Abstract
Regulation of mtDNA expression is critical for maintaining cellular energy homeostasis and may, in principle, occur at many different levels. The leucine-rich pentatricopeptide repeat containing (LRPPRC) protein regulates mitochondrial mRNA stability and an amino-acid substitution of this protein causes the French-Canadian type of Leigh syndrome (LSFC), a neurodegenerative disorder characterized by complex IV deficiency. We have generated conditional Lrpprc knockout mice and show here that the gene is essential for embryonic development. Tissue-specific disruption of Lrpprc in heart causes mitochondrial cardiomyopathy with drastic reduction in steady-state levels of most mitochondrial mRNAs. LRPPRC forms an RNA-dependent protein complex that is necessary for maintaining a pool of non-translated mRNAs in mammalian mitochondria. Loss of LRPPRC does not only decrease mRNA stability, but also leads to loss of mRNA polyadenylation and the appearance of aberrant mitochondrial translation. The translation pattern without the presence of LRPPRC is misregulated with excessive translation of some transcripts and no translation of others. Our findings point to the existence of an elaborate machinery that regulates mammalian mtDNA expression at the post-transcriptional level.
Keywords: Leigh syndrome French Canadian variant, LRPPRC, mitochondria, RNA stability, SLIRP
Introduction
The regulation of mammalian oxidative phosphorylation capacity in response to physiological demands and disease states is complex and requires the concerted action of both nuclear and mtDNA-encoded genes (Scarpulla, 2008). The mtDNA genome only encodes 13 proteins, but these are essential for the oxidative phosphorylation system (Larsson et al, 1998). Reduced mtDNA expression is a well-recognized cause of human mitochondrial disease (Tuppen et al, 2010) and is heavily implicated in age-associated diseases and ageing (Larsson, 2010; Wallace, 2010). Nuclear genes are necessary for maintenance and expression of mtDNA, for example, by controlling mtDNA copy number (Ekstrand et al, 2004), transcription initiation (Falkenberg et al, 2002) and translation (Metodiev et al, 2009; Camara et al, 2011).
Control of mtDNA transcription initiation is thought to have a key role in regulation of oxidative phosphorylation capacity. The basal machinery for transcription of mtDNA consists of the nuclear-encoded mitochondrial RNA polymerase (POLRMT), mitochondrial transcription factor A (TFAM; Parisi and Clayton, 1991) and mitochondrial transcription factor B2 (TFB2M; Bogenhagen, 1996; Falkenberg et al, 2002), which together are sufficient and necessary for in vitro transcription initiation from mtDNA fragments containing the heavy and light strand promoter (HSP and LSP; Falkenberg et al, 2002). Mitochondrial transcription generates large polycistronic transcripts, which undergo RNA processing to release 13 mRNAs, 2 rRNAs and 22 tRNAs. In the polycistronic transcripts, mRNAs are often flanked by tRNAs and endonucleolytic processing to release tRNAs will therefore also release mRNAs, according to the so-called tRNA punctuation model (Ojala et al, 1981). The enzymatic excision of tRNAs involves two enzymatic activities, that is, RNase P at the 5′ end (Holzmann et al, 2008) and RNase Z suggested to process the 3′ end (Takaku et al, 2003; Dubrovsky et al, 2004). Most mRNAs are subsequently polyadenylated by the mitochondrial polyA polymerase (mtPAP; Tomecki et al, 2004) and polyadenylation is often necessary to generate the stop codon at the 3′ end of the open reading frame encoded by the mRNA. A number of enzymes are involved in rRNA (Metodiev et al, 2009; Camara et al, 2011) and tRNA modification (Nagaike et al, 2001; Suzuki et al, 2011). The function of polyadenylation, besides generating stop codons in some transcripts, is not fully understood. Polyadenylation is implicated in regulation of mitochondrial mRNA stability (Nagaike et al, 2005; Slomovic and Schuster, 2008; Wydro et al, 2010) and a mutation in the mtPAP gene has been reported to cause impaired mitochondrial function and ataxia in humans (Crosby et al, 2010).
The mechanism whereby mature mRNAs are recognized by the ribosome for subsequent translation initiation is well characterized in prokaryotes. Most prokaryotic mRNAs have an untranslated region (UTR) upstream of the start codon containing a so-called Shine–Dalgarno (SD) sequence. This SD sequence is complementary to a sequence in the 16S rRNA of the 30S bacterial ribosomal subunit and allows the mRNA start codon to find the correct position at the P site of the ribosome (Shine and Dalgarno, 1974). In yeast mitochondria, mRNA recognition by the ribosome takes advantage of the affinity between the 5′ UTR of the mRNA and transcript-specific translational activators. One such example is PET309, a proposed homologue of leucine-rich pentatricopeptide repeat containing (LRPPRC), which acts as a specific translational activator for the COXI mRNA to promote translation initiation (Tavares-Carreon et al, 2008). Mammalian mitochondrial mRNAs do not have 5′ UTRs and an alternate mechanism must therefore be responsible for mRNA recognition by mammalian ribosomes.
The pentatricopeptide repeat (PPR) protein family was first discovered in plants and is characterized by a canonical, often repeated, 35 amino acid motif involved in RNA binding. A surprisingly large number of PPR proteins have been reported in plants, where they are implicated in regulating processing, editing and stability of organelle genome transcripts in chloroplasts and mitochondria (Schmitz-Linneweber and Small, 2008; Zehrmann et al, 2011). Mammals have only seven PPR proteins and while the function of some has been at least partly elucidated (Holzmann et al, 2008; Xu et al, 2008; Davies et al, 2009; Rackham et al, 2009), the molecular mechanisms remain unclear. One of the mammalian PPR proteins, LRPPRC, was first discovered as being highly expressed in hepatoma cancer cell lines (Hou et al, 1994). Subsequent papers have associated LRPPRC with a ribonucleoprotein complex responsible for shuttling mature mRNAs from the nucleus to the cytosol (Mili and Pinol-Roma, 2003). LRPPRC has also been proposed to be a cofactor of the eukaryotic translation initiation factor 4E, which is involved in control of nuclear gene expression by regulating the export of specific mRNAs from the nucleus to the cytosol (Topisirovic et al, 2009). In addition, a nuclear role for LRPPRC has been reported as it has been shown to interact with the co-activator PGC-1α to regulate the expression of nuclear genes involved in mitochondrial biogenesis (Cooper et al, 2006). Recessive mutations of Lrpprc cause the French-Canadian type of Leigh syndrome (LSFC; Mootha et al, 2003), a mitochondrial disease which is characterized by infantile onset of severe neurodegeneration in the brain stem and a profound cytochrome c oxidase deficiency in liver and brain (Merante et al, 1993; Debray et al, 2011). Studies of the subcellular distribution of LRPPRC have demonstrated that it is mainly present in mitochondria (Tsuchiya et al, 2002; Mili and Pinol-Roma, 2003; Xu et al, 2004; Cooper et al, 2006; Sasarman et al, 2010; Sterky et al, 2010). We recently reported that transcription of the Lrpprc gene only seems to produce a single mRNA isoform, which is encoding an LRPPRC protein with a mitochondrial targeting sequence that is cleaved after import to the mitochondrial matrix (Sterky et al, 2010). Results from studies of cell lines indeed suggest that LRPPRC has an intramitochondrial role in regulation of mtDNA expression (Gohil et al, 2010; Sasarman et al, 2010), although the mechanism by which it acts is controversial (Sondheimer et al, 2010). On the one hand, it has been shown that knockdown of LRPPRC in tissue culture cells causes a general decrease in mitochondrial mRNA levels, impaired translation and a general decrease of the respiratory chain complexes (Sasarman et al, 2010). On the other hand, fibroblasts from LSFC patients have a respiratory chain deficiency mainly affecting complex IV. Immunoprecipitation experiments and blue native polyacrylamide gel electrophoresis (BN–PAGE) gel analyses have demonstrated that LRPPRC interacts with the stem-loop interacting RNA binding protein (SLIRP) in an RNA-independent way (Sasarman et al, 2010). SLIRP was initially described as a protein binding the nuclear RNA augmenting co-activation of nuclear receptors, but recent results suggest it is mainly present in mitochondria and has a role in maintaining mitochondrial mRNAs (Baughman et al, 2009). Recent studies have implicated LRPPRC in apoptosis (Michaud et al, 2011) and in autophagocytosis (Xie et al, 2011); however, these effects may be secondary because deficient oxidative phosphorylation is known to increase apoptosis (Wang et al, 2001; Kujoth et al, 2005) and has been reported to induce autophagy (Narendra et al, 2008).
We have characterized the in vivo function of LRPPRC by generating and characterizing conditional knockout mice. We report here that LRPPRC is essential for embryonic survival and that loss of LRPPRC in the heart leads to a drastic reduction in steady-state levels of all mitochondrial mRNAs, except ND6. LRPPRC forms an RNA-dependent complex with SLIRP, which is necessary for maintaining a pool of non-translated mitochondrial mRNAs. Loss of LRPPRC does not only lead to decreased mRNA stability but also causes loss of mRNA polyadenylation and the appearance of a misregulated mitochondrial translation pattern. Thus, LRPPRC has important roles in post-transcriptional regulation of mtDNA expression and is an essential regulator of oxidative phosphorylation capacity in mammals.
Results
LRPPRC is essential for embryonic development in the mouse
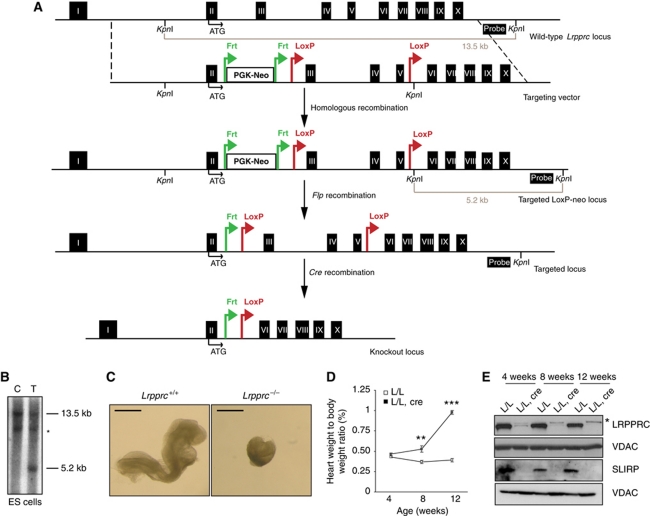
To determine the in vivo function of LRPPRC in mammals, we generated a conditional knockout allele of the mouse Lrpprc gene (Figure 1A and B). Lrpprc was targeted in embryonic stem (ES) cells and the mutated locus was transmitted through the germline to obtain heterozygous Lrpprc+/loxP−neo animals, which in turn, were crossed with transgenic mice expressing the Flp recombinase to excise the neomycin cassette (Figure 1A). The resulting Lrpprc+/loxP mice were mated to mice expressing cre recombinase under the control of the β-actin promoter to generate heterozygous Lrpprc knockout mice (Lrpprc+/−). Intercrossing of Lrpprc+/− mice produced no viable homozygous knockouts (Lrpprc−/−), whereas the other genotypes were recovered at expected Mendelian ratios (genotyped pups n=111; Lrpprc+/+ n=41, Lrpprc+/− n=70). We proceeded to dissect staged embryos derived from intercrossing of Lrpprc+/− mice. We analysed embryos at embryonic day (E) 8.5 and found ∼25% embryos with a mutant appearance (Figure 1C). Genotyping of these embryos (n=38) showed that all mutant embryos were homozygous knockouts (Lrpprc−/−, n=11) whereas the remaining normally appearing embryos had other genotypes (Lrpprc+/−, n=17 or Lrpprc +/+, n=10). These results show that loss of LRPPRC causes embryonic lethality at ∼E8.5, which is consistent with results from analyses of other mouse genes that are essential for mtDNA expression, for example, TFAM (Larsson et al, 1998), MTERF3 (Park et al, 2007), TFB1M (Metodiev et al, 2009) and MTERF4 (Camara et al, 2011).
Figure 1.
Disruption of Lrpprc in the germline and heart. (A) Targeting strategy for the conditional disruption of the Lrpprc gene. Probe and restriction sites used for the screening of the ES cells are shown in the picture. (B) Southern blot analysis of genomic DNA digested with KpnI in control (C) and targeted (T) ES cells. *Crossreacting band. (C) Morphology of wild-type (Lrpprc+/+) and homozygous knockout (Lrpprc−/−) embryos at day E8.5. Scale bar, 0.5 mm. (D) Heart weight to body weight ratio in control (L/L) and knockout mice (L/L, cre) at different ages. At 4 weeks L/L n=5, L/L, cre n=5; at 8 weeks L/L n=8, L/L, cre n=8; at 12 weeks L/L n=6, L/L, cre n=12. Error bars indicate s.e.m.; **P<0.01; ***P<0.001, Student's t-test. (E) Western blot analysis of LRPPRC and SLIRP levels in heart mitochondrial extracts from control (L/L) and knockout mice (L/L, cre) at different ages. VDAC was used as a loading control. *Crossreacting band.
Next, we performed a genetic rescue experiment by introducing a bacterial artificial chromosome (BAC) clone encoding LRPPRC with a C-terminal Flag tag (Supplementary Figure S1A). This clone was modified by introduction of a silent mutation, which eliminated a BglII site in exon 3, thereby enabling the discrimination of LRPPRC mRNAs expressed from the transgene or the endogenous gene (Supplementary Figure S1B). We obtained viable and normally appearing Lrpprc germline knockout mice containing the BAC clone (genotype Lrpprc−/−, +/BAC-LRPPRC–Flag) (Supplementary Figure S1B). The homozygous Lrpprc knockout can, thus, be rescued by the BAC transgene, showing that the knockout of the Lrpprc gene is causing the observed embryonic lethality.
Tissue-specific disruption of Lrpprc in heart causes mitochondrial cardiomyopathy
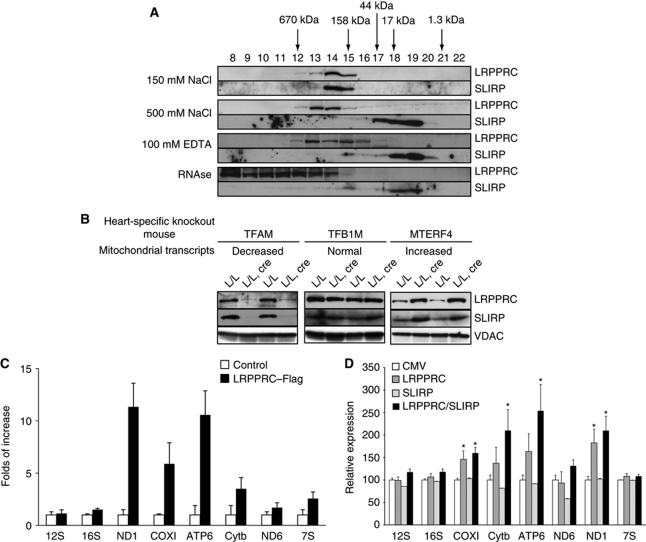
We proceeded to cross Lrpprc+/loxP mice with transgenic mice expressing cre recombinase under the control of muscle creatinine kinase promoter (Ckmm-cre), in order to generate mice with tissue-specific knockout of Lrpprc in heart and skeletal muscle. These conditional knockout mice had a drastically shortened lifespan and all of them died before 16 weeks of age (Supplementary Figure S1C). We calculated the ratio of the heart weight to body weight at different ages and found a progressive enlargement of the heart in the knockouts (Figure 1D). Western blot analyses of heart mitochondria showed that the LRPPRC protein was present at very low levels in 4-week-old heart knockouts and it could thereafter not be detected (Figure 1E). We found an accompanying absence of the SLIRP protein in Lrpprc heart knockouts at all investigated ages (Figure 1E), despite normal SLIRP mRNA levels (Supplementary Figure S1D). These results are consistent with reports by others that LRPPRC and SLIRP form a complex (Sasarman et al, 2010), and additionally show that the stability of the SLIRP protein depends on the presence of the LRPPRC protein.
Loss of LRPPRC in heart causes severe cytochrome c oxidase deficiency
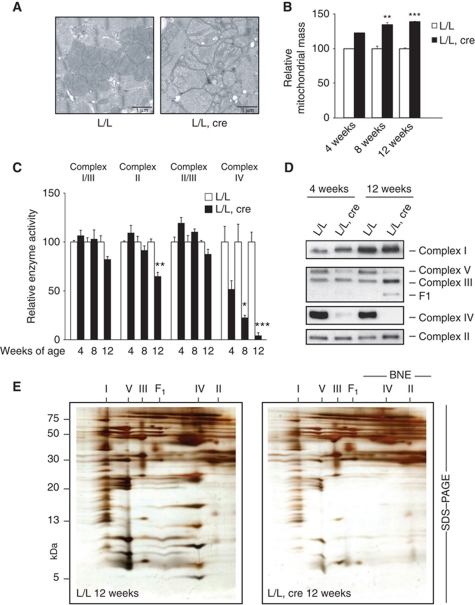
Electron micrographs of knockout heart tissue showed a progressive increase of mitochondrial mass and the presence of mitochondria with abnormally appearing cristae (Figure 2A and B). Consistently, we found increased enzyme activities of citrate synthase and glutamate dehydrogenase, two matrix proteins often used as markers for mitochondrial mass (Supplementary Figure S2A). The levels of mtDNA were normal in Lrpprc knockout hearts (Supplementary Figure S2B), demonstrating that LRPPRC is dispensable for mtDNA maintenance, despite being implicated as a component of the mitochondrial nucleoid (Bogenhagen et al, 2008). Measurement of respiratory chain function showed a profound reduction in complex IV activity in Lrpprc knockout hearts, whereas the activities of the other complexes were unaffected or showed a moderate decrease (Figure 2C). Also the nucleus-encoded complex II showed moderately decreased enzyme activity in end-stage knockout animals and this is likely a secondary phenomenon. We have previously seen a similar reduction of complex II activity in other mouse knockouts with disrupted mtDNA expression (Park et al, 2007; Metodiev et al, 2009). Complex II is dependent of FeS clusters for its function and superoxide-induced damage or impaired synthesis of FeS clusters caused by the deficient oxidative phosphorylation may provide an explanation for the observed enzyme deficiency.
Figure 2.
Loss of LRPPRC in heart causes mitochondrial dysfunction. (A) Electron micrographs of heart tissue from 12-week-old control (L/L) and knockout mice (L/L, cre). Scale bar, 1 μm. (B) Quantification of the relative mitochondrial mass obtained by electron microscopy analysis of control (L/L) and Lrpprc knockout (L/L, cre) hearts at 4 weeks (L/L n=2, L/L, cre n=2), 8 weeks (L/L n=3, L/L, cre n=3) and 12 weeks (L/L n=5, L/L, cre n=5). Error bars indicate s.e.m.; **P<0.01; ***P<0.001, Student's t-test. (C) Relative activities of respiratory chain enzyme complexes in heart mitochondria from control (L/L) and Lrpprc knockout mice (L/L, cre) at 4, 8 and 12 weeks of age. The analysed enzyme activities are NADH cytochrome c oxidoreductase (complex I/III), succinate dehydrogenase (complex II) and cytochrome c oxidase (complex IV). All activities are referred to milligrams of mitochondrial protein. The number of analysed animals at each time point were L/L (n=3) and L/L, cre (n=3). Error bars indicate s.e.m.; *P<0.05, **P<0.01, ***P<0.001, Student's t-test. (D) BN–PAGE analysis of the assembled respiratory chain complexes in heart mitochondria from control (L/L) and Lrpprc knockout (L/L, cre) mice at 4 and 12 weeks of age. Immunoblotting was performed to detect nuclear-encoded subunits of complex I (NDUFA9), complex II (SDHA), complex III (UQCRC2), complex IV (COXIV) and complex V (ATP5A1). The second panel from top shows a blot first hybridized with antibodies to detect complex III and then rehybridized with antibodies to detect complex V (without prior removal of complex III antibodies). (E) Heart mitochondria from 12-week-old control (L/L; left panel) and Lrpprc knockout mice (L/L, cre; right panel) were solubilized with DDM and separated by 2D BN/SDS–PAGE. Gels were silver stained. Assignment of oxidative phosphorylation complexes: complex I (I), complex V or ATP synthase (V), complex III (III), subcomplexes of ATP synthase (F1), complex IV (IV) and complex II (II).
We proceeded to analyse levels of assembled respiratory chain complexes by using BN–PAGE and observed a profound reduction of complex IV and a moderate reduction of ATP synthase (complex V) in Lrpprc knockout hearts from age 4 weeks and onwards (Figure 2D). In 12-week-old knockouts, an antibody against the α-subunit of the F1 portion of complex V detected an abnormal complex which had a size corresponding to an F1 subcomplex (Figure 2D). We have previously reported a similar subcomplex in other mouse knockouts with impaired mtDNA expression (Park et al, 2007; Metodiev et al, 2009; Camara et al, 2011). We performed further characterization of respiratory chain complexes by two-dimensional electrophoresis (BN–PAGE followed by SDS–PAGE) and found profound reduction of complex IV, moderate reduction in complexes I and V, and increased levels of the F1 subcomplex (Figure 2E). The levels of complex I were normal or moderately decreased and the levels of complex III were normal or moderately increased in Lrpprc knockout hearts at age 12 weeks (Figure 2D and E). In conclusion, we report here that the activity of complex IV is much more impaired than the activities of the other complexes in Lrpprc knockout hearts, which is in good agreement with the observation that LFSC patients have a profound complex IV deficiency (Merante et al, 1993; Debray et al, 2011). Knockdown of LRPPRC in cell lines has been reported to cause a generalized respiratory chain deficiency (Sasarman et al, 2010), which suggests that the biochemical phenotype in cell lines is at least partly different from the effects we have observed in differentiated tissues in vivo, perhaps due to the continuous proliferation and glycolytic metabolism of tissue culture cells.
LRPPRC regulates mitochondrial mRNA stability
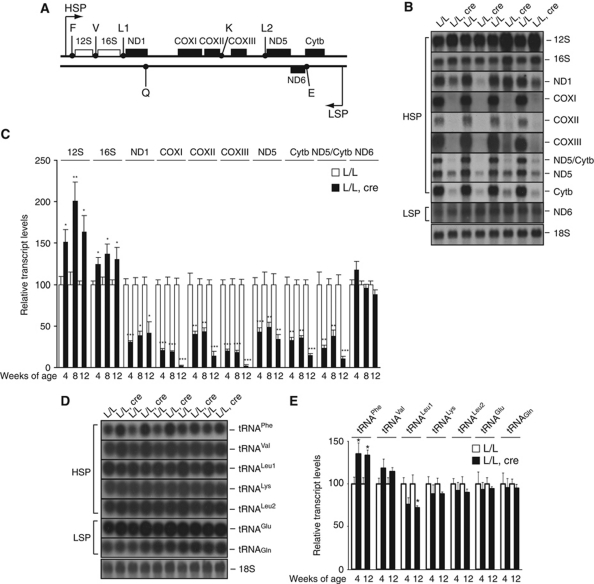
We performed northern blot analyses of mitochondrial transcripts in Lrpprc knockout hearts and found a profound decrease of most mRNAs already at 4 weeks of age (Figure 3A–C). The steady-state levels of the COXI, COXII, COXIII and Cytb mRNAs were only 2–20% of the levels in control hearts at age 12 weeks. These data are in agreement with the results obtained by knocking down LRPPRC in cell lines, which have shown decrease in steady-state levels of mRNAs (Cooper et al, 2006; Gohil et al, 2010; Sasarman et al, 2010). Interestingly, the levels of ND6, the only L strand-encoded mRNA, were not affected in the knockout hearts, demonstrating that LRPPRC is not important for stability of all mRNA species. In addition, we found that the levels of 12S rRNA and 16S rRNA were increased in the Lrpprc knockout hearts. We observed no significant change in the steady-state levels of a precursor transcript containing 16S and ND1 (RNA19; Supplementary Figure S3), suggesting that RNA processing is normal in the absence of LRPPRC.
Figure 3.
Steady-state levels of mitochondrial mRNAs, rRNAs and tRNAs in the absence of LRPPRC. (A) Linear map of mouse mtDNA indicating the relative position of each transcript analysed by northern blot. (B) Northern blot analysis of RNA isolated from heart tissues of control (L/L) and knockout (L/L, cre) mice at 12 weeks of age. A separate autoradiograph is shown for every analysed transcript. Nuclear ribosomal RNA (18S) was used as a loading control. (C) Quantification of steady-state levels of the transcripts from control (L/L) and knockout (L/L, cre) mice at different ages. At 4 weeks n=5, at 8 weeks n=5 and at 12 weeks n=4. Error bars indicate s.e.m.; *P<0.05, **P<0.01, ***P<0.001, Student's t-test. (D) Northern blot analysis of RNA isolated from heart tissue of control (L/L) and knockout (L/L, cre) mice at 12 weeks of age. Nuclear ribosomal RNA (18S) was used as a loading control. (E) Quantification of steady-state levels of mitochondrial tRNAs from control (L/L) and knockout (L/L, cre) mice at different ages. At 4 weeks n=5 and at 12 weeks n=5. Error bars indicate s.e.m.; *P<0.05, Student's t-test.
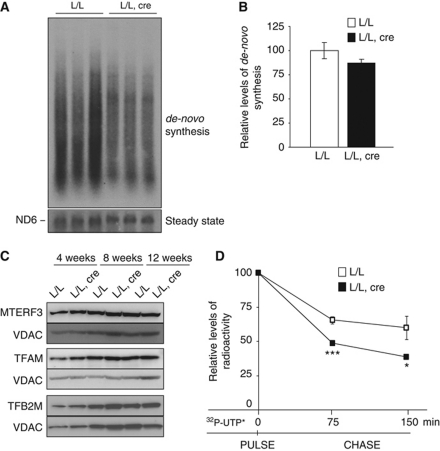
In previous studies, we have reported a good correlation between levels of tRNAs and de novo transcription activity (Park et al, 2007; Metodiev et al, 2009; Camara et al, 2011). Therefore, the finding of normal steady-state levels of tRNAs (Figure 3D and E) and low steady-state levels of mRNAs (Figure 3B and C) is likely not due to inhibition of transcription in Lrpprc knockout hearts. We verified this prediction by performing in organello transcription experiments, which showed normal de novo transcription of mtDNA in knockout mitochondria (Figure 4A and B). In additional support for unaltered transcription, we found normal levels of the transcriptional activators TFAM (Parisi and Clayton, 1991) and TFB2M (Falkenberg et al, 2002) and normal levels of the transcriptional repressor MTERF3 (Park et al, 2007; Figure 4C). Next, we studied the stability of newly synthesized transcripts by performing in organello transcription assays after labelling mitochondrial transcripts with 32P-UTP followed by a cold chase. The stability of newly synthesized mitochondrial transcripts was significantly lower in LRPPRC-deficient heart mitochondria than in control mitochondria (Figure 4D). We conclude that LRPPRC is not a critical transcriptional regulator, but rather plays a role in the regulation of mitochondrial mRNA stability.
Figure 4.
Loss of LRPPRC does not affect mitochondrial transcription, but reduces stability of mitochondrial transcripts. (A) In organello transcription in heart mitochondria isolated from 12-week-old control (L/L) and knockout (L/L, cre) mice. ND6 was used as a loading control, due to its unchanged steady-state levels. (B) Quantification of the bulk of newly synthesized mitochondrial transcripts in heart mitochondria of control (L/L) and knockout (L/L, cre) mice. L/L n=3 and L/L, cre n=3. Error bars indicate s.e.m. (C) Western blot analysis of the steady-state levels of proteins involved in regulation of mitochondrial transcription, MTERF3, TFAM and TFB2M, in heart mitochondrial extracts from control (L/L) and knockout (L/L, cre) mice at different ages. VDAC was used as a loading control. (D) Pulse-chase analysis of the stability of newly synthesized mitochondrial transcripts in heart mitochondria from control (L/L) and knockout (L/L, cre) mice. Mitochondrial transcription products were pulse labelled for 1 h with 32P-UTP. Their degradation was monitored 75 and 150 min after the pulse labelling had ended. tRNAQ was used as a loading control after autoradiographic detection using a γ-32P-labelled oligonucleotide probe. L/L n=3 and L/L, cre n=3 for each time point. Error bars indicate s.e.m.; *P<0.05, ***P<0.001, Student's t-test.
LRPPRC forms an RNA-dependent complex with SLIRP
We proceeded to study the role of the LRPPRC–SLIRP complex in regulation of mRNA stability. To this end, we performed size-exclusion chromatography on mitochondrial extracts from mouse liver and found that LRPPRC migrated with a higher apparent molecular weight than the one predicted for LRPPRC monomers (Figure 5A). We found SLIRP in the same fractions (numbers 14 and 15) as LRPPRC supporting that they are part of a complex with an apparent molecular weight ∼250 kDa (Figure 5A). Others have reported that the interaction between LRPPRC and SLIRP is independent of mitochondrial RNA based on association of the two proteins in cell lines lacking mtDNA (Sasarman et al, 2010). We nevertheless proceeded to investigate the nature of the LRPPRC–SLIRP complex by using different conditions to prepare mitochondrial extracts followed by gel filtration analysis (Figure 5A). We used different conditions to analyse the complex, that is, high salt (500 mM NaCl) to dissociate the complex, 100 mM EDTA to abolish protein–RNA interactions and RNase A to degrade any accompanying mitochondrial RNAs. Under all of these conditions, the location of SLIRP shifted from fractions 14 and 15 to fractions 18 and 19, which correspond well to the predicted molecular weight of ∼12 kDa of monomeric SLIRP (Figure 5A). In contrast, the location of LRPPRC shifted to higher molecular weight fractions, particularly in the RNAse-treated fractions, which indicate that LRPPRC becomes unstable and aggregates in the absence of RNA or becomes part of another protein complex (Figure 5A). The size-exclusion chromatography procedure we used likely causes partial RNA degradation and an in vivo complex containing full-length incorporated RNAs may therefore be substantially larger than the 250-kDa complex we observed. Despite these limitations, the biochemical fractionation experiments we present here strongly suggest that LRPPRC and SLIRP form an RNA-dependent complex. We obtained further support for this conclusion by investigating levels of LRPPRC and SLIRP in mouse models with altered steady-state levels of mitochondrial transcripts. Tissue-specific Tfam-knockout mice have low steady-state levels of transcripts in heart mitochondria (Wang et al, 1999), tissue-specific Tfb1m knockout mice have normal steady-state levels of most transcripts in heart mitochondria (Metodiev et al, 2009) and tissue-specific Mterf4 knockout mice have high steady-state levels of most transcripts in heart mitochondria (Camara et al, 2011). We found that the protein levels of LRPPRC and SLIRP correlate with mitochondrial RNA abundance in all of these models (Figure 5B).
Figure 5.
LRPPRC and SLIRP are part of an RNA-dependent ribonucleoprotein complex. (A) Size-exclusion chromatography of wild-type liver mitochondrial extracts under different buffer conditions. LRPPRC and SLIRP were detected using specific antibodies. (B) Western blot analysis of LRPPRC and SLIRP levels in heart mitochondrial extracts from mouse knockout models with different steady-state levels of mitochondrial RNAs. VDAC was used as a loading control. (C) Isolation and identification of mitochondrial transcripts interacting with hLRPPRC–SLIRP complex. hLRPPRC and SLIRP were co-immunoprecipitated using anti-Flag antibody. Transcripts, bound to the LRPPRC–Flag/SLIRP complex, were detected and quantified using real-time PCR and their abundance is shown as percentage of levels in non-transfected controls. (D) Real-time PCR quantification of steady-state levels of mitochondrial transcripts after overexpression of LRPPRC and/or SLIRP in HeLa cells. Nucleus-encoded 18S rRNA was used as an internal control for normalization of samples. Statistical comparisons are made between transfected cells with the empty vector (CMV) and cells overexpressing LRPPRC, SLIRP and LRPPRC or SLIRP. Experiments were performed in triplicates and results are representative of n=3 independent biological experiments. Error bars indicate s.e.m.; *P<0.05, Student's t-test.
The LRPPRC–SLIRP complex binds mitochondrial mRNAs
We further characterized RNA interactions of the LRPPRC/SLIRP complex by generating human cell lines with either stable or inducible expression of human LRPPRC with a Flag tag at the carboxy terminus (hLRPPRC–Flag). The Flag-tagged mouse LRPPRC can rescue the LRPPRC germline knockout (Supplementary Figure S1A and B), indicating that the addition of a Flag tag has no functional consequences for the LRPPRC protein. We purified hLRPPRC–Flag from mitochondrial extracts and determined the N-terminal sequence by Edman degradation and found that the mitochondrial isoform of hLRPPRC lacks the first 59 amino-terminal amino acids (Supplementary Figure S4A), consistent with previous results showing cleavage of the mitochondrial targeting sequence after mitochondrial import (Sterky et al, 2010). We performed immunoprecipitation experiments with anti-Flag resin to determine optimal conditions for isolation of the LRPPRC–SLIRP complex (Supplementary Figure S4B) and then proceeded with RNA immunoprecipitation experiments. We found that the LRPPRC–SLIRP complex preferentially binds different mitochondrial mRNAs, whereas 12S rRNA, 16S rRNA and 7S RNA are absent in the complex (Figure 5C). The LRPPRC/SLIRP complex thus has a fairly broad spectrum of interacting RNAs, which is unexpected as the related plant PPR proteins have been reported to have a rather narrow substrate specificity (Delannoy et al, 2007; Zehrmann et al, 2011).
Based on the results above, we investigated whether individual or combined overexpression of LRPPRC and SLIRP can influence the steady-state levels of mitochondrial transcripts. HeLa cells were transiently transfected with a plasmid encoding hLRPPRC and the levels of mitochondrial transcripts were determined by real-time PCR. We found that all mRNA transcripts, except ND6, were slightly increased whereas rRNA levels remained unchanged (Figure 5D). Overexpression of hSLIRP alone had no effect on the steady-state levels of mitochondrial RNAs. Combined overexpression of hLRPPRC and hSLIRP had essentially the same effect as isolated overexpression of hLRPPRC (Figure 5D). These results show that LRPPRC is the critical component of the LRPPRC–SLIRP complex that is necessary for regulation of mRNA stability by this complex in mammalian mitochondria.
Loss of LRPPRC leads to a misregulated mitochondrial translation pattern
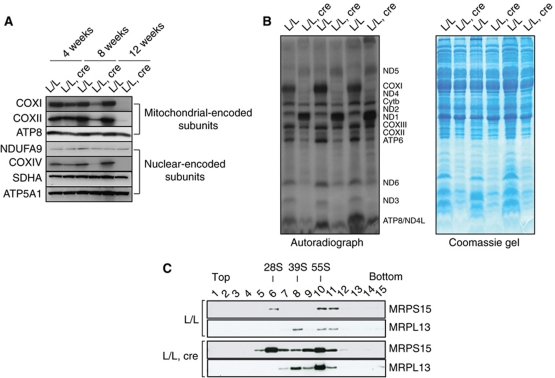
The presence of decreased steady-state levels of several mRNAs (Figure 3B and C) suggests that reduction in the levels of mtDNA-encoded polypeptides can explain the observed severe respiratory chain deficiency in heart mitochondria lacking LRPPRC (Figure 2A–E). Western blot analyses of mitochondrial extracts from control and knockout mice showed a gradual decrease in the steady-state levels the COXI and COXII subunits of complex IV, whereas subunits of complexes I, II and V were present at normal levels (Figure 6A). In order to understand why a general drop in the steady-state levels of mRNAs (Figure 3B and C) causes a profound defect in complex IV and only a mild-to-moderate defect in complexes I and V (Figure 2C–E), we tested whether the synthesis of the 13 mitochondrial translation products was affected in the absence of LRPPRC (Figure 6B and Supplementary Figure S6). Using in vitro labelling of mitochondrial translation products, we found a misregulated pattern of mitochondrial protein synthesis in the absence of LRPPRC, which has not been observed in previous studies of cell lines (Xu et al, 2004; Sasarman et al, 2010). Remarkably, the de novo translation of ND1, ND2 and ND5 was drastically increased, whereas the synthesis of ND3, ND6, COXI and ATP6 was very low or absent in knockout mitochondria (Figure 6B). Additionally, we analysed the stability of each newly synthesized polypeptide by using pulse-chase experiments (Supplementary Figure S5A). In control mitochondria, there was a faster turnover of COXI than of other newly synthesized polypeptides, which may explain how decreased translation of COXI results in such a drastic complex IV deficiency in LRPPRC-deficient heart mitochondria.
Figure 6.
Misregulated mitochondrial translation and ribosomal biogenesis in Lrpprc knockout heart mitochondria. (A) Western blot analysis of steady-state levels of mitochondrial and nucleus-encoded subunits of the respiratory chain complexes in heart mitochondria from control (L/L) and Lrpprc knockout (L/L, cre) mice at different ages. (B) In organello translation in isolated heart mitochondria from control (L/L) and Lrpprc knockout (L/L, cre) mice at 12 weeks of age. Coomassie blue staining of proteins after SDS–PAGE is shown as a loading control. (C) Sedimentation analysis of the small (28S) and large (39S) ribosomal subunit and the assembled (55S) ribosome in heart mitochondria from control (L/L) and Lrpprc knockout (L/L, cre) mice of 12 weeks of age. Migration of the small (28S) and the large (39S) ribosomal subunits as well as of the assembled ribosome was detected by immunoblotting with MRPS15- and MRPL13-specific antibodies.
The aberrant de novo synthesis of mitochondrial translation products in Lrpprc heart knockout mitochondria prompted us to test the integrity of the ribosomes by performing gradient sedimentation analyses of mitochondrial extracts. We found an increase in the steady-state levels of the 28S and 39S ribosomal subunits and an increase of the assembled 55S ribosomes in the absence of LRPPRC (Figure 6C), which is in good agreement with the observation that the levels of 12S and 16S rRNA are increased (Figure 3B and C). In further support for increased biogenesis of mitochondrial ribosomes, we found elevated levels of the ribosomal proteins MRPS15 (present in the 28S subunit), MRPL13 (present in the 39S subunit) and TFB1M (required for 12S rRNA methylation and stability of the 28S subunit) (Metodiev et al, 2009), in Lrpprc knockout heart mitochondria (Supplementary Figure S5B). The increased biogenesis of mitochondrial ribosomes in the absence of LRPPRC may be a component of the observed mitochondrial biogenesis response induced by the profound respiratory chain deficiency.
Aberrant polyadenylation of mitochondrial mRNAs in the absence of LRPPRC
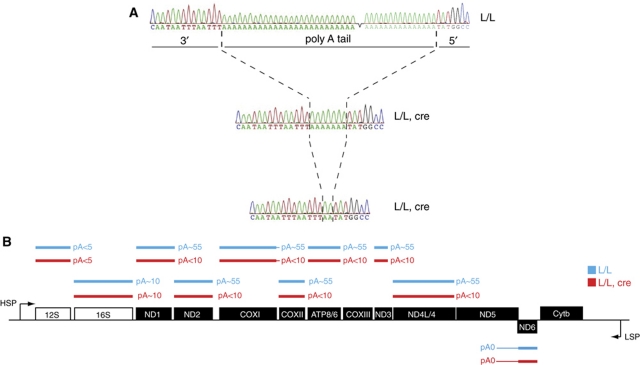
The finding of a decrease of most mitochondrial mRNAs (Figure 3B and C), a strong increase of assembled ribosomes (Figure 6C; Supplementary Figure S5B) and a misregulated translation pattern (Figure 6B) prompted us to closer investigate the nature of the mitochondrial mRNA species interacting with the LRPPRC–SLIRP complex. We were especially puzzled by the finding that a general decrease of mRNA levels could lead to strong increase in steady-state levels of certain mitochondrial translation products (e.g., ND1, ND2 and ND5) whereas others were absent or present at low levels (e.g., ND3, ND6, COXI and ATP6). We analysed the 5′ and 3′ ends of mitochondrial transcripts by using RNA circularization and sequence analysis of the corresponding cDNAs (Stewart and Beckenbach, 2009; Figure 7A and B). Interestingly, the polyA tails of mRNAs encoded on the H strand were substantially shorter in LRPPRC-deficient (pA<10) than in control (pA∼50–60) heart mitochondria (Figure 7A and B). The ND6 mRNA had a long 3′ region, corresponding to part of the adjacent antisense region of ND5, and was not polyadenylated in controls or in knockouts (Figure 7B). It should be noted that ND6 is the only mRNA encoded by the L strand (Figure 3A) and absence of polyadenylation of this transcript has also been reported in human tissue culture cells (Temperley et al, 2010). Surprisingly, the finding that ND6 lacks a polyA tail in controls shows that polyadenylation is dispensable for both stability and translation of this mRNA. We found no significant binding of the ND6 mRNA to the LRPPRC–SLIRP complex (Figure 5C) and ND6 was the only mRNA that maintains normal steady-state levels in the absence of LRPPRC (Figure 3B and C). We found an identical polyadenylation status of the 12S rRNA (pA<5) and 16S rRNA (pA∼10) in heart mitochondria from control and Lrpprc knockout animals (Figure 7B). The polyadenylation defect in the absence of LRPPRC is thus specific to a subset of the mRNAs and does not affect rRNAs. The 5′ ends of all investigated mitochondrial mRNAs and rRNAs were unaltered in the heart mitochondria from knockouts in comparison with controls. The results from sequence analysis of 5′ and 3′ ends of mitochondrial transcripts (Supplementary Table 1) were consistent with available mtDNA sequence data and transcript maps from human cells (Temperley et al, 2010) and fish (Coucheron et al, 2011).
Figure 7.
Loss of mRNA polyadenylation in Lrpprc knockout heart mitochondria. (A) Examples of electropherograms showing the polyadenylation status of the COXII mRNA in control (L/L) and Lrpprc knockout (L/L, cre) heart mitochondria. (B) A summary of the polyadenylation status of mRNAs in controls (L/L) and Lrpprc knockout (L/L, cre) heart mitochondria.
LRPPRC and SLIRP stabilize a pool of translationally inactive mRNAs
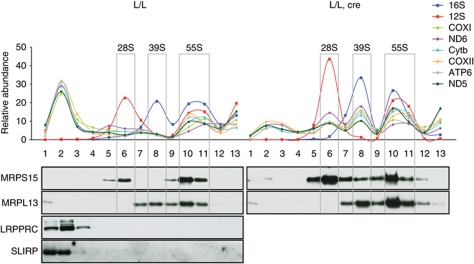
We further investigated the interaction between the LRPPRC/SLIRP complex, the mitochondrial mRNAs and the mitochondrial ribosome by performing sucrose gradient sedimentation analyses of mitochondrial extracts from LRPPRC knockout mitochondria. We used TaqMan probes to measure levels of 12S and 16S rRNA in different gradient fractions to determine the sedimentation profile of the small (28S) and large (39S) ribosomal subunits and the fully assembled (55S) ribosomes (Figure 8). The 12S rRNA co-migrated with the MRPS15 protein and the 16S rRNA with the MRPL13 protein (Figure 8), thus confirming that the 28S subunit was mainly present in fraction 6, the 39S subunit in fraction 8 and the assembled 55S ribosome in fractions 10 and 11 of the sucrose gradient. Next, we proceeded to analyse the migration of six different mRNAs and found two clearly defined sedimentation peaks (Figure 8). The first peak was in the low-molecular weight region of the gradient (fraction 2) and an additional peak co-migrated with the assembled 55S ribosome (fractions 10 and 11) in wild-type mitochondria (Figure 8, left panels). The mRNA abundance was clearly higher in fraction 2 than in fractions 10 and 11, showing that only a portion of all mRNAs is translated at a given time. Interestingly, LRPPRC and SLIRP co-migrated with the mRNA pool in fraction 2 of the gradient, which is in line with the results above (Figure 5), showing that both proteins form mRNA-containing complexes. In further support of the role for LRPPRC in maintaining a non-translated pool of mRNAs, we found that this mRNA pool was almost completely absent in Lrpprc knockout heart mitochondria and the residual levels of mRNAs were found mostly associated with the ribosomal subunits or the fully assembled ribosome. We observed two mRNA peaks in association with the free large (39S) and small (28S) ribosomal subunits only in the LRPPRC knockout mitochondria (Figure 8, L/L, cre panel). This association differs from the current model for translation initiation in mammalian mitochondria, which postulates that the mRNA binds the 28S ribosomal subunit prior to the assembly of 55S ribosome and onset of translation (Haque et al, 2008). Therefore, the findings of abnormal interaction between ribosomal subunits and mRNAs further strengthen the conclusion that translation is misregulated in the absence of LRPPRC.
Figure 8.
Sedimentation profile of mitochondrial transcripts and the LRPPRC/SLIRP complex in sucrose density gradients. Sedimentation profiles of mitochondrial transcripts and the LRPPRC/SLIRP complex in heart mitochondrial lysates from 12-week-old control (L/L) and Lrpprc knockout (L/L, cre) mice. Individual mitochondrial transcripts were detected using TaqMan-specific probes. The abundance of mRNA in each fraction is shown as percentage from the sum of the abundance in all fractions. The migration of 28S, 39S and 55S is assessed using subunit-specific antibodies and is shown for reference.
Discussion
We report here a number of new and unexpected insights into the function of LRPPRC in the post-transcriptional regulation of mitochondrial expression in vivo. Studies of knockdown cell lines have previously indicated that decrease of LRPPRC levels causes a global decrease in mRNA stability and translation. Surprisingly, we show here that loss of LRPPRC in the mouse in vivo causes an unexpectedly complex phenotype. LRPPRC is necessary for stability of all mtDNA-encoded mRNAs except ND6. Without LRPPRC proper coordination of translation is lost and translation pattern becomes aberrant. Some mRNAs are translated at much higher levels than others and in several cases the newly produced translation products are unstable. We also report that LRPPRC is necessary for mRNA polyadenylation and in its absence mRNAs are only oligoadenylated. Studies of cell lines have indicated that LRPPRC forms an RNA-independent complex with SLIRP. At variance with these results, we show here that the LRPPRC/SLIRP complex is RNA dependent and that it maintains a pool of extra-ribosomal non-translated transcripts. Without LRPPRC the extra-ribosomal pool of mRNAs disappears and mRNAs are found aberrantly bound to free ribosomal subunits, in addition to the normally occurring binding to the assembled ribosome. Taken together, these results define LRPPRC as a key post-transcriptional regulator of mtDNA expression. The use of knockout mice has the advantage that insights are provided into the physiological function of LRPPRC in a differentiated tissue. Continuously dividing transformed or primary culture cells are mainly glycolytic and are usually grown at much higher oxygen tensions than those present in real tissues, which may explain why previous in vitro studies have not identified a role for LRPPRC in translation coordination (Xu et al, 2004; Sasarman et al, 2010).
Regulation of mRNA stability is important in respiratory chain dysfunction (Wang et al, 1999; Wredenberg et al, 2002), but the molecular mechanisms of this process are largely unknown. Our findings demonstrate that LRPPRC is required for the regulation of mRNA levels, but its importance differs between the individual mitochondrial transcripts. Among the most LRPPRC-dependent transcripts are COXI, COXII and COXIII, which are dramatically reduced upon loss of LRPPRC. Patients with LSFC have a profound COX deficiency, similarly to the Lrpprc knockout mice, and our findings argue that this is due to the drastic decrease of COXI–III mRNA levels causing reduced synthesis of these subunits. In addition, we report a high turnover of newly synthesized COXI subunits, which may make complex IV especially vulnerable to decreased synthesis of its subunits.
The levels of ND6 mRNA, the only mRNA transcribed from LSP, remain normal in the absence of LRPPRC, whereas the levels of all other mRNAs are decreased. The ND6 mRNA is interesting as it is normally not polyadenylated in heart mitochondria, but instead it contains a long 3′ UTR. Surprisingly, there is thus no need for polyadenylation to regulate stability and translation of the ND6 mRNA. LRPPRC forms a physical complex with SLIRP and mitochondrial mRNAs. The mRNA component is essential for the stability of the complex because RNase A treatment leads to disruption of the LRPPRC–SLIRP interactions. The levels of the LRPPRC–SLIRP complex correlates nicely with total mRNA levels in different mouse models and in human cell lines. The only mRNA not found to be associated with the LRPPRC–SLIRP complex is ND6 and changes in LRPPRC levels do not affect ND6 mRNA stability, again demonstrating the unique nature of this transcript.
There is a dramatic drop in mRNA levels upon loss LRPPRC, but no compensatory increase of mitochondrial transcription. This finding stands in stark contrast to previous reports of other mutants, where impaired translation leads increased de novo transcription (Metodiev et al, 2009; Camara et al, 2011; Kolanczyk et al, 2011). The lack of a stimulatory effect on transcription could be explained if LRPPRC is itself directly involved in sensing mRNA levels in mammalian mitochondria. Results from studies of mouse knockouts for TFAM, TFB1M and MTERF4, which have decreased, normal and increased steady-state levels of mtDNA transcripts, respectively, show that the LRPPRC levels follow the mRNA levels. In addition, we show that knockout of Lrpprc leads to a rapid decrease of mRNA levels and that LRPPRC overexpression in cell lines increases steady-state levels of mRNAs. Thus, the LRPPRC protein binds and stabilizes mRNAs and the LRPPRC protein may under normal physiological circumstances be saturated with mRNA molecules. A reduction in mRNA levels, for example, due to decreased transcription, may decrease the mRNA saturation of LRPPRC and thereby elicit a response that increases mtDNA transcription. This hypothetical model would explain why loss of LRPPRC, due to knockout of its gene, leads to decreased mRNA levels without a concomitant increase of mtDNA transcription.
Mutations that inactivate the yeast mitochondrial degradosome, responsible for mRNA degradation cause an accumulation of transcripts, which in turn impairs mitochondrial gene expression (Rogowska et al, 2006). This effect can be suppressed by partial loss-of-function mutations in genes that encode for the yeast homologues of POLRMT and TFB2M, thus suggesting that the absolute levels of transcripts must be tightly regulated in yeast mitochondria. If transcript levels need to be tightly controlled also in mammalian mitochondria remains to be established, but based on our findings, LRPPRC could be a component of a system that senses transcript levels and signals to the transcription apparatus in order to maintain a balance between RNA synthesis and degradation.
LRPPRC does not only bind to mitochondrial mRNA species, but the protein also influences polyadenylation as its loss causes a substantial shortening of polyA tails. In mitochondria, polyadenylation is performed by a mitochondrial isoform of the polyA polymerase (mtPAP) (Tomecki et al, 2004; Nagaike et al, 2005). There have been reports that polyadenylation is a two-step process, with oligoadenylation preceding the addition of long polyA tails, but the mechanisms behind this effect and what constitutes the switch between oligoadenylation and polyadenylation remain obscure. Based on our findings, it is possible that LRPPRC plays a role in helping to extend the polyA tail of oligoadenylated mitochondrial transcripts. In some support of this suggestion is the finding that oligoadenylation of the rRNAs is normal. In nuclear mRNA processing, efficient polyadenylation is not only dependent on polyA polymerase (PAP), but also requires a polyA binding protein (PABP), which helps recruit and stabilize PAP to its substrate RNA. In fact, the polyA tail length in human cells is controlled via the stabilization or destabilization of a ternary complex containing PAP and PABP. It is possible that LRPPRC performs related roles in mitochondria, perhaps by stabilizing an interaction between mtPAP and the oligoadenylated transcript. In support of this model is the finding that the non-polyadenylated ND6 mRNA is the only mRNA not bound by LRPPRC and not affected by loss of LRPPRC. Interestingly, PPR proteins in trypanosomes have recently been shown to affect polyadenylation of mitochondrial mRNA transcripts. In this unicellular protozoa, the PPR proteins KPAF1 and KPAF2 associate with the mtPAP and stimulate mRNA polyadenylation, thereby coordinating stability and translation of mRNA (Aphasizheva et al, 2011).
The polyA tail is required for translation of nuclear transcripts. In contrast, our data suggest that mitochondrial translation can be very effective even in the absence of mRNA polyadenylation. However, LRPPRC does seem to have an important role in coordinating the translation of different transcripts. In the Lrpprc knockout heart, translation is misregulated with massive translation of some transcripts and no translation of others. Interestingly, our results demonstrate that neither LRPPRC nor SLIRP associates directly with the ribosome. Instead, the two proteins maintain an extra-ribosomal pool of mRNAs, which may be presented to the ribosome in an orderly fashion. The LRPPRC–SLIRP–mRNA complex is lost in the Lrpprc knockout tissues and the remaining levels of mRNAs are bound to the ribosomal subunits or the assembled ribosome. Thus, a pool of translationally inactive mRNAs is maintained and stabilized by the LRPPRC–SLIRP complex. Controlled release of mRNA from this pool may be necessary for coordinated translation of different respiratory chain components in mammalian mitochondria. In the absence of LRPPRC, the pool is lost and the translation becomes misregulated as mRNAs may enter the translation machinery at random. In addition, the lack of polyadenylation also has different effects on the individual mRNA transcripts, with levels of some mRNAs decreasing much faster than others. A combination of these effects, that is, loss of the non-translated mRNA pool and loss of polyadenylation, may cause a drastically increased translation of some mRNA molecules, which blocks the translation machinery and thereby prevents translation of other transcripts. This effect may explain why ND6 translation and protein levels are severely decreased in Lrpprc knockout hearts despite the observation that the levels of the ND6 transcript are unaffected by the loss of LRPPRC.
In summary, our findings define a novel role for LRPPRC and shows that this protein is necessary for mRNA stability and polyadenylation. In addition, we report that LRPPRC is necessary for maintaining a pool of non-translated transcripts and for coordination of mitochondrial translation. These findings point to the existence of an elaborate machinery that is regulating mammalian mtDNA gene expression at the post-transcriptional level.
Materials and methods
Generation of LRPPRC knockout mice
The targeting vector for disruption of Lrpprc in embryonic stem cells (ESCs) was generated using isogenic 129R1 DNA. A genomic clone containing the Lrpprc gene was identified in a 129Sv RPCI-22M BAC library (Invitrogen) using a partial cDNA probe covering the 5′ region of the gene. A 14-kb BAC fragment, containing exons 2–10, was cloned into pBluescript II SK+ (Stratagene) by ET recombination to generate pBS-LRPPRC. Next, the pDELBOY-3X plasmid, containing a loxP sequence and an Frt-PGK-neomycin-Frt cassette, was modified by introduction of MluI and BamHI sites in the XhoI site and an MluI site in the KpnI site. These modifications allowed the excision of the loxP sequence and the Frt-PGK-neo-Frt cassette as an MluI fragment, which was inserted into an MluI site of pBS-LRPPRC to create the plasmid pBS-LRPPRC-Neo. Subsequently, a fragment containing loxP sequence and a KpnI restriction site was excised from the pST7 plasmid and inserted into the unique PacI site of the pBS-LRPPRC-Neo plasmid, thereby creating the targeting vector pBS-LRPPRC-TV. In this vector, an Frt-PGK-neomycin-Frt cassette and a loxP site were present between exons 2 and 3, whereas a second loxP site was present between exons 5 and 6 (Figure 1A). The pBS-LRPPRC-TV vector was linearized by NotI digestion and electroporated into 129R1 cells and ESC clones were selected with gentamicin and analysed by Southern blotting. A total of 196 ESC clones were analysed by KpnI digestion and only 2 clones that had undergone homologous recombination were identified. Chimeras were generated by blastocyst injection and germline transmission was obtained from both clones. The PGK-Neomycin cassette was removed by mating of Lrpprc+/loxP−neo mice with transgenic mice ubiquitously expressing Flp recombinase. The resulting Lrpprc+/loxP mice were mated with mice ubiquitously expressing cre recombinase to generate heterozygous knockout Lrpprc+/− mice (Figure 1A).
Tissue-specific disruption of Lrpprc
Heart- and skeletal muscle-specific knockout mice were generated as described previously (Park et al, 2007; Metodiev et al, 2009; Camara et al, 2011). Basically, LrpprcloxP/loxP mice were crossed with transgenic mice expressing cre recombinase under the control of the muscle creatinine kinase promoter (Ckmm-cre). The resulting double heterozygous mice (LrpprcloxP/+, +/Ckmm-cre) were mated to LrpprcloxP/loxP mice to generate tissue-specific knockout (LrpprcloxP/loxP, +/Ckmm-cre) and control (LrpprcloxP/loxP) mice.
Generation of Lrpprc–Flag transgenic mice
A BAC clone of 241 kb (RP24-100M10) containing the whole Lrpprc gene was obtained from Children's Hospital Oakland-BAC-PAC Resources. The BAC was modified by ET recombination to allow discrimination between transcripts expressed from the endogenous Lrpprc gene and the introduced BAC clone. A silent mutation, which did not alter the encoded amino acids but did eliminate a BglII site was introduced in exon 3. Additionally, the stop codon in exon 38 was removed and a Flag sequence was cloned in frame to allow expression of LRPPRC–Flag (Supplementary Figure S1A). The modified BAC was purified by caesium chloride gradient centrifugation and injected into the pronucleus of fertilized oocytes. Founders (+/BAC–LRPPRC–Flag) were identified by PCR and restriction enzyme analysis of genomic DNA to detect loss of the BglII site in the Lrpprc gene and the presence of sequences encoding the Flag tag.
Supplementary Material
Acknowledgments
This study was supported by Swedish Research Council grants (2010-2766 to NGL and 2009-4848 to CG), Leducq Foundation to NGL, Lundberg foundation to CG, an ERC Advanced Investigator joint grant to NGL and CG, Deutsche Forschungsgemeinschaft, SFB 829 (NGL), SFB815 (Z1 to UB and ZI 552/3-1 to VZ) and the Cluster of Excellence ‘Macromolecular Complexes’ EXC 115 (UB) at the Goethe University. We thank the Karolinska Center for Transgenic Technologies (KCTT) for help with production of knockout mice. We thank Ilka Wittig for valuable help and Vahid Edrisi, Karin Siegmund and Andrea Duchene for excellent technical assistance. AW is funded by a FEBS Long-Term Fellowship.
Author contributions: BR, CMG, UB and NGL designed the experiments; BR, MDM, AW, AB, JBS, CBP, YC, DM, VZ, RW, KH, HEB and PT performed the experiments; BR and NGL wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R (2011) Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol Cell 42: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman JM, Nilsson R, Gohil VM, Arlow DH, Gauhar Z, Mootha VK (2009) A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet 5: e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF (1996) Interaction of mtTFB and mtRNA polymerase at core promoters for transcription of Xenopus laevis mtDNA. J Biol Chem 271: 12036–12041 [PubMed] [Google Scholar]
- Bogenhagen DF, Rousseau D, Burke S (2008) The layered structure of human mitochondrial DNA nucleoids. J Biol Chem 283: 3665–3675 [DOI] [PubMed] [Google Scholar]
- Camara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, Franz T, Erdjument-Bromage H, Tempst P, Hallberg BM, Gustafsson CM, Larsson NG (2011) MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab 13: 527–539 [DOI] [PubMed] [Google Scholar]
- Cooper MP, Qu L, Rohas LM, Lin J, Yang W, Erdjument-Bromage H, Tempst P, Spiegelman BM (2006) Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1alpha/LRP130 complex. Genes Dev 20: 2996–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucheron DH, Nymark M, Breines R, Karlsen BO, Andreassen M, Jorgensen TE, Moum T, Johansen SD (2011) Characterization of mitochondrial mRNAs in codfish reveals unique features compared to mammals. Curr Genet 57: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby AH, Patel H, Chioza BA, Proukakis C, Gurtz K, Patton MA, Sharifi R, Harlalka G, Simpson MA, Dick K, Reed JA, Al-Memar A, Chrzanowska-Lightowlers ZM, Cross HE, Lightowlers RN (2010) Defective mitochondrial mRNA maturation is associated with spastic ataxia. Am J Hum Genet 87: 655–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SM, Rackham O, Shearwood AM, Hamilton KL, Narsai R, Whelan J, Filipovska A (2009) Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett 583: 1853–1858 [DOI] [PubMed] [Google Scholar]
- Debray FG, Morin C, Janvier A, Villeneuve J, Maranda B, Laframboise R, Lacroix J, Decarie JC, Robitaille Y, Lambert M, Robinson BH, Mitchell GA (2011) LRPPRC mutations cause a phenotypically distinct form of Leigh syndrome with cytochrome c oxidase deficiency. J Med Genet 48: 183–189 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Stanley WA, Bond CS, Small ID (2007) Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans 35: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A (2004) Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res 32: 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG (2004) Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet 13: 935–944 [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM (2002) Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet 31: 289–294 [DOI] [PubMed] [Google Scholar]
- Gohil VM, Nilsson R, Belcher-Timme CA, Luo B, Root DE, Mootha VK (2010) Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J Biol Chem 285: 13742–13747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Grasso D, Spremulli LL (2008) The interaction of mammalian mitochondrial translational initiation factor 3 with ribosomes: evolution of terminal extensions in IF3 mt. Nucleic Acids Res 36: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W (2008) RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135: 462–474 [DOI] [PubMed] [Google Scholar]
- Hou J, Wang F, McKeehan WL (1994) Molecular cloning and expression of the gene for a major leucine-rich protein from human hepatoblastoma cells (HepG2). In Vitro Cell Dev Biol Anim 30A: 111–114 [DOI] [PubMed] [Google Scholar]
- Kolanczyk M, Pech M, Zemojtel T, Yamamoto H, Mikula I, Calvaruso MA, van den Brand M, Richter R, Fischer B, Ritz A, Kossler N, Thurisch B, Spoerle R, Smeitink J, Kornak U, Chan D, Vingron M, Martasek P, Lightowlers RN, Nijtmans L et al. (2011) NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol Biol Cell 22: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA (2005) Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309: 481–484 [DOI] [PubMed] [Google Scholar]
- Larsson NG (2010) Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem 79: 683–706 [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 18: 231–236 [DOI] [PubMed] [Google Scholar]
- Merante F, Petrova-Benedict R, MacKay N, Mitchell G, Lambert M, Morin C, De Braekeleer M, Laframboise R, Gagne R, Robinson BH (1993) A biochemically distinct form of cytochrome oxidase (COX) deficiency in the Saguenay-Lac-Saint-Jean region of Quebec. Am J Hum Genet 53: 481–487 [PMC free article] [PubMed] [Google Scholar]
- Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG (2009) Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab 9: 386–397 [DOI] [PubMed] [Google Scholar]
- Michaud M, Barakat S, Magnard S, Rigal D, Baggetto LG (2011) Leucine-rich protein 130 contributes to apoptosis resistance of human hepatocarcinoma cells. Int J Oncol 38: 169–178 [PubMed] [Google Scholar]
- Mili S, Pinol-Roma S (2003) LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol Cell Biol 23: 4972–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, Mitchell GA, Morin C, Mann M, Hudson TJ, Robinson B, Rioux JD, Lander ES (2003) Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA 100: 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Katoh T, Ueda T (2005) Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem 280: 19721–19727 [DOI] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Tomari Y, Takemoto-Hori C, Negayama F, Watanabe K, Ueda T (2001) Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J Biol Chem 276: 40041–40049 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474 [DOI] [PubMed] [Google Scholar]
- Parisi MA, Clayton DA (1991) Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252: 965–969 [DOI] [PubMed] [Google Scholar]
- Park CB, Asin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, Falkenberg M, Gustafsson CM, Larsson NG (2007) MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell 130: 273–285 [DOI] [PubMed] [Google Scholar]
- Rackham O, Davies SM, Shearwood AM, Hamilton KL, Whelan J, Filipovska A (2009) Pentatricopeptide repeat domain protein 1 lowers the levels of mitochondrial leucine tRNAs in cells. Nucleic Acids Res 37: 5859–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowska AT, Puchta O, Czarnecka AM, Kaniak A, Stepien PP, Golik P (2006) Balance between transcription and RNA degradation is vital for Saccharomyces cerevisiae mitochondria: reduced transcription rescues the phenotype of deficient RNA degradation. Mol Biol Cell 17: 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA (2010) LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol Biol Cell 21: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88: 611–638 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Shine J, Dalgarno L (1974) The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA 71: 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Schuster G (2008) Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA 14: 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Fang JK, Polyak E, Falk MJ, Avadhani NG (2010) Leucine-rich pentatricopeptide-repeat containing protein regulates mitochondrial transcription. Biochemistry 49: 7467–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky FH, Ruzzenente B, Gustafsson CM, Samuelsson T, Larsson NG (2010) LRPPRC is a mitochondrial matrix protein that is conserved in metazoans. Biochem Biophys Res Commun 398: 759–764 [DOI] [PubMed] [Google Scholar]
- Stewart JB, Beckenbach AT (2009) Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene 445: 49–57 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miyauchi K, Yokobori SI, Shigi N, Kondow A, Takeuchi N, Yamagishi A, Watanabe K (2011) Taurine-containing uridine modifications in tRNA anticodons are required to decipher non-universal genetic codes in ascidian mitochondria. J Biol Chem 286: 35494–35498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku H, Minagawa A, Takagi M, Nashimoto M (2003) A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res 31: 2272–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares-Carreon F, Camacho-Villasana Y, Zamudio-Ochoa A, Shingu-Vazquez M, Torres-Larios A, Perez-Martinez X (2008) The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J Biol Chem 283: 1472–1479 [DOI] [PubMed] [Google Scholar]
- Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM (2010) Human mitochondrial mRNAs--like members of all families, similar but different. Biochim Biophys Acta 1797: 1081–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP (2004) Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res 32: 6001–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Siddiqui N, Lapointe VL, Trost M, Thibault P, Bangeranye C, Pinol-Roma S, Borden KL (2009) Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J 28: 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya N, Fukuda H, Sugimura T, Nagao M, Nakagama H (2002) LRP130, a protein containing nine pentatricopeptide repeat motifs, interacts with a single-stranded cytosine-rich sequence of mouse hypervariable minisatellite Pc-1. Eur J Biochem 269: 2927–2933 [DOI] [PubMed] [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW (2010) Mitochondrial DNA mutations and human disease. Biochim Biophys Acta 1797: 113–128 [DOI] [PubMed] [Google Scholar]
- Wallace DC (2010) Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen 51: 440–450 [DOI] [PubMed] [Google Scholar]
- Wang J, Silva JP, Gustafsson CM, Rustin P, Larsson NG (2001) Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc Natl Acad Sci USA 98: 4038–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson NG (1999) Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet 21: 133–137 [DOI] [PubMed] [Google Scholar]
- Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, Larsson NG (2002) Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci USA 99: 15066–15071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydro M, Bobrowicz A, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM (2010) Targeting of the cytosolic poly(A) binding protein PABPC1 to mitochondria causes mitochondrial translation inhibition. Nucleic Acids Res 38: 3732–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Nguyen S, McKeehan K, Wang F, McKeehan WL, Liu L (2011) Microtubule-associated protein 1S (MAP1S) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation. J Biol Chem 286: 10367–10377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Ackerley C, Maj MC, Addis JB, Levandovskiy V, Lee J, Mackay N, Cameron JM, Robinson BH (2008) Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem J 416: 15–26 [DOI] [PubMed] [Google Scholar]
- Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH (2004) The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem J 382: 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehrmann A, Verbitskiy D, Hartel B, Brennicke A, Takenaka M (2011) PPR proteins network as site-specific RNA editing factors in plant organelles. RNA Biol 8: 67–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.