed phosphorylation of BAF60c promotes the formation of one complex containing both MyoD and Brg1.
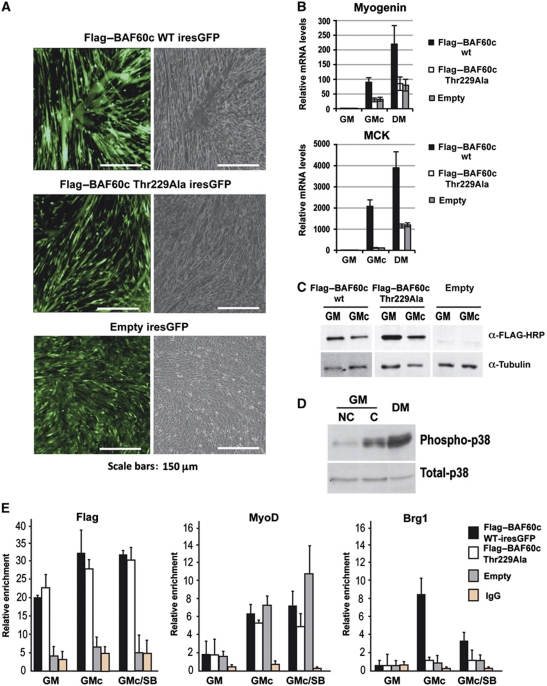
We next investigated the biological and biochemical properties of BAF60c wt versus Thr229Ala mutant in C2C12 myoblasts. Using retroviral infection to express proteins from GFP-ires containing transcript, we observed that overexpression of BAF60c wt by retroviral infection greatly accelerated the differentiation of confluent C2C12 myoblasts cultured in GM. BAF60c wt-expressing C2C12 cells showed an increased formation of multinucleated myotubes (Figure 4A) and expressed higher levels of the differentiation markers, such as myogenin and MCK (Figure 4B), as compared with C2C12 cells expressing the BAF60c Thr229Ala mutant (Figure 4A and B), despite comparable levels of ectopically expressed proteins (Figure 4C). Because cell confluence induces p38 activity in myoblasts cultured in GM conditions (Figure 4D), this experimental setting is instrumental to investigate the functional relationship between p38 activation, BAF60c phosphorylation and Brg1-based SWI/SNF recruitment to the myogenin promoter. ChIP analysis was used to determine whether the differential ability of BAF60c wt versus Thr229Ala mutant to promote differentiation and myogenin expression in C2C12 myoblasts was due to their different ability to bind to the chromatin of myogenin promoter and recruit MyoD and Brg1. Both BAF60c wt and Thr229Ala mutant bound to the myogenin promoter, and MyoD was detected on the same promoter in the presence of either of the proteins (Figure 4E). In confluent myoblasts (GMc) expressing empty vector, Brg1 recruitment to myogenin promoter was barely detectable; however, Brg1 chromatin binding was enhanced in C2C12 confluent myoblasts in which BAF60c wt was overexpressed, but not in BAF60c Thr229Ala-expressing cells (Figure 4E). This evidence further supports the conclusion that BAF60c phosphorylation by differentiation-activated p38α/β promotes the incorporation of MyoD-associated BAF60c into a Brg1-containing complex.
Figure 4.
Overexpression of BAF60c promotes myogenic differentiation. (A, B) Overexpression of BAF60c wt-iresGFP (top panel), BAF60c Thr229Ala iresGFP (middle panel) or the empty vector (lower panel) in confluent C2C12 myoblasts. (A) Effect on morphology and myotube formation. (B) Effect on the expression of muscle differentiation genes, myogenin and MCK in GMnc (growth medium, non-confluent cells) or GMc (growth medium, confluent cells). (C) Western blot analysis of ectopically expressed Flag–BAF60c wt, Flag–BAF60c Thr229Ala mutant or empty vector in C2C12 cells. (D) The activation status of endogenous p38 was monitored by western blot (using antibodies against phosphorylated p38, which indicates the active form, and total p38α) in C2C12 myoblasts growing in GMnc, GMc or DM conditions. (E) ChIP analysis of myogenin promoter in C2C12 cells expressing BAF60c wt (black bars), BAF60c Thr229Ala mutant (white bars) or empty vector (grey bars), using Flag–BAF60c, MyoD and Brg1 antibodies (the levels of IgG refer to BAF60c wt-infected cells and are taken as background for all experimental points). ChIP values are normalized against the input and expressed as relative enrichment of the material precipitated by the indicated antibody on myogenin promoter (relative quantification using the comparative Ct method (2−(Ct sample−Ct input))). Error bars indicate mean±s.d. The graphic shown is representative of at least two independent experiments.
A two-step model of signal-dependent SWI/SNF recruitment target genes during MyoD-directed myogenic conversion and in-vitro gene transcription
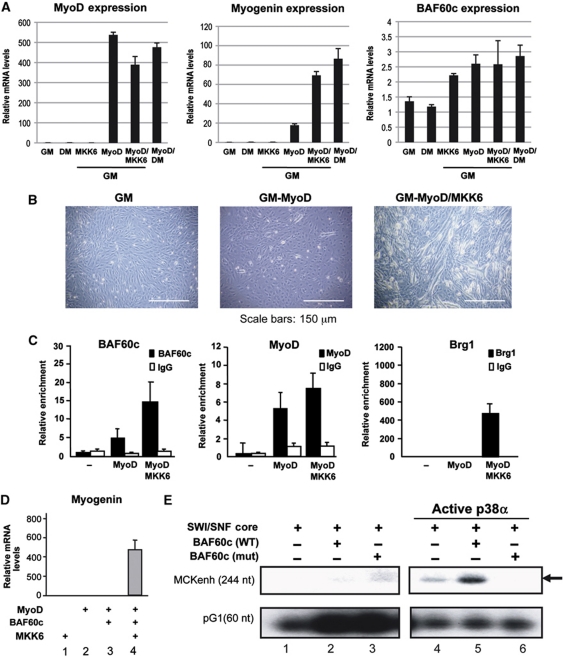
We used MyoD-mediated myogenic conversion of 10T1/2 fibroblasts to dissect in vivo the stepwise recruitment of Brg1 on the myogenin promoter by pre-assembled MyoD–BAF60c complex. 10T1/2 fibroblasts express BAF60c, but not MyoD. Therefore, myogenin is not expressed (Figure 5A). Ectopic expression of MyoD does not promote expression of myogenin, unless cells are exposed to differentiation medium (DM) (Figure 5A). However, deliberate activation of p38 kinases by ectopic expression of MKK6EE is sufficient to induce myogenin transcription (Figure 5A) and myotube formation (Figure 5B) in MyoD-expressing fibroblasts cultured in GM. Thus, this experimental setting provides an optimal system to delineate the individual role of MyoD and BAF60c phosphorylation in the recruitment of Brg1 to the myogenin promoter for gene activation. In the absence of MyoD, BAF60c does not occupy the myogenin promoter in 10T1/2 fibroblasts (Figure 5C). MyoD expression promotes myogenin promoter occupancy by MyoD itself and BAF60c, indicating that MyoD is required for chromatin recruitment of BAF60c at muscle genes. However, Brg1 recruitment to the myogenin promoter was only observed following MKK6EE-mediated activation of p38 kinases (Figure 5C).
Figure 5.
BAF60c and p38-dependent SWI/SNF recruitment to MyoD-target genes during MyoD-activated gene expression. (A–C) 10T1/2 myogenic conversion induced by ectopic expression of MyoD and a constitutively active form of MKK6. 10T1/2 cells were transduced with adenovirus for mock, MKK6EE and MyoD in the presence of growth media (GM) or differentiation media (DM). (A) Quantitative RT–PCR normalized for GAPDH expression shows an upregulation of Myogenin and MCK in the cells overexpressing MyoD and MKK6EE. MyoD expression was monitored for infection control purposes. (B) Phase contrast photographs show myotube formation when 10T1/2 cells are overexpressing MyoD and MKK6EE or MyoD alone in the presence of DM. (C) Chip analysis monitoring the recruitment of BAF60c, Brg1 and MyoD to the Myogenin promoter in the indicated treatments. Ectopic expression of MyoD and MKK6EE induces the recruitment of the indicated factors to the myogenin promoter. ChIP values are normalized against the input and expressed as relative enrichment of the material precipitated by the indicated antibody on myogenin promoter (relative quantification using the comparative Ct method (2−(Ct sample−Ct input))). Error bars indicate mean±s.d. The graphic shown is representative of at least two independent experiments. (D) Hela-MyoD cells were infected with a retroviral BAF60c in the presence or absence of adenoviral MKK6EE. Myogenin expression was measured by RT–PCR and normalized for GAPDH expression. (E) In-vitro transcription from ‘chromatinized’ MyoD-responsive pMCKenh-AL3 template. All lanes contain the MyoD∼E12 forced heterodimer, p300, PCAF and MEF2D and Hela-purified SWI/SNF complex (using Flag–Ini1 expressing HeLa cells and anti-Flag affinity purification). Active p38α kinase wt or mutant (mt) were included in the reaction when indicated.
Finally, we used in-vitro transcription from an MyoD-activated template that was incorporated into nucleosomal arrays to determine the importance of BAF60c phosphorylation by p38 kinases in mediating the functional interactions between the ATPase-containing SWI/SNF complex and MyoD. We used the natural mouse MCK enhancer (−1250 to −1000) fused to the minimal MCK promoter (−35 to +150)—pMCKenh-AL3 as an MyoD-responsive template. This gene regulatory region contains two characterized E-box sequences as well as other DNA-binding elements. Because of this, the activation of this template is more complex, when compared with an artificial promoter that contained four copies of the MCK right E-box (4R-MCK), which was used in previous studies (Dilworth et al, 2004). Indeed, a combination of MyoD-E12 forced heterodimers, with Mef2d, p300 and pCAF-purified proteins were sufficient to activate transcription from the 4R-MCK template (Dilworth et al, 2004), but not the ‘chromatinized’ pMCKenh-AL3. We therefore sought to use this template to investigate the role of the SWI/SNF complex in the activation of MyoD-mediated transcription. Surprisingly, the inclusion in the reaction of the SWI/SNF complex purified from HeLa cells (HeLa SWI/SNF) could not activate transcription (Figure 5E, lane 1). An explanation for this lack of effect was provided by the lack of BAF60c expression in HeLa cells (Wang et al, 1996). Indeed, HeLa cells are among the few cell types that show resistance to myogenic conversion following ectopic expression of MyoD (Weintraub et al, 1989; Figure 5D, lane 2). Reintroduction of BAF60c in HeLa cells did not rescue MyoD-mediated activation of myogenin transcription, unless the p38 activator MKK6EE was also expressed in these cells (Figure 5D, lanes 3 and 4). This evidence demonstrates that BAF60c deficiency is responsible for the resistance of HeLa cells to MyoD-mediated myogenic conversion, and that the activation of the p38 pathway is necessary for BAF60c to support MyoD activation of myogenin transcription in HeLa cells. Based on these findings, we sought to complement the HeLa SWI/SNF complex with purified BAF60c to activate MyoD-mediated transcription from pMCKenh-AL3 in our in-vitro transcription system. Inclusion of BAF60c wt alone could not rescue pMCKenh-AL3 activation (Figure 5E, lane 2); however, when an activated form of p38α kinase (Khokhlatchev et al, 1997) was included in the reaction, together with BAF60c wt and SWI/SNF, a strong activation of pMCKenh-AL3 was observed (Figure 5E, lane 5). By contrast, phosphorylation-resistant BAF60c Thr229Ala mutant (mut) could not promote MyoD-mediated activation of pMCKenh-AL3, either alone (Figure 5E, lane 3) or in the presence of active p38α kinase (Figure 5E, lane 6).
This result conclusively establishes that BAF60c phosphorylation by p38 kinases mediates the recruitment of SWI/SNF to MyoD-target genes.
BAF60c is required for SWI/SNF-mediated activation of MyoD-target genes
The data presented so far indicate that MyoD and BAF60c are present on myogenin promoter before myogenin transcription, and that a BAF60c phosphorylation by differentiation-induced p38α mediates the incorporation of MyoD and BAF60c into a Brg1-based SWI/SNF complex. Therefore, MyoD-associated BAF60c could provide the signal-responsive element for SWI/SNF recruitment to the myogenin promoter. According to this model, MyoD–BAF60c would function as ‘pioneer’ complex that primes MyoD-target genes for recruitment of Brg1-based SWI/SNF complex, upon differentiation signals.
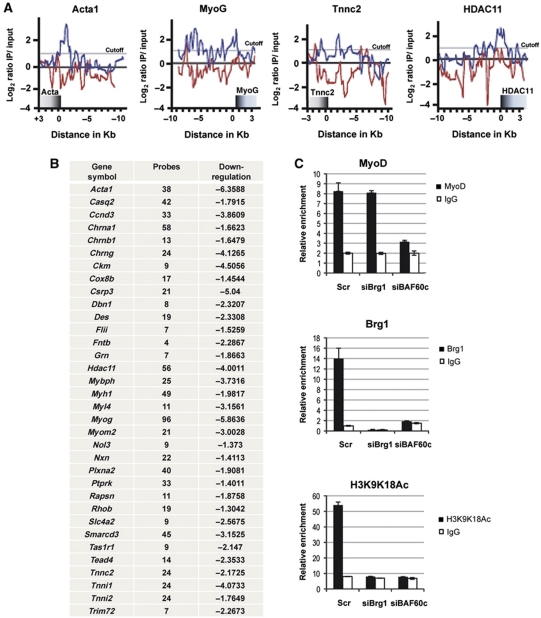
We investigated the effect of BAF60c depletion on the assembly of MyoD-associated chromatin-modifying enzymes by ChIP analysis as compared with normal C2C12. In the absence of BAF60c, the myogenin promoter was devoid of MyoD, Brg1 and several transcriptional stimulatory marks, which typically reflect the productive engagement of chromatin-modifying enzymes, such as p300 and PCAF acetyltransferases (acetyl-H3) (Puri et al, 1997) and the Ash2L-containing methyltransferase complex (H3K4 tri-methylation) (Rampalli et al, 2007) (Supplementary Figure S6). This result emphasizes the importance of BAF60c as a key pioneer factor for the assembly of the chromatin-remodelling machinery on the myogenin promoter. Interestingly, the absence of MyoD on the myogenin promoter in BAF60c-depleted cells suggests that BAF60c can also facilitate MyoD binding to its consensus elements.
We used ChIP-chip technology to further establish the importance of BAF60c in the regulation of MyoD–chromatin binding at target genes. We prepared high-density arrays containing MyoD-target genes that were identified by previous ChIP-chip studies (Blais et al, 2005; Cao et al, 2006). Recent work showed overlapping data from ChIP-chip and ChIP-seq analysis with anti-MyoD antibodies (Lei et al, 2010). Our ChIP-chip analysis confirmed the enrichment in MyoD binding on the large majority of the genes previously annotated (Blais et al, 2005; Cao et al, 2006, 2010) when the chromatin was prepared from control C2C12 myoblasts (Supplementary Table SII). However, in C2C12 cells in which the expression of BAF60c was downregulated by siRNA, we observed a significant reduction of MyoD binding in most of its target genes (Figure 6A and B; Supplementary Table SII). As we expected, genes that showed reduced binding of MyoD in the absence of BAF60c, presented a reduced expression (Figure 6B, right column). Notably, the reduced binding of MyoD in BAF60c-depleted cells was not a general outcome of SWI/SNF disruption, but was specific for BAF60c downregulation; siRNA-mediated downregulation of Brg1 did not affect MyoD binding on the myogenin promoter (Figure 6C), although both BAF60c and Brg1 are required for local hyperacetylation and expression of the myogenin gene (Figure 6C).
Figure 6.
BAF60c is required for MyoD recruitment to target genes. (A, B) MyoD binding to the chromatin of target genes was evaluated by ChIP-chip performed in C2C12 depleted of BAF60c by siRNA, as compared with control C2C12. (A) Differences in MyoD binding on representative genes. (B) List of genes in which MyoD binding was reduced by BAF60c depletion—left vertical row indicates the name of the genes in which MyoD chromatin binding is affected by BAF60c depletion; middle row indicates the number of probes for which reduced MyoD binding was observed upon BAF60c depletion; right vertical row indicates the extent of downregulation (cutoff >1.3-fold downregulation) for those genes in which MyoD binding was induced at least two two-fold during GM–DM transition (the number indicates the fold decrease of expression siBAF60c DM versus ctrl DM). (C) ChIP analysis of MyoD and Brg1 chromatin recruitment and H3-acetylation on myogenin promoter in C2C12 depleted of either BAF60c or Brg1 by siRNA, as compared with control C2C12. ChIP values are normalized against the input and expressed as relative enrichment of the material precipitated by the indicated antibody on myogenin promoter (relative quantification using the comparative Ct method (2−(Ct sample−Ct input))). Error bars indicate mean±s.d. The graphic shown is representative of at least two independent experiments.
This evidence, together with the data shown in Figure 5C, reveals positive reciprocal interactions between MyoD and BAF60c on the myogenin promoter, with MyoD directing BAF60c recruitment and BAF60c facilitating MyoD binding.
Discussion
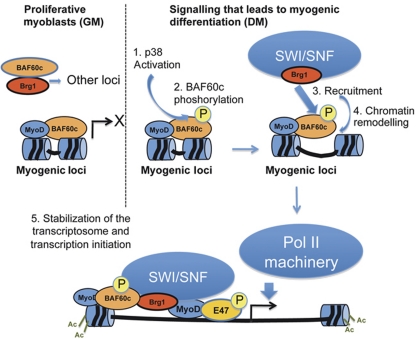
Our findings reveal an essential role for BAF60c during skeletal myogenesis and illustrate a previously undescribed mechanism for ‘core SWI/SNF’ recruitment to target genes during cellular differentiation. This model postulates that a pre-assembled, BAF60c–MyoD complex marks the chromatin of target genes for signal-dependent recruitment of ATPase-containing SWI/SNF remodelling complex. This signal is provided by differentiation-activated p38 kinases α/β and allows the formation of a remodelling competent SWI/SNF complex on MyoD-target promoters to displace the nucleosome, thereby permitting full access by MyoD to the promoter and the loading of MyoD-associated co-activator complexes (see model in Figure 7). This evidence explains the mechanism by which tissue-specific transcriptional activators overcome the barrier to activation of transcription at previously silent loci imposed by the presence of the nucleosome (Cairns, 2009; Radman-Livaja and Rando, 2010). Thus, the BAF60c–MyoD complex plays a pioneering role in marking the loci susceptible for chromatin remodelling on reception of differentiation cues.
Figure 7.
Illustration of the stepwise recruitment of SWI/SNF to MyoD-target genes by pre-assembled BAF60c–MyoD complex. In undifferentiated myoblasts, myogenin promoter is bound by MyoD and BAF60c, in the absence of Brg1-based SWI/SNF core complex. SWI/SNF core is recruited to the promoter by differentiation-activated p38 kinase α/β, through phosphorylation of BAF60c on threonine 229, thereby promoting chromatin remodelling and assembly of the transcriptosome for initiation of transcription.
BAFs (for Brg1/Brm-associated factors) are conventionally viewed as structural components of the SWF/SNF complex. BAF variants have been proposed as an interface for SWI/SNF interactions with a variety of transcriptional activators (Chi, 2004). For instance, BAF60c was previously implicated as a mediator of SWI/SNF interactions with nuclear receptors (Debril et al, 2004). We have found that structural components of the SWI/SNF complex, such as BAF60c, interact with transcriptional activators outside the SWI/SNF complex core and are only incorporated into ATPase-based SWI/SNF in response to extrinsic signals. This might explain the functional relationship between BAF variants, heterogeneous SWI/SNF composition, cell type-specific differentiation and stage-specific activation of restricted subsets of genes (Kadam and Emerson, 2003; Wu et al, 2007, 2009; Ryme et al, 2009). It will be of interest to know if some additional SWI/SNF components associate with BAF60c–MyoD in undifferentiated myoblasts and whether this complex might have repressive activity towards MyoD. In this respect, Flowers et al (2009) have shown that BRM-containing complexes can be detected on repressed promoters during osteoblast differentiation and are required for association of the co-repressor HDAC1, a well-know repressor of MyoD (Puri et al, 2001). However, we have failed so far to detect Brm on the chromatin of myogenin in undifferentiated myoblasts, using commercially available antibodies (data not shown).
The pioneering role of BAF60c in the assembly of the transcriptosome, via facilitation of TF binding and promoter recruitment of Brg1-based SWI/SNF, was recently suggested by Takeuchi and Bruneau (2009), who showed need for BAF60c in the chromatin recruitment of GATA4 and Brg1 on target genes in cardiac progenitors. Our study significantly extends this finding and provides mechanistic detail on BAF60c-mediated recruitment of ‘core SWI/SNF’ in response to differentiation signals, such as those that activate the p38 pathway. As such, our data provide an important framework that elucidates the mechanism that regulates SWI/SNF chromatin re-distribution in response to extrinsic signals.
In accordance with Takeuchi and colleagues, our data also indicate that BAF60c facilitates MyoD binding to target genes (see Figure 5). Thus, it is likely that BAF60c facilitates MyoD recognition of (or access to) target sequences that are masked by nucleosomes (Cairns, 2009). Previous studies showed that initial recruitment of MyoD to the myogenin promoter is mediated by the homeodomain factor Pbx1 on a distal non-canonical E-box (Berkes et al, 2004; de la Serna et al, 2005). It is possible that BAF60c mediates interactions between Pbx1 and MyoD (Berkes et al, 2004; Heidt et al, 2007), thereby facilitating MyoD access to early target genes/previously silent loci, such as myogenin.
Recent evidence by ChIP-seq demonstrated that MyoD–chromatin interactions in myoblasts coincide with peaks of histone acetylation (Cao et al, 2010). In previous studies, de La Serna et al (2005) also found that histone hyperacetylation preceded Brg1 binding to myogenin promoter in an MyoD-dependent manner. Thus, transient histone acetylation might link initial positioning of the MyoD/BAF60c complex, possibly through interaction with Pbx1. Interestingly, de La Serna et al (2005) detected MyoD binding only subsequent to histone acetylation and Brg1 interaction, supporting a two-step model whereby MyoD initially interacts indirectly with the myogenin promoter, and attracts chromatin-remodelling enzymes, which facilitate direct binding by MyoD and other regulatory proteins. Our data support an essential two-step role for BAF60c in this model. First, it mediates the initial recognition of the myogenin promoter by MyoD in myoblasts. Second, BAF60c phosphorylation mediated by p38α/β recruits the ATPase-containing SWI/SNF complex. Future studies are required to further characterize the mechanism by which BAF60c facilitates MyoD (or other TFs, such as GATA4) promoter recognition of target genes.
Materials and methods
Two-hybrid assay
C-terminus of mouse MyoD (aa 217–318) was used to screen mouse embryonic cDNA library in yeast two-hybrid system. In all, 1.5 million clones were selected and positive interactions were confirmed by transformation of yeast and by co-immunoprecipitation.
Cell cultures
Satellite cells were isolated by standard procedures (Serra et al, 2007). C2C12 myoblasts, 10T1/2 fibroblasts and HeLa cells were propagated in growth medium (GM): DMEM high glucose, without pyruvate and supplemented with 15% FBS (Omega). L8 myoblasts and HuSK were propagated in DMEM high glucose supplemented with 10% FBS. At 80% of confluence, differentiation was induced by placing the cultures in differentiation medium (DM)—2% horse serum supplemented with 1 × ITS (Sigma, I3146).
MyoD-mediated myogenic conversion assays
10T1/2 cells were infected with Adeno-Mock or Adeno-MyoD and incubated for 24 h in GM, prior to subsequent infection with Adeno-MKK6EE (or Adeno-Mock) (Simone et al, 2004). Cells were harvested for RT–PCR and ChIP analysis after additional 48 h in either GM or DM. HeLa and MyoD-expressing HeLa cells (Caretti et al, 2006) were infected with a lentivirus expressing BAF60c (or empty lentivirus) and then infected with Adeno-MKK6EE (or Adeno-Mock). After 5 days, cells were harvested for RT–PCR.
Viral infections
All retroviruses (retro-shRNAi BAF60c; retro-Flag–BAF60c) were generated by transfecting retroviral vectors into Phoenix-Ampho- packaging cells with Fugene6 reagent following the manufacturer's protocol. Virus-containing supernatants were collected at 48 h post-transfection and passed through a 0.45-μm filter. Infection of C2C12 was performed diluting the supernatants 1:5 and in the presence of 8 μg/ml of polybrene.
RNA interference
Downregulation of gene expression was obtained by transfecting with Dharmafect3 reagent (Dharmacon) C2C12, satellite cells and L8 with 100 nM final concentration of siGenome Smart pool collections (Dharmacon, for BAF60c #M-063444, BAF60b #M-048262, BAF60a #M-046893, Brg1 # L-041135, non-targeting pool # D-001810-10-20 and control siGlo #D-001620-03) following the manufacturer's protocol. Cells were induced to differentiate for 36 h in DM 36 h post-transfection. Stable downregulation of BAF60c expression in C2C12 was performed by standard transfection protocol using Fugene 6 (Roche) of a retro-pSUPER-shBAF60c. Selection with 4 mg/ml of puromycin (Sigma) was made after titration, and clones were picked and amplified.
Gene expression analysis (affymetrix)
Duplicates of growing C2C12 cells were transfected with siRNA oligonucleotides specifically targeting BAF60c, BAF60b, BAF60a and Brg1 (siGenome Smart pool collections, Dharmacon, for BAF60c #M-063444, BAF60b #M-048262, BAF60a #M-046893, Brg1 # L-041135, non-targeting pool #D-001810-10-20 and control siGlo #D-001620-03) at the final concentration of 100 nM with DharmaFECT3 reagent according to the manufacturer's instructions (Thermo Scientific). An siRNA against a scrambled sequence was used as control. After 36 h from the onset of transfection, cells were shifted in differentiation medium (ITS) and harvested after 18 h for gene expression analysis. For affymetrix analysis, total RNA was purified with TRIZOL Reagent (Invitrogen) and labelled cRNA was prepared from 500 ng RNA using the Illumina® RNA Amplification Kit from Ambion (San Diego, USA). The labelled cRNA (1500 ng) was hybridized overnight at 58°C to the Sentrix® MouseWG-6 Expression BeadChip (>46 000 gene transcripts; Illumina, San Diego, CA, USA) according to the manufacturer's instructions. BeadChips were subsequently washed and developed with fluorolink streptavidin-Cy3 (GE Healthcare). BeadChips were scanned with an Illumina BeadArray Reader. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE24573 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24573).
Expression data analysis
We analysed illumina MouseWG-6 Expression BeadChips using the manufacturers BeadArray Reader and collected primary data using the supplied Scanner software. Data analysis was done in three stages. First, expression intensities were calculated for each gene probed on the array for all hybridizations using illumina's GenomeStudio software. Second, intensity values were quality controlled and normalized: quality control was carried out by using the illumina Beadstudio detection P-value set to <0.05 as a cutoff. This procedure removed genes, which were effectively absent from the array (i.e., were not detected). After this step, the initial ∼46 000 genes were reduced to ∼22 000 (∼50% reduction). All the arrays were then normalized using the normalize.quantiles routine from the Affy package in Bioconductor. This procedure accounted for any variation in hybridization intensity between the individual arrays.
Finally, normalized data were imported into GeneSpring and analysed for differentially expressed genes. Only those genes with at least two-fold change during GM-to-DM transition were considered. Among these genes only those with a fold change of at least 1.3 annotated in duplicate experiments of RNAi for BAF60b, c or Brg1 were considered. These genes were analysed for the presence of overrepresented GO categories (Biological Process) and GeneGo processes using MetaCore software from GeneGo Inc.
Proximity ligation assay
C2C12 cells were plated on Labtek II chambers, placed in growth medium for 24 h, and then shifted to differentiation medium for 48 h. Cells were fixed with 4% paraformaldehyde for 10 min at RT, followed by an incubation of 6 min in 100% methanol at −20°C and an incubation with 0.1% Triton X in Tris-buffered saline (TBS) for 10 min. Rehydration was performed with TBS washes. Blocking was performed using the Duolink blocking solution following the manufacturer’s indications (Duolink™ in situ PLA kit, Olink) and published information (Fredriksson et al, 2002). The primary antibodies used in this assay are listed in Supplementary data. Combinations of antibodies were incubated with the cells in a humidified chamber overnight at 4°C. The incubation with the PLA probes, hybridization, ligation, amplification, detection and mounting was performed following Duolink kit indications.
Images were acquired with a Nikon microscope, edited using Image J and analysed with the BlobFinder software (Allalou and Wählby, 2009) that was used to localize and quantify the blobs from images acquired with fluorescent microscopy. This program allows quantifications for the number of blobs per nuclei. The average of blobs/nuclei in at least 10 cells from three independent experiments was reported in the graphics of Figures 1C, D, E and 3E.
Immunofluorescence
C2C12 cells wt or knockdown for BAF60c were fixed with 4% PFA for 10 min, permeabilized with 0.25% triton and blocked with 4% BSA in PBS 1 h at room temperature. Immunostaining with MyHC (MF20, DSHB) was performed O/N at 4°C and Alexa 555-conjugated secondary antibody was used (Invitrogen).
Western blots
Total cell extracts (lysis in 50 mM Tris–HCl (pH 8.0), 125 mM NaCl, 1 mM DTT, 5 mM MgCl2, 1 mM EDTA, 10% glycerol and 0.1% NP-40 supplemented with 1 mM PMSF and protease inhibitor cocktail (Roche)) were resolved in SDS–polyacrylamide gels and transferred to nitrocellulose membranes (Hybond-XL, Amersham). Primary antibodies are listed in Supplementary data. Secondary antibodies were revealed with the ECL chemoluminescence kit (Amersham).
RT–PCR
Total RNA was extracted with Trizol, and real-time quantitative PCR was performed using Power SYBR Green Master mix (Applied Biosystems) following the manufacturer's indications. Primers sequences are listed in Supplementary Table SIII. All quantitative PCR were performed following the indications of the manufacturer and relative expressions calculated by the comparative Ct method for relative quantification (Applied Biosystems, Real-Time PCR Systems) using GAPDH (for SYBR Green) or B2 microglobulin (for TaqMan) as normalizing genes.
Co-immunoprecipitation studies
C2C12 nuclear extracts prepared as described in Simone et al (2004), for each 150 cm2 dish, supernatants containing the nuclear extracts were diluted in 0.5 ml of dilution buffer, 20 mM Tris pH 7.9, 20% (vol/vol) Glycerol, 2 mM EDTA, 0.5 mM DTT, pre-cleared with protein A beads for 1 h at 4°C in rotation and immunoprecipitated (IP) with 50 ml of anti-FLAG M2 affinity gel (Sigma), in rotation for 2 h at 4°C. Immunoprecipitated material was washed extensively with dilution buffer plus increasing concentrations of KCl (150 mM twice, 175 mM twice and one 250 mM) then twice with TBS and once with 50 mM Tris pH 7.5. Protein complexes were eluted with 1 mg/ml FLAG peptide (Sigma) in 50 mM Tris pH 7.5.
Flag-tagged MyoD and Xp-tagged BAF60c were transfected into COS7 cells and allowed for growth and gene expression for 24 h. Cell extracts were harvested in 0.1% triton lysis buffer. In all, 1 μg anti-express antibody (Santa Cruz) and 20 μl protein A sepharose beads were used to immunoprecipitate XP-BAF60C from whole-cell extract with rotation in 4°C for overnight.
Antibodies for co-immunoprecipitations are listed in Supplementary data.
GST pull downs
GST proteins were purified from BL21 bacteria following standard procedures. The GST pull-down assays were performed as follows; 200 ng of magnet-GST–BAF60c beads were incubated ON in rotation at 4°C with 200 ng of flag-tagged recombinant proteins or 10 μg of nuclear extracts in 500 μl of IP buffer (50 mM Tris–HCl pH 7.9, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 5 mM MgCl2, 0.1% NP-40 plus protease inhibitors). Beads were washed three times with the IP buffer and the associated proteins eluted with 20 μl of Laemmli SDS sample buffer and analysed by 4–12% SDS–PAGE. The gels were transferred to a PVDF membrane and analysed by standard WB procedures (anti-Flag-HRP, Sigma, A8592; anti-Brg1 (Santa Cruz, sc-17796X)).
In-vitro kinase assay and phospho-peptide mapping
In all, 200 ng of sepharose-GST–BAF60 proteins were incubated with 100 ng of recombinant active p38 (Cell Signaling) in the presence of 60 mM MgCl2, 60 μM ATP, 50 mM Tris–HCl pH 7.5, 12 mM DTT and phosphatase inhibitors (Calbiochem), supplemented with 0.7 μCi of γ-(32P)ATP at room temperature for 20 min in continuous shacking. Reactions were stopped with Laemmli Buffer and resolved by SDS–PAGE; phosphorylated proteins were visualized by autoradiography. Protein loading was checked by Coomassie staining. γ-(32P) incorporation indicates p38-dependent phosphorylation background; however, γ-(32P) incorporation was enhanced in BAF60c wt and specifically eliminated by the inclusion of the p38α/β inhibitor SB203580. Phosphorylated bands were excised and subjected to phospho-peptide mapping.
ChIP and ChIP-Chip
ChIP
ChIP assay was performed as previously described (Simone et al, 2004) with some modifications. Briefly, DNA was double-crosslinked to proteins with 1.5 mM EGS (Pierce) and 1% formaldehyde (Sigma). After incubation for 10 min at 37°C, glycine was added to give a final concentration of 0.125 M for 5 min. The cells were washed twice with PBS containing 1 mM PMSF, scraped and pelleted. Lysis buffer (1% Triton-100, 0.1% deoxycholate, 50 mM Tris 8.1, 150 mM NaCl, 5 mM EDTA) was added to the samples and incubated on ice for 30 min. Sonication to obtain chromatin fragments of around 200–300 bp was performed using a Misonix 3000 sonicator (15″ON, 30″ OFF, for a total time of 4 min at output 4). For each immunoprecipitation, 3 mg of antibodies were conjugated to magnetic beads (G-protein magnetic Beads, Invitrogen). Antibodies used for ChIP are listed in Supplementary data. After extensive washing, bound DNA fragments were eluted and analysed by quantitative PCR using the SYBR Green Master Mix (Applied Biosystems). ChIP values are normalized against the input and expressed as relative enrichment of the material precipitated by the indicated antibody on myogenin promoter (relative quantification using the comparative Ct method (2−(Ct sample−Ct input))). Error bars indicate mean±s.d. The graphics shown in Figures 2A, B, 4E, 5C and 6C are representative of at least two independent experiments.
Chip primers are listed in Supplementary Table SIII.
ChIP-Chip
The IP DNA samples were amplified using the WGA2 whole genome amplification system (Sigma-Aldrich) according to the manufacturer's protocol. Amplification products were purified using the PCR purification columns (Qiagen). DNA concentration was measured at 260 nm. The quality of the amplified DNA samples was confirmed at OD260/280 and by agarose gel-electrophoresis. The amplified IP DNA and whole-cell extract DNA (WCE DNA) samples (1 μg) were labelled with Cy5-dUTP and Cy3-dUTP, respectively, using the Genomic DNA Enzymatic Labeling kit (Agilent). Labelled DNAs were hybridized with custom microarrays for 40 h at 65°C and then washed according to the manufacturer's instructions. Hybridization images were obtained using an Agilent DNA microarray scanner, and intensity data were extracted using Feature Extraction software Version 10.5 (Agilent). Genomic regions enriched by ChIP were identified using DNA Analytics version 4 software (Agilent).
ChIP-Chip data are available on the GEO database with the accession number.
In-vitro transcription
The plasmid pMCKenh-AL3 was generated by cloning the mouse MCK upstream enhancer (−1250 to −1000) and minimal promoter (−48 to +150) into the plasmid pAL3. The plasmid was incorporated into nucleosomal arrays using purified human histone octamers, and recombinant NAP-1 and ACF complex as previously described (Dilworth et al, 2004). Micrococcal nuclease digestion was performed as control to show the expected digestion pattern. Chromatinized pMCKenh-AL3 was incubated with 1 μM ATP, 1 μM acetyl CoA, Flag–MyoD-E12, Mef2d-Flag, His-p300 and Flag–pCAF in the presence or absence of His-p38α at 27°C. GST–BAF60c (wt or the T229A mutant) was included in reactions as indicated in Figure 5E. After 20 min, reactions were supplemented with 60 μg of HeLa nuclear extract and purified SWI/SNF complex, and allowed to incubate for 10 min at 30°C. Transcription was then initiated by the addition of rNTPs and the internal control plasmid template pG1 and allowed to proceed for 45 min at 30°C. Transcription reactions were then processed and analysed as previously described (Dilworth et al, 2004). S1 nuclease analysis of the transcription reactions provided a protected fragment of 244 nt when correctly transcribed from the pMCKenh-AL3 plasmid, and 60 nt when transcribed from the internal control pG1 vector.
Description of plasmids, cloning and retroviral vectors; expression and purifications of recombinant TFs are provided in Supplementary data.
Supplementary Material
Acknowledgments
We thank Dr T Sudo for anti-p38α antibodies, Dr J Auwerx for anti-BAF60c antibodies and BAF60c constructs, Dr Imbalzano for anti-Ini1 antibodies and Dr Sartorelli for providing Hela-MyoD cells. We also thank Dr S Williams for assistance on phospho-peptide mapping of BAF60c. We thank Ianessa Morantte for technical assistance and Harvey Evans for editing help. PLP is an Associate Telethon Scientist of the Dulbecco Telethon Institute (DTI) and Associate Investigator of Sanford Children's Health Research Center. This work was supported by the following grants to PLP: R01 AR056712 and AR053779 from the National Institute of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), Associazione Italiana Ricerca sul Cancro (AIRC), Sanford Children Health Research award. This work was benefited from research funding from the European Community's Seventh Framework Programme in the project FP7-Health—2009 ENDOSTEM 241440 (Activation of vasculature associated stem cells and muscle stem cells for the repair and maintenance of muscle tissue). SA was supported by a CIRM training grant. SF was supported by Instituto de Salud Carlos III (PI09/2444).
Author contributions: PLP designed most of the experiments of the manuscript, in collaboration with SVF. ZW provided the two-hybrid data. FJD provided the in-vitro transcription data. SVF executed most of the experiments, with the collaboration of SA, LG, BM, LC, AC, PC, VS, SC and KW. SB and AM (Agilent) performed the ChIP-Chip hybridization. RW uploaded the expression arrays on the GEO systems. PLP wrote the manuscript, in collaboration with SVF. All the authors discussed, commented the results and read the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albini S, Puri PL (2010) SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: it's time to exchange!. Exp Cell Res 316: 3073–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allalou A, Wählby C (2009) BlobFinder, a tool for fluorescence microscopy image cytometry. Comput Methods Programs Biomed 94: 58–65 [DOI] [PubMed] [Google Scholar]
- Berghella L, De Angelis L, De Buysscher T, Mortazavi A, Biressi S, Forcales SV, Sirabella D, Cossu G, Wold BJ (2008) A highly conserved molecular switch binds MSY-3 to regulate myogenin repression in postnatal muscle. Genes Dev 22: 2125–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ (2002) Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell 9: 587–600 [DOI] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ (2004) Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 14: 465–477 [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD (2005) An initial blueprint for myogenic differentiation. Genes Dev 19: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH (2009) Transcriptional memory at the nuclear periphery. Curr Opin Cell Biol 1: 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR (2009) The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ (2006) Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J 25: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, Gentleman RC, Tapscott SJ (2010) Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V (2006) The RNAhelicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell 4: 547–560 [DOI] [PubMed] [Google Scholar]
- Chi T (2004) A BAF-centred view of the immune system. Nat Rev Immunol 12: 965–977 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN (2001) Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet 27: 187–190 [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN (2005) MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol 25: 3997–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN (2006) Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet 7: 461–473 [DOI] [PubMed] [Google Scholar]
- Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J (2004) Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem 279: 16677–16686 [DOI] [PubMed] [Google Scholar]
- Di Padova M, Caretti G, Zhao P, Hoffman EP, Sartorelli V (2007) MyoD acetylation influences temporal patterns of skeletal muscle gene expression. J Biol Chem 282: 37650–37659 [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ (2004) In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc Natl Acad Sci USA 101: 11593–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Cheng TC, Cserjesi P, Chakraborty T, Olson EN (1992) Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol Cell Biol 12: 3665–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers S, Nagl NG Jr, Beck GR Jr, Moran E (2009) Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J Biol Chem 15: 10067–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, Ostman A, Landegren U (2002) Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 5: 473–477 [DOI] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ (1997) Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev 11: 436–450 [DOI] [PubMed] [Google Scholar]
- Gong F, Fahy D, Smerdon MJ (2006) Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol 13: 902–907 [DOI] [PubMed] [Google Scholar]
- Hall TA (1997) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98 [Google Scholar]
- Heidt AB, Rojas A, Harris IS, Black BL (2007) Determinants of myogenic specificity within MyoD are required for noncanonical E box binding. Mol Cell Biol 16: 5910–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM (2003) Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell 11: 377–389 [DOI] [PubMed] [Google Scholar]
- Khokhlatchev A, Xu S, English J, Wu P, Schaefer E, Cobb MH (1997) Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. Efficient synthesis of active protein kinases. J Biol Chem 272: 11057–11062 [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Bergstrom DA, Uetsuki T, Dac-Korytko I, Sun YH, Wright WE, Tapscott SJ, Kamps MP (1999) A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx-Meis/Prep1. Nucleic Acids Res 27: 3752–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Fukushige T, Niu W, Sarov M, Reinke V, Krause M (2010) A widespread distribution of genomic CeMyoD binding sites revealed and cross validated by ChIP-Chip and ChIP-Seq techniques. PLoS One 5: e15898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Crabtree GR (2010) Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol 10: 503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FQ, Coonrod A, Horwitz M (1996) Preferential MyoD homodimer formation demonstrated by a general method of dominant negative mutation employing fusion with a lysosomal protease. J Cell Biol 135: 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG (2004) Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432: 107–112 [DOI] [PubMed] [Google Scholar]
- Lluis F, Ballestar E, Suelves M, Esteller M, Munoz-Canoves P (2005) E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. EMBO J 24: 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal AK (2006) Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J 25: 3323–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M (2001) When the SWI/SNF complex remodels…the cell cycle. Oncogene 20: 3067–3075 [DOI] [PubMed] [Google Scholar]
- Palacios D, Summerbell D, Rigby PW, Boyes J (2010) Interplay between DNA methylation and transcription factor availability: implications for developmental activation of the mouse Myogenin gene. Mol Cell Biol 15: 3805–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J (2006) Mammalian SWI/SNF complexes facilitate DNA-double strand break repair by promoting gamma-H2AX induction. EMBO J 25: 3986–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Iezzi S, Stiegler P, Chen TT, Shiltz L, Muscat G, Giordano A, Wang JYJ, Sartorelli V (2001) Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell 8: 885–897 [DOI] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V (2000) Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol 185: 155–173 [DOI] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, Levrero M (1997) Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell 1: 35–45 [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ (2010) Nucleosome positioning: how is it established, and why does it matter? Dev Biol 339: 258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, Dilworth FJ (2007) p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol 14: 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson EA (2009) The SWI/SNF and cancer. Oncogene 28: 1653–1668 [DOI] [PubMed] [Google Scholar]
- Ryme J, Asp P, Böhm S, Cavellán E, Farrants AK (2009) Variations in the composition of mammalian SWI/SNF chromatin remodelling complexes. J Cell Biochem 108: 565–576 [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Caretti G (2005) Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev 15: 528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Huang J, Hamamori Y, Kedes L (1997) Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol 17: 1010–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ (2010) UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J 29: 1401–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, Jones DR, Du K, Jhala US, Simone C, Puri PL (2007) Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell 28: 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL (2004) p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 36: 738–743 [DOI] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR (2010) Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141: 943–955 [DOI] [PubMed] [Google Scholar]
- Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL (2009) Recombinational repair within heterochromatin requires AP-dependent chromation remodeling. Cell 138: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG (2009) Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459: 708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ (2005) The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132: 2685–2695 [DOI] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR (1996) Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev 10: 2117–2130 [DOI] [PubMed] [Google Scholar]
- Weintraub H (1993) The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell 75: 1241–1244 [DOI] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD (1989) Activation of muscle-specific genes in pigment, nerve, fat, liver and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA 86: 5434–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR (2009) Understanding the words of chromatin regulation. Cell 136: 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR (2007) Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56: 94–108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.