Abstract
Ubiquitin-conjugating enzymes (E2s) coordinate distinct types of ubiquitination via specific E3 ligases, to a large number of protein substrates. While many E2 enzymes need only the presence of an E3 ligase for substrate ubiquitination, a number of E2s require additional, non-canonical binding partners to specify their function. Here, we have determined the crystal structure and function of an E2/co-activator assembly, the Pex4p:Pex22p complex. The peroxisome-associated E2 enzyme Pex4p binds the peroxisomal membrane protein Pex22p through a binding site that does not overlap with any other known interaction interface in E2 enzymes. Pex22p association enhances Pex4p's ability to transfer ubiquitin to a substrate in vitro, and Pex22p binding-deficient forms of Pex4p are unable to ubiquitinate the peroxisomal import receptor Pex5p in vivo. Our data demonstrate that the Pex4p:Pex22p assembly, and not Pex4p alone, functions as the E2 enzyme required for Pex5p ubiquitination, establishing a novel mechanism of E2 enzyme regulation.
Keywords: E2 co-activator complex, peroxisomal receptor recycling, Pex22p, Pex4p, ubiquitin-conjugating enzyme structure
Introduction
The ubiquitin (Ub) cascade sequentially requires the activity of three different enzymes to attach Ub to a substrate. First, Ub is activated in an ATP-dependent manner by a ubiquitin-activating enzyme (E1), then transferred to the active site cysteine residue of a ubiquitin-conjugating enzyme (E2) and finally, with the aid of a ubiquitin ligase (E3), attached to the substrate. E2s play a coordinating role in the cascade, through highly dynamic interactions with many E3 ligases, ultimately determining the fate of hundreds of substrates for functional regulation, or alternatively for degradation by the proteasome (Capili and Lima, 2007; Dye and Schulman, 2007; Kleiger et al, 2009; Ye and Rape, 2009). Since E2 enzymes are largely conserved in structure and sequence, a number of common principles are shared by all members, including conserved binding sites for the E1 and E3 (Wenzel et al, 2011). However, many E2 enzymes are required to perform specific ubiquitination events in the cell, such as the attachment of ubiquitin to a certain side chain within a given substrate. While our understanding of how E2s coordinate these events is still incomplete, recent data may suggest an important role for non-canonical binding partners. For example, the ERAD-associated E3 ligase gp78 binds the ‘backside’ of Ube2g2 (an E2) through a region distinct from its ligase domain, and this binding event enhances Ube2g2's ubiquitin-conjugating activity (Das et al, 2009). Two additional examples include the non-catalytic ubiquitin E2 variant (UEV) protein Mms2, which binds to the E2 Ube2n, allowing it to specifically build lysine 63-linked polyubiquitin chains (VanDemark et al, 2001), and the non-covalent binding of ubiquitin itself to a number of E2s, which may play a role in processive Ub chain formation (Brzovic et al, 2006; Knipscheer et al, 2007).
While the above mentioned examples demonstrate regulation of an E2 by a co-factor already implicated in the Ub cascade (Ub, UEV, E3), the peroxisome-associated E2 enzyme Pex4p (Ubc10p) requires the presence of an additional co-factor, the trans-membrane (TM) domain-containing protein Pex22p (Koller et al, 1999; Collins et al, 2000; Zolman et al, 2005). Pex4p is an unusual E2 enzyme in that it facilitates site-specific cysteine ubiquitination of its substrate, the cycling receptor Pex5p (Wiebel and Kunau, 1992; Platta et al, 2007; Williams et al, 2007). Pex5p, required for import of proteins bearing a Peroxisomal Targeting Signal type-1 (PTS1) into peroxisomes, is modified on a conserved cysteine residue at its N-terminus by the addition of two Ub moieties (Kragt et al, 2005; Williams et al, 2007). Pex4p-dependent ubiquitination of Pex5p (sometimes referred to as Pex5p monoubiquitination), which occurs on the peroxisomal membrane and at a point after the receptor has delivered its cargo into the peroxisomal lumen, regulates the dissociation of Pex5p from the peroxisomal membrane (Williams et al, 2007; Platta et al, 2008). However, this is not the only form of Pex5p ubiquitination. Under certain conditions, for example in the absence of Pex4p or in other mutants disturbed in the membrane dissociation step, Pex5p is modified by another E2 enzyme, Ubc4p (Platta et al, 2004; Kragt et al, 2005). Ubc4p attaches between 1 and 4 Ub moieties to conserved lysine residues in Pex5p (sometimes referred to as Pex5p polyubiquitination), which targets the receptor for proteasome-mediated degradation (Williams et al, 2007; Platta et al, 2008). For clarity, we will refer to the two forms of Pex5p modification as Pex4p- and Ubc4p-dependent ubiquitination.
Both the Pex4p- and Ubc4p-dependent ubiquitination of Pex5p require Pex2p, Pex10p and Pex12p, three RING domain-containing proteins that form a complex at the peroxisomal membrane (Kragt et al, 2005). Each RING domain exhibits E3 ligase activity in vitro (Williams et al, 2008; Platta et al, 2009) yet the situation in vivo is complicated by the requirement of each RING domain for complex formation (Eckert and Johnsson, 2003) and consequently, for Pex5p modification (Kragt et al, 2005). While Pex10p's E3 ligase activity is required for Ubc4p-dependent ubiquitination of Pex5p in vivo, loss of this activity does not affect the Pex4p-dependent modification (Williams et al, 2008). Consequently, the identity of the E3 ligase functioning with Pex4p remains elusive.
Despite recent advances in the field, a number of key issues remain concerning Pex4p-dependent ubiquitination of Pex5p including crucially, the role of Pex4p's binding partner Pex22p in this process. Here, we report that Pex22p not only provides a docking platform for Pex4p on the peroxisomal membrane but also functions as an E2 co-activator, required for Pex4p's ability to ubiquitinate its substrate, the PTS1 receptor Pex5p. The crystal structure of the Pex4p:Pex22p complex from Saccharomyces cerevisiae reveals a previously unobserved mode of binding for an E2 enzyme and, together with our functional data, provides important insights into how a non-canonical binding partner regulates the activity of an E2 enzyme.
Results
Pex22p specifically stimulates Pex4p's ubiquitin-conjugating activity in vitro
Pex22p contains an N-terminal TM segment and a large cytosol-exposed region of unknown fold. Using secondary structure and domain predictions, a number of truncated versions of Pex22p were constructed and their ability to interact with Pex4p was tested using the yeast two-hybrid system (Supplementary Table S1). The minimal region required for Pex4p binding consists of residues 54–180, corresponding to the soluble region of Pex22p (referred to as Pex22S). Likewise, we observed that the first 14 amino acids of Pex4p were not required for Pex22S binding and consequently, we have used this truncated version, termed Pex415–183, in subsequent experiments. We quantified the Pex415–183:Pex22S interaction using isothermal titration microcalorimetry (ITC), confirming it to be direct, with a 1:1 stoichiometry and to be of high affinity, with a measured dissociation constant (Kd) of 2 nM (Table I; Supplementary Figure S1). In contrast, we were unable to detect an interaction between Pex22S and the other ubiquitin-conjugating enzyme involved in Pex5p ubiquitination, Ubc4p (Table I).
Table 1. Summary of the ITC data.
| Complex | Kd (nM) | ΔH (kcal mol−1) | TΔS (kcal mol−1) | ΔG (kcal mol−1) | n |
|---|---|---|---|---|---|
| Pex415−183:Pex22S | 2.0±0.08 | −18.2±1.6 | −6.3±1.6 | −11.9±0.1 | 1.16±0.01 |
| Ubc4p:Pex22S | No binding detectable | ||||
| Pex415−183 Y172A:Pex22S | No binding detectable | ||||
| Pex415−183 Y172F:Pex22S | 25.2±2.8 | −6.4±0.9 | 3.9±0.6 | −10.4±0.06 | 1.13±0.03 |
| Pex415−183:Pex22S K133A | 18.4±5.0 | −6.9±0.9 | 3.7±1.1 | −10.6±0.1 | 0.92±0.05 |
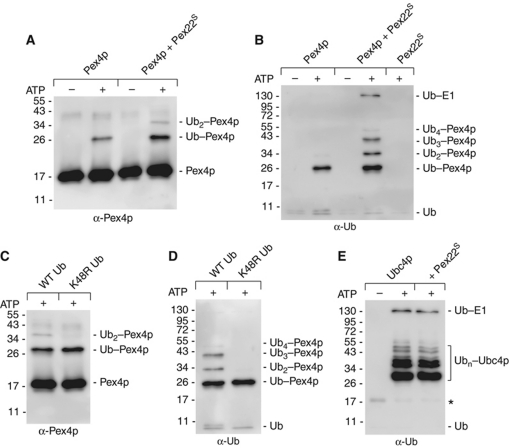
To test the effect such a tight binding partner would have on Pex4p's activity in vitro Pex415–183, together with E1 and Ub was incubated in the presence or absence of ATP and Pex22S and ubiquitin-conjugating activity was followed using an E2 self-ubiquitination assay (Figure 1; Supplementary Figure S2A). In line with our previous results (Williams et al, 2008), purified Pex415–183 exhibited ubiquitin-conjugating activity, as seen by the formation of an anti-Ub and anti-Pex4p reactive species in reactions containing ATP (Figure 1A and B). The estimated molecular mass of this species (26 kDa) correlates well with that of Pex415–183 attached to Ub (19.5+8 kDa). Interestingly, the addition of Pex22S to the reaction resulted not only in an enhanced production of Ub–Pex415–183, but also in the formation of additional, higher molecular mass species, corresponding to Pex415–183 attached to 2 (35.5 kDa), 3 (43.5 kDa) or 4 (51.5 kDa) Ub moieties (Figure 1A and B). All these species are lysine linked, as they are resistant to the reducing agent β-mercaptoethanol. Pex22S alone exhibited no activity (Figure 1B, last lane). Next, we analysed Pex415–183's ubiquitin-conjugating activity in the presence of a mutant form of Ub where the lysine at position 48 was mutated to an arginine (K48R). Interestingly, inclusion of K48R Ub effectively blocked the formation of the additional, higher molecular mass Ub species, indicating that Pex22S binding allows Pex415–183 to make lysine 48 linked Ub chains (Figure 1C and D). The ubiquitin-conjugating activity of Ubc4p remained unaffected upon addition of Pex22S (Figure 1E), confirming Pex22S's specificity for Pex415–183.
Figure 1.
Pex22p acts as a specific co-activator for Pex4p in vitro. (A–E) E2 self-ubiquitination assays containing some of the following: 0.2 μM E1 enzyme, 11 μM wild type (WT) or a lysine 48 to arginine mutant form (K48R) of Ub (Sigma), 5 mM ATP, 1.2 μM Pex415–183, 1.2 μM Ubc4p and 1.7 μM Pex22S, probed with either anti-Pex4p (A, C) or anti-Ub antibodies (B, D and E). *Defines an anti-Ub crossreactive species, likely to be Ubc4p. The presence of Pex22S stimulates the ubiquitin-conjugating activity of Pex415–183 and additionally allows Pex415–183 to produce Ub chains (A, B), linked through lysine 48 in Ub (C, D) but does not affect Ubc4p's activity (E).
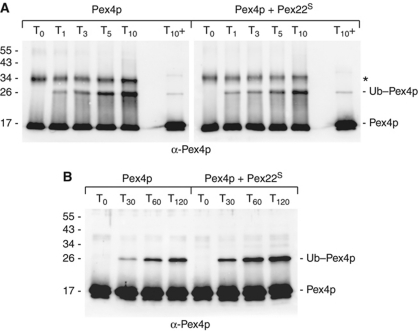
To gain further insight into the role of Pex22S binding, we examined the rate of Pex415–183 charging by E1 in the presence and absence of Pex22S. Samples of the reactions were taken soon after initiation (1, 3, 5 and 10 min) and were subjected to non-reducing SDS–PAGE, western blotting and anti-Pex4p immunoblotting (Figure 2A; Supplementary Figure S2B). As the presence of Pex22S is not required for the production of thioester-linked Ub–Pex415–183, nor does its presence enhance production of this species (Figure 2A), we conclude that Pex22S does not stimulate Pex4p's activity by enhancing Ub transfer from E1 to Pex4p.
Figure 2.
Pex22p binding stimulates transfer of Ub to a substrate. (A) Pex415–183 charging assays, performed in the absence (left panel) or presence (right panel) of Pex22S, with protein concentrations as follows: 0.27 μM E1, 29 μM Ub, 10 mM ATP, 8.3 μM Pex415–183 and 11 μM Pex22S. Charging of E2 was assessed by following the formation of thioester-linked Ub–Pex415–183. Reactions were quenched in non-reducing loading buffer at the indicated times, subjected to SDS–PAGE and probed with anti-Pex4p antibodies. To gauge background levels of lysine-linked Ub–Pex415–183, samples taken after 10 min were also analysed after treatment with β-mercaptoethanol (T10+). *Represents a dimeric form of Pex4p visible when DTT is absent from the reaction. (B) E2 self-ubiquitination assay, with protein concentrations as in Figure 1, using the K48R form of Ub. Samples of reactions were taken at the time points indicated and subjected to SDS–PAGE, western blotting and anti-Pex4p staining. Pex22S binding is not involved in promoting the charging of Pex4p by E1 (A) but enhances the transfer of Ub to a substrate (B).
Consequently, our results led us to believe that the role of Pex22S involves the Ub-to-substrate transfer step. To test this hypothesis, we examined the rate of Pex415–183 self-ubiquitination using the K48R form of Ub (Figure 2B; Supplementary Figure S2C), which blocks the formation of Ub chains, allowing us to gauge the rate of formation of a single Ub–Pex4p species. We observed that the presence of Pex22S enhances production of Ub–Pex415–183 by around 2–3-fold (Figure 2B), confirming the theory that Pex22p binding influences the transfer of Ub to a substrate.
Overall structure of the Pex415–183:Pex22S complex
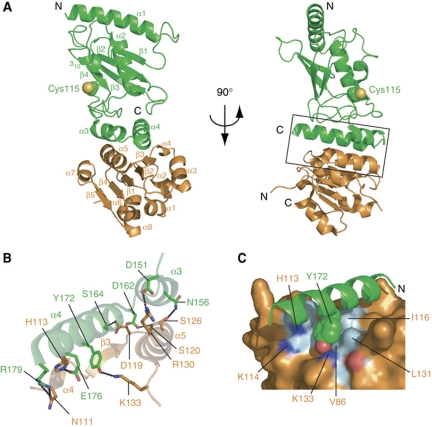
To unravel the molecular basis of Pex4p binding to Pex22p, we determined the crystal structure of Pex415–183 in complex with Pex22S at 2.6 Å (Figure 3; Table II). The structure is complete, with the exception of the termini of Pex415–183 and the three N-terminal residues and one loop (residues 145–151) in Pex22S. Pex415–183 adopts a typical UBC fold (Cook et al, 1997; Worthylake et al, 1998; Merkley and Shaw, 2004), containing a core domain of four anti-parallel β-strands (β1–β4) with one α-helix (α2), a small 310 helix (310), an N-terminal helix (α1) and a helix-turn-helix (α3–α4) at the C-terminus (Figures 3A and 4). As in other E2 structures, the active site cysteine (Cys115, Figure 3A) is situated in a cleft formed by the 310-helix and the loop region between α2 and α3.
Figure 3.
Molecular basis of Pex4p:Pex22p complex formation. (A) Overall structure of Pex415–183 (green) in complex with Pex22S (orange) in two different orientations. The interface is formed by the C-terminal helices α3 and α4 from Pex415–183 and helices α4 and α5 and β-strand β3 from Pex22S (boxed). Pex4p's active site (Cys115) is indicated in sphere presentation (carbon, green; sulphur, yellow). The termini of the Pex415–183 and Pex22S sequences and secondary structural elements are labelled. (B) Specific residue interactions found in the Pex415–183:Pex22S interface. Residues that participate in hydrogen bonding (dotted lines) are shown in stick representation and labelled. Nitrogen and oxygen atoms are coloured blue and red, respectively, with carbon atoms coloured as in (A). (C) Surface representation of Pex22S (orange) showing residues involved in hydrophobic contacts (cyan) with Tyr172 of Pex415–183 (green). Nitrogen and oxygen atoms are coloured blue and red, respectively.
Table 2. Data collection and refinement statistics.
| Data collection | Native 1 | Native 2 | 4 wavelength MAD | |||
|---|---|---|---|---|---|---|
| Space group | P21 | C2 | C2 | |||
| Cell dimensions | ||||||
| a, b, c (Å) | 35.7, 81.3, 112.8 | 139.3, 43.1, 60.4 | 138.8, 43.1, 60.2 | |||
| α, β, γ (deg) | 90, 98.9, 90 | 90, 100.6, 90 | 90, 100.7, 90 | |||
| Peak | Inflection | High energy remote | Low energy remote | |||
| Wavelength (Å) | 0.872 | 0.933 | 0.9776 | 0.9782 | 0.9747 | 0.960 |
| Resolution (Å) | 20.0–2.9 (3.0–2.9)a | 34.7–2.6 (2.74–2.6) | 20.0–2.7 (2.9–2.7) | 20.0–2.7 (2.9–2.7) | 20.0–2.7 (2.8–2.7) | 20.0–2.7 (2.85–2.7) |
| Rmerge (%) | 12.0 (62.0) | 6.0 (47.0) | 7.2 (26.8) | 7.1 (27.5) | 6.8 (27.5) | 7.2 (29.6) |
| I/σI | 9.9 (2.6) | 26.7 (4.8) | 20.8 (5.9) | 20.7 (5.7) | 21.8 (5.8) | 21.3 (5.6) |
| Completeness (%) | 97.4 (96.2) | 99.4 (99.1) | 99.6 (98.8) | 99.4 (97.7) | 99.0 (95.0) | 99.4 (97.7) |
| Redundancy | 2.7 (2.6) | 7.5 (7.6) | 7.3 (6.7) | 7.3 (6.8) | 7.3 (6.8) | 7.3 (6.9) |
| Refinement | ||||||
| Resolution (Å) | 20.0–2.6 | |||||
| Unique reflections | 10450 | |||||
| Rwork/Rfree | 20.8/25.9 | |||||
| No. of atoms | ||||||
| Protein | 2247 | |||||
| Ligandb | 16 | |||||
| Water | 9 | |||||
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.012 | |||||
| Bond angles (deg) | 1.33 | |||||
| Ramachandran plot (%) | ||||||
| Favoured | 94.2 | |||||
| Allowed | 5.8 | |||||
| Disallowed | 0 | |||||
| Each data set was collected from a single crystal. | ||||||
| aValues in parentheses are for the highest resolution shell. | ||||||
| bEthylene glycol. | ||||||
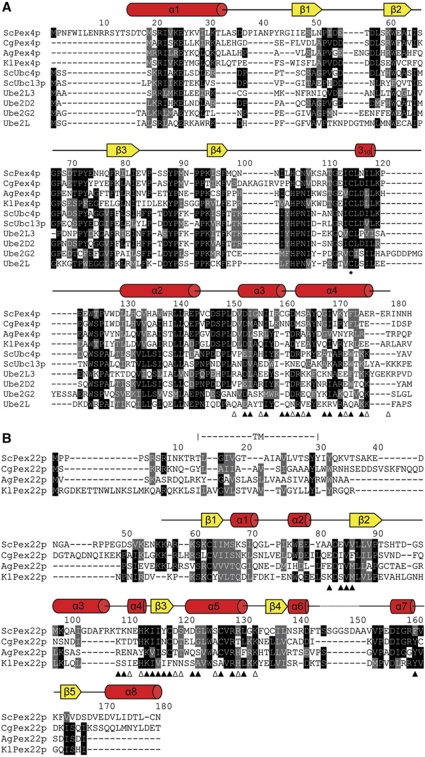
Figure 4.
Sequence alignment of Saccharomyces cerevisiae Pex4p and Pex22p. (A) ScPex4p aligned with a selection of homologous Pex4p sequences and a number of other ubiquitin-conjugating enzymes, allowing comparison with E2s whose functions are not dependent on Pex22p. Secondary structure elements in Pex415–183 are shown above the alignment. The active site cysteine is indicated with an asterisk. Pex415–183 residues involved in interface formation with Pex22S are indicated with triangles below the alignment, with open triangles denoting residues involved in residue-specific polar interactions. Black shading indicates identity and grey shading similarity when present in at least five of the ten sequences aligned. Sc, Saccharomyces cerevisiae; Cg, Candida glabrata; Ag, Ashbya gossypii; Kl, Kluyveromyces lactis. (B) Sequence alignment of Pex22p with homologous sequences. Secondary structure elements, as well as the trans-membrane segment (TM) in ScPex22p, are indicated above the rows. Residues involved in interface formation with Pex415–183 are indicated as in (A). When present in at least three of the four sequences aligned, conservation is indicated as in (A). The four organisms selected in the Pex22p alignment have unambiguously identified Pex4p sequences (A).
Pex22S also comprises a mixed β-sheet/α-helical fold, consisting of five parallel β-strands in the order β3–β2–β1–β4–β5, which form a central β-sheet that is tightly sandwiched on both sides by eight α-helices (Figures 3A and 4; Supplementary Figure S3). At first glance, the secondary structural topology of Pex22S is suggestive of a Rossmann-like fold, particularly as each β-strand is linked (N- and C-terminally) to two α-helices (Supplementary Figure S3), a topology typical for Rossmann and Rossmann-like folds (Rao and Rossmann, 1973). However, although a similarity search using PDBeFOLD (Krissinel and Henrick, 2004) did identify a number of potential homologous Rossmann-like fold structures, the observation that homology was, for the most part, confined to the central β-sheet, coupled with the low Q-scores (highest Q-score: 0.193), leads us to the conclusion that the overall fold of Pex22S is novel.
Examination of the residue-average B-factors in the co-crystal structure, an evaluation of the fluctuation of atoms in a protein structure relative to their average positions (Supplementary Figure S4) suggests the presence of some highly flexible ‘hotspots’ in Pex415–183—the α1–β1 loop, 310–α2 loop and α2–α3 loop. It could be suggested that these flexible loops effectively segment Pex415–183 into three, loosely associated subdomains, a ‘cap’ comprising α1, a ‘core’ domain consisting of β1–β2–β3–β4–310–α2 and an interaction motif formed by α-helices α3 and α4. These subdomains could form the basis of the series of folding/unfolding intermediates seen using CD (see Supplementary Data).
Molecular details of the Pex4p–Pex22p interface
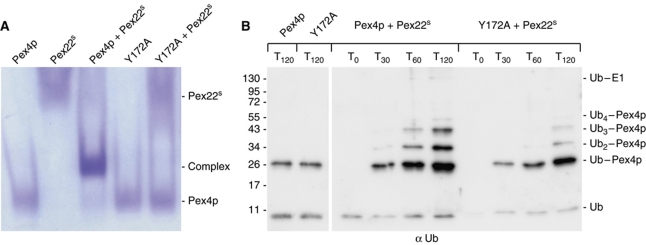
The Pex415–183:Pex22S high affinity binding site is established by the insertion of helix α5 from Pex22S into the V-shaped arrangement of the C-terminal helices α3 and α4 of Pex415–183 (Figures 3 and 5). Additional interactions are formed by the exposed surface of helix α4 from Pex415–183 and strand β3, helix α4 and the joining loop region from Pex22S, effectively burying around 750 Å2 of the solvent-accessible interface area. In Pex415–183, a total of 19 residues participate in interface formation, as determined by PISA (Krissinel and Henrick, 2007). Interface formation involves both polar and hydrophobic contacts (Figures 3, 4 and 5) and a number of interfacing residues, specifically Asp151, Ala165, Gly168, Ile169 and Tyr172, are conserved in homologous Pex4p sequences, but not in other ubiquitin-conjugating enzymes (Figures 4 and 5B). Particularly, the contribution of Tyr172 to the interaction, beyond the hydrogen bond to Lys133 from Pex22S, extends into extensive Van-der-Waals interactions, involving Val86, His113, Lys114, Leu116 and Leu131 from Pex22S, residues that are, for a large part conserved within the Pex22p family (Figures 3C, 4 and 5D). To test the prominent role of Tyr172 in complex formation, we mutated this residue to both an alanine and a phenylalanine and quantitatively measured the effect on Pex22S binding with ITC. The Y172F mutation reduced the ability of Pex415–183 to interact with Pex22S by a factor of 12 and likewise, mutation of Lys133 in Pex22S to an alanine reduced Pex415–183 binding by a factor of 9 (Table I; Supplementary Figure S1). Interestingly, we were unable to detect an interaction between Pex22S and the Y172A form of Pex415–183 using ITC (Table I). Therefore, in order to assess further the affect of the Y172A mutation on complex formation, we performed native gel electrophoresis and in-vitro ubiquitination assays (Figure 6). We saw no evidence of complex formation between Pex415–183 Y172A and Pex22S (Figure 6A) and additionally, the ubiquitin-conjugating activity of the mutant in the presence of Pex22S was considerably reduced, although not abolished (Figure 6B). This low level of Pex22S-induced activity may be due to a weak and/or transient interaction between Pex415–183 Y172A and Pex22S that is below the detection levels of our assays. Importantly, Pex415–183 Y172A retained levels of ubiquitin-conjugating activity similar to the wild-type protein in the absence of Pex22s (Figure 6B).
Figure 5.
Interfacing residues are conserved within homologous Pex4p and Pex22p sequences. (A) View of the Pex415–183 (surface representation) Pex22S complex (cartoon), indicating the region of Pex22S (shown in red and labelled) that forms the interface with Pex415–183. (B) Surface representation of the Pex22S binding site in Pex415–183, showing residues involved in interface formation which are invariant (blue), partially conserved (cyan) or not-conserved (grey) within homologous Pex4p sequences. (C) Mode of binding of Pex415–183 (cartoon) to Pex22S (surface representation) indicating the interface forming helices α3 and α4 in Pex415–183 (blue). (D) Surface conservation within the Pex415–183 binding site in homologous Pex22p sequences, indicating residues which are invariant (blue), partially conserved (cyan) or not-conserved (grey).
Figure 6.
In-vitro characteristics of the Y172A mutant form of Pex415–183. (A) Native gel electrophoresis analysis of the ability of wild-type and Y172A forms of Pex415–183 to bind Pex22S. (B) E2 self-ubiquitination assay performed as in Figure 2B and containing E1, ATP, wild-type or Y172A forms of Pex415–183 in the presence or absence of Pex22S. Samples of the reactions were taken at the time points indicated and probed with anti-Ub antibodies. The Y172A mutation, which disturbs Pex22S binding (A), does not affect the intrinsic activity of Pex415–183 (B, left panel) but inhibits the Pex22S-dependent enhancement of Pex4p activity (B, right panel).
Pex22S binding is an essential requirement for Pex4p's activity in vivo
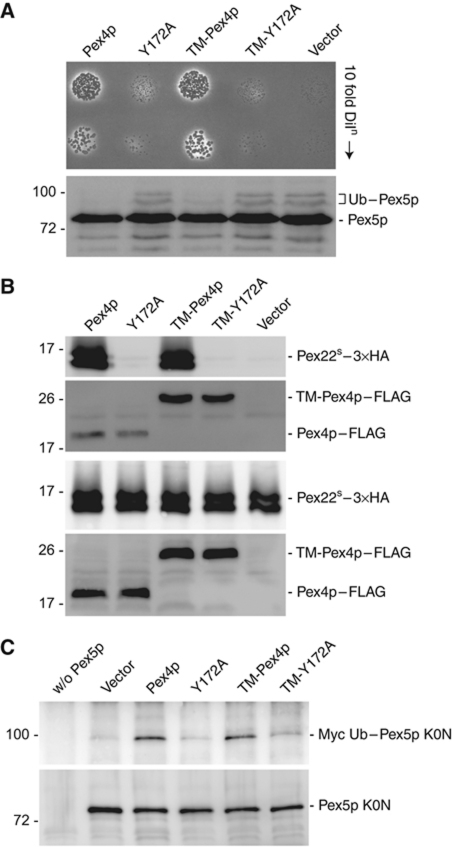
Our data indicate that the function of Pex22p goes beyond simply targeting Pex4p to the peroxisomal membrane (Koller et al, 1999). To investigate the additional role(s) of Pex22p, we carried out an in-vivo growth assay, testing the ability of pex4Δ yeast cells expressing Pex4p variants disturbed in Pex22p binding to grow on oleic acid, a carbon source requiring functional peroxisomes for its metabolism (Williams et al, 2007). Furthermore, we analysed the status of Ubc4p-dependent Pex5p ubiquitination, a phenotype indicative of a loss of Pex4p activity and consequently, a deficiency in Pex5p recycling (Platta et al, 2007; Williams et al, 2007). To separate Pex22p's role in Pex4p targeting from its hypothesised role as E2 co-activator, we tethered Pex4p (complete with C-terminal FLAG tag) to the peroxisomal membrane by fusing the first 54 amino acids of Pex22p, containing the TM segment, to its N-terminus. Expression of this chimeric protein, referred to as TM-Pex4p–FLAG, under control of the Pex4p promoter rescues the growth of pex4Δ cells on oleic acid (Figure 7A, upper panel), indicating that TM-Pex4p–FLAG is a functional protein. However, pex4Δ cells expressing the Y172A form of TM-Pex4p–FLAG are severely disturbed in their ability to grow under these conditions and additionally, exhibit Ubc4p-dependent ubiquitination of Pex5p (Figure 7A, lower panel), an observation that signifies a loss of Pex4p function. To confirm that the effect we observed with the Y172A mutant is indeed due to a loss of interaction, we assessed its ability to bind Pex22p in vivo (Figure 7B). Unfortunately, we are unable to detect Pex4p in cells when expressed under control of its own promoter, employing either anti-Pex4p or anti-FLAG antibodies. Therefore, we placed our constructs under control of the catalase promoter and subjected cells overexpressing either wild-type or Y172A forms of Pex4p–FLAG and TM-Pex4p–FLAG, together with 3 × HA-tagged Pex22S, to co-immunoprecipitation with anti-FLAG antibodies. Wild-type, but not the Y172A mutant form of Pex4p–FLAG and TM-Pex4p–FLAG was able to co-immunoprecipitate Pex22S (Figure 7B), confirming that the Y172A mutant is indeed unable to bind Pex22p in vivo and accordingly, that Pex22p binding plays an essential role in Pex4p's activity in vivo.
Figure 7.
In vivo analysis of the Pex4p:Pex22p complex. (A) Phenotypical analysis of Pex4p mutants disturbed in Pex22p binding. pex4Δ cells expressing Pex4p variants or an empty vector were grown on media containing oleic acid as sole carbon source for 6 days at 28°C (upper panel) or lysed and samples subjected to SDS–PAGE, western blotting and staining with anti-Pex5p antibodies (lower panel). Ub–Pex5p denotes Ubc4p-modified forms of Pex5p. (B) Pex22p binding capabilities of Pex4p in vivo. Lysates of cells (lower panels) expressing wild-type or mutant forms of Pex4p–FLAG and TM-Pex4p–FLAG, together with 3 × HA-tagged Pex22S were subjected to co-immunoprecipitation with anti-FLAG antibodies and samples were probed with anti-FLAG and anti-HA (upper panels). (C) Ability of Pex4p mutants disturbed in Pex22p binding to modify Pex5p. pex4Δpex5Δ cells, co-expressing Myc-tagged Ub, the Pex5p K0N variant and different Pex4p mutants, were lysed and subjected to immunoprecipitation using Pex5p antibodies. Immunoprecipitates were probed with SDS–PAGE, western blotting and staining with anti-Myc (upper panel) or anti-Pex5p (lower panel) antibodies. Disturbing the interaction between Pex4p and Pex22p (B) results in a growth defect on oleic acid (A) and Ubc4p-dependent ubiquitination of Pex5p (A), a phenotype caused by the inability of Pex4p to ubiquitinate Pex5p on its conserved cysteine residue (C).
Taking our analysis one step further, we directly assayed the effect of disturbing the Pex4p:Pex22p interaction on Pex4p's ability to attach Ub to its substrate, the cycling receptor Pex5p (Figure 7C). Since the Y172A mutation in TM-Pex4p–FLAG causes Ubc4p-dependent ubiquitination of Pex5p, we used the Pex5p K0N variant, in which all lysine residues in the N-terminal domain of Pex5p are mutated to arginines. This Pex5p mutant, which is severely restricted in Ubc4p-dependent but unaffected in Pex4p-dependent ubiquitination, allows us to assess the ability of our constructs to ubiquitinate Pex5p on its conserved cysteine residue in vivo (Williams et al, 2007). Using immunoprecipitation analysis, we could confirm that TM-Pex4p–FLAG is able to complement the pex4Δ knockout phenotype (Figure 7A) due to its ability to modify Pex5p, in a manner comparable to that of the wild-type protein (Figure 7C, compare lanes 3 and 5). Significantly, the ability of TM-Pex4p–FLAG Y172A to modify Pex5p was strongly reduced, confirming the vital role Pex22p binding plays in Pex4p's ability to transfer ubiquitin to a substrate and ultimately, its cellular role.
Discussion
In this contribution, we have presented the crystal structure of an E2/co-activator complex, consisting of the ubiquitin-conjugating enzyme Pex4p and the peroxisomal membrane protein Pex22p from S. cerevisiae. When comparing the Pex4p:Pex22p interface with other E2-complex structures, there is no overlap with established interfaces of any E3 ligase category (both canonical and non-canonical), as well as known E1, ubiquitin and UEV binding sites (Dye and Schulman, 2007; Ye and Rape, 2009; van Wijk and Timmers, 2010). This observation even holds for complexes of more distantly related SUMO- and NEDD8-conjugating enzymes. However, a partial overlap is seen with one of the interfaces used by the SUMO conjugating enzyme Ube2L when binding to Importin13 (Grünwald and Bono, 2010), as well as when Ube2L binds to its substrate, RanGAP1 (Reverter and Lima, 2005). In both cases, Ube2L utilises elements of α-helix 3 in the interaction. While the Ube2L:Importin13 interaction plays a role in the nuclear targeting, rather than the SUMO conjugating activity of Ube2L (Grünwald and Bono, 2010), contacts made by α-helix 3 allow the target lysine in RanGAP1 to come into close proximity of Ube2L's active site cysteine (Reverter and Lima, 2005). Taking into account that many functional features of E2 enzymes with different substrate specificities can be conserved (Dye and Schulman, 2007; Ye and Rape, 2009; van Wijk and Timmers, 2010), this observation may suggest that Pex22p binding is involved in positioning Pex4p close to its target, the conserved cysteine residue in the N-terminal region of Pex5p. Cadwell and Coscoy (2008) reported that attachment of Ub to a substrate can be dependent upon the position of the target residue in the substrate, rather than the type of residue, since both cysteine and lysine residues could be utilised when present at equivalent positions in a substrate. Recently, Grou et al (2009) showed that replacement of the conserved cysteine in human Pex5p with a more conventional lysine residue does not affect ubiquitination or function of the protein, suggesting that positioning is more important than the type of target residue for Pex5p ubiquitination. The presence of Pex22p may therefore allow for site-specific ubiquitination of the conserved cysteine, by correct positioning of Pex4p in relation to the exposed N-terminus of Pex5p.
As Pex22p stimulates the intrinsic Ub-to-substrate transfer activity of Pex4p, but does not contain a RING/U-box or HECT domain, it would be possible to describe the protein as non-canonical E3 ligase, similar to the SUMO E3 RanBP2 or the Replication Transcriptional Activator (RTA) Ub E3 ligase (Pichler et al, 2004; Yu et al, 2005). These proteins lack recognisable E3 ligase domains but can catalyse the transfer of Ub/SUMO from E2 to substrates. In the case of RanBP2, the comparison is particularly relevant since RanBP2 binding to Ube2L, as with Pex22p binding to Pex4p, causes changes in intrinsic E2 activity, probably through an allosteric mechanism, rather than by providing a bridge between E2 and substrate (Pichler et al, 2004). However, in vivo, Pex4p-dependent ubiquitination of Pex5p still requires the presence of three RING domain-containing proteins, all of which exhibit E3 ligase activity in vitro (Williams et al, 2008; Platta et al, 2009), suggesting that the role of Pex22p is more fundamental to Pex4p's activity and that additionally, Pex22p's function also precedes the step requiring an E3 ligase.
In vitro, binding of Pex22p allows Pex4p to produce lysine 48 linked Ub chains and in vivo, Pex22p binding is crucial for Pex4p's ability to attach two Ub moieties to the cycling receptor Pex5p, a modification required for Pex5p recycling (Platta et al, 2007; Williams et al, 2007). Although currently, the type of linkage used to attach the second Ub to Pex5p has not been established in vivo, observations from the related yeast H. polymorpha are worthy of note (Kiel et al, 2005). Expression of the K48R mutant form of Ub in this organism blocks peroxisomal matrix protein import, but only in the presence of Pex4p, providing a link between the synthesis of K48-linked Ub chains and the function of Pex4p. Whether K48-linked Ub chains are formed on the target Pex5p in vivo remains to be investigated.
Interestingly, the ‘α3–α4’ binding site (employing the surface exposed regions of α-helices α3 and α4) represents the only surface on an E2 for which to date, no binding partner has been identified (Wenzel et al, 2011). This mode of binding to an E2 would not occlude binding of E1 and/or E3, therefore negating a requirement for the complex to dissociate in order to allow procession of the Ub cascade. Consequently, the α3–α4 binding site provides an additional level of specificity for ubiquitin-conjugating enzymes, potentially allowing them to associate with proteins regulating their cellular localisation and/or activity, without compromising E1/E3 binding. Pex22p binding does not appear to alter the overall secondary structure of Pex4p (Supplementary Table S2) but, as our efforts to gain structural data on Pex4p alone have so far been unsuccessful, we cannot rule out that subtle changes in conformation do occur upon Pex22p binding. Indeed, two recent reports on E2 activity regulation offer interesting comparisons. Binding of the E3 ligase gp78 to Ube2g2 induces allosteric changes within the E2, which enhance E2:RING-domain interactions and in turn, ubiquitin conjugation (Das et al, 2009). Equally, Ceccarelli et al (2011) reported on an inhibitor of the ubiquitin-conjugating enzyme Cdc34, the binding of which causes a subtle repositioning of secondary structure elements within the E2, leading to an inhibition of activity. In both cases, the overall secondary structure of the E2 was not altered by ligand binding. Therefore, we propose that the influence Pex22p exerts on Pex4p is comparable and consists of a general ‘rigidification’ or ‘tightening up’ of the loosely associated Pex4p structure, effectively locking Pex4p in a conformation favourable for Ub-to-substrate transfer. However, Pex22p binding may also convey a signal through Pex4p to additional peroxisomal membrane components, such as the RING proteins. Therefore, an attractive model arises where the presence of Pex22p would not only secure the E2 in a conformation favourable for transferring Ub to a substrate, but would also play a role in recruiting the E3 ligase platform and in positioning Pex4p correctly in relation to its target, ultimately leading to site-specific ubiquitination of Pex5p. Accordingly, an important part of future research will be to address the affect Pex22p binding has on Pex4p:RING protein interactions.
Our ITC analysis indicates that the Pex4p:Pex22p interaction is of a high affinity and the considerable free energy change upon association (ΔG=−11.9 kcal mol–1) points towards a highly stable complex. Once bound, Pex4p will very likely remain in complex with Pex22p at the peroxisomal membrane, a situation that would increase the local concentration of Pex4p while concomitantly ensuring that active Pex4p is only present at its site of action. In line with this, Pex4p from the yeast P. pastoris is found predominantly associated with peroxisomes in vivo (Crane et al, 1994). This, coupled with the central role Pex22p plays in Pex4p's activity, leads us to conclude that the E2 required for cysteine-specific ubiquitination of Pex5p is best described in terms of the Pex4p:Pex22p complex, rather than Pex4p alone.
Here, we present data showing how a ubiquitin-conjugating enzyme can be regulated by an interacting partner not previously implicated in the Ub cascade. While the Pex4p:Pex22p complex represents the first structure of an E2/co-activator complex, the requirement of additional, non-canonical binding partners in the Ub cascade has been known for some time. One well-characterised example is the ERAD-specific E2 enzyme Ubc7p, which requires binding of the ER membrane protein Cue1p for full ERAD to occur (Bazirgan and Hampton, 2008; Kostova et al, 2009). Interestingly, as with Pex22p, Cue1p does not affect the activity of other E2 enzymes (Bazirgan and Hampton, 2008; Kostova et al, 2009). Another protein in this class is RSUME (Carbia-Nagashima et al, 2007). This RWD domain-containing protein binds to Ube2L and stimulates the SUMOylation of a number of substrates through an unknown mechanism. Again, RSUME's action is specific, as it does not influence the activity of other E2s (Carbia-Nagashima et al, 2007). Currently, our understanding of how such a small number of E2 enzymes can control substrate-specific ubiquitination in the presence of the huge number of E3 ligases present in the cell is incomplete. We propose, based on our own results and those previously reported that, for at least a subset of E2 enzymes, non-canonical binding partners not previously linked to the Ub cascade play a crucial role in substrate-specific ubiquitination, by regulating E2 activity and/or localisation. A number of large-scale two-hybrid and proteomics approaches have identified numerous E2 binding partners (Ewing et al, 2007; van Wijk et al, 2009), many of which are not E3 ligases. Therefore, a challenge of future research will be to define the role these interacting partners play in site-specific and target-specific ubiquitination and the mechanistic details thereof.
Materials and methods
Strains, growth conditions, plasmids and protein preparation
Plasmids are described in Supplementary Table S3. Yeast strains used in this study are S. cerevisiae BJ1991 pex4Δ (Hettema et al, 2000) and BJ1991 pex4Δpex5Δ (MATα, pex5∷KanMX4, pex4∷loxP, leu2, ura3-251, trp1, prb1-1122, pep4-3, gal2). For (co-) immunoprecipitations and trichloroacetic acid (TCA) lysates, yeast cells were grown on 0.67% YNB containing 0.3% glucose for 24 h and then shifted to 0.1% oleate medium containing 0.5% potassium phosphate buffer pH 6.0, 0.3% yeast extract, 0.5% peptone, 0.2% Tween-40 and grown for between 7 and 16 h. Mutant plasmids were constructed using the Quikchange™ site directed mutagenesis kit (Stratagene). Ubc4p and wild-type and mutant forms of Pex415–183 and Pex22S were expressed and purified as described in Williams et al (2008). Selenomethionine (SeMet) incorporation into Pex22S was achieved using the B834 pRARE E. coli strain (Novagen) and confirmed using mass spectrometry. Protein concentration was determined by specific absorbance at 280 nm using a NanoDrop™ (http://www.nanodrop.com).
Crystallisation
Proteins were mixed and gel filtrated into GF buffer (25 mM Tris pH 7.5, 100 mM NaCl, 1% glycerol, 3 mM β-mercaptoethanol) using a Superdex 75 (10/300) column (GE Healthcare). Crystals of the resulting complex (at 10 mg ml–1) were obtained using hanging drop vapour diffusion at 20°C: native Pex4:Pex22S complex 1 (Native 1) 1.5 M sodium malonate pH 7; native Pex4:Pex22S complex 2 (Native 2); SeMet-labelled Pex4:Pex22S complex, Native 2 condition plus 10 mM 5-Amino-2,4,6-triiodoisophthalic acid (I3C, Sigma). Cryoprotectant was 6 M sodium formate or 12% (v/v) ethylene glycol.
X-ray data collection and structure determination
Single wavelength X-ray data sets were collected on ID23eh2 (Native 1) and ID14eh1 (Native 2) at the ESRF, Grenoble. The multiple wavelength, anomalous diffraction (MAD) X-ray data set was collected on X12 at EMBL/DESY, Hamburg. All data were processed using XDS (Kabsch, 1988) and COMBAT and scaled using SCALA (Evans, 1993). Using the MAD data set, three selenium sites were located with autoSHARP (Vonrhein et al, 2007) and AUTORICKSHAW (Panjikar et al, 2005) and the phases were improved using RESOLVE (Terwilliger, 2000). Further improvements were made using DMMULTI (Collaborative Computational Project Number 4, 1994), carrying out multi-crystal averaging with the MAD and Native 1 data set phases. The resulting model was enhanced manually using COOT (Emsley and Cowtan, 2004) and used as molecular replacement template for the Native 2 data set, from which the final model is derived. Refinement was carried out using CNS (Brunger, 2007) and REFMAC (Murshudov et al, 1997), applying TLS parameters (Winn et al, 2001). Model quality was assessed with Molprobity (Chen et al, 2010). The interface between Pex415–183 and Pex22S was analysed using PISA (Krissinel and Henrick, 2007). All structure representations were made with PyMOL (http://www.pymol.org).
Isothermal titration microcalorimetry
Prior to measurement, proteins were dialysed against GF buffer without glycerol. Measurements with Ubc4p or Pex415–183 at 10 μM as a sample and Pex22S at 100 μM as the titration ligand were carried out at 25°C on a MicroCal VP-ITC and data were fitted using MicroCal Origin 7.0.
In-vitro ubiquitination assays
In-vitro ubiquitination reactions, performed as described in Williams et al (2008), contained 25 mM Tris pH 7.5, 75 mM NaCl, 5 mM MgCl2, 1 mM DTT, 5 mM ATP and proteins at the concentrations indicated. E2 charging experiments (Figure 2A) were performed in 25 mM Tris pH 7.5, 75 mM NaCl, 10 mM MgCl2, 10 mM ATP. Reactions were initiated by the addition of ATP and incubated at 28°C with gentle shaking. Samples were taken after 60 min, unless otherwise stated, quenched with SDS–PAGE loading buffer containing 5% β-mercaptoethanol (except in E2 charging experiments) and subjected to SDS–PAGE. Western blots were probed with anti-Pex4p (Rabbit, polyclonal) and anti-Ub (FK2 clone, Biotrend).
Co-immunoprecipitation assays
Oleate grown cells (20 A600 units) were lysed with glass beads in 300 μl lysis buffer (PBS pH 7.4, containing 0.5% NP-40, 1 mM PMSF, 0.5 mM DTT and yeast Protease Inhibitor Cocktail; Sigma) at 4°C. Debris was removed by centrifugation and lysates were incubated end over end with 1 μl of mouse monoclonal FLAG antiserum (M2 monoclonal F3165, Sigma) at 4°C. After 2 h, 100 μl of Protein G-Sepharose beads (GE Healthcare) was added and lysates were incubated for 1 h at 4°C. Precipitates were washed two times with lysis buffer and once with PBS and elution was carried out by heating the beads in 50 μl of IP-elution buffer (125 mM Tris, pH 6.8, 1.5% SDS, 6 M urea and 20% glycerol) for 10 min at 37°C. Samples were analysed by SDS–PAGE and immunoblotting, using antibodies raised against the FLAG or HA (Mouse monoclonal 12CA5) tags.
Native gel electrophoresis
Prior to loading, purified proteins (1:1 molar ratio for complex analysis) were incubated end over end for 5 min at 21°C in 25 mM Tris pH 7.5, 75 mM NaCl. Samples were subjected to native electrophoresis using 4–20% TBE gels (Invitrogen) according to the manufacturer's instructions and proteins were visualised with Coomassie brilliant blue staining.
Miscellaneous
Protocols used for yeast two-hybrid analysis (Williams et al, 2005), TCA lysates, immunoprecipitation and growth analysis experiments (Williams et al, 2007) are described elsewhere.
Supplementary Material
Acknowledgments
We thank Stan Fields for the Ubc4p plasmid, Jörg Höhfeld for the E1 enzyme and Krisztian Fodor for helpful suggestions. We acknowledge the European Synchrotron Facility (ESRF) for provision of synchrotron radiation facilities and for assistance in using beamlines ID14eh1 and ID23eh2. The project has been supported by a Rubicon Fellowship from the Netherlands Organisation for Scientific Research to CW (825.08.023), and by the Framework VI project ‘3D Repertoire’ from the European Commission to MW (LSHG-CT-2005-512028). The coordinates and structure factors of the Pex4p–Pex22p complex have been deposited in the protein data bank under the accession code 2y9m.
Author contributions: CW, BD and MW designed the study; CW, BD and MB made the clones; CW expressed and purified the proteins, performed ITC, crystallised the complex, collected the X-ray data and performed CD measurements; CW and SP solved and refined the structure; WAS analysed and processed the CD measurements; MB performed in-vitro ubiquitination assays and in-vivo experiments; MW and BD supervised the project; CW wrote the paper, with contributions from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bazirgan OA, Hampton RY (2008) Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem 283: 12797–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT (2007) Version 1.2 of the crystallography and NMR system. Nat Protoc 2: 2728–2733 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell 21: 873–880 [DOI] [PubMed] [Google Scholar]
- Cadwell K, Coscoy L (2008) The specificities of Kaposi's sarcoma-associated herpesvirus-encoded E3 ubiquitin ligases are determined by the positions of lysine or cysteine residues within the intracytoplasmic domains of their targets. J Virol 82: 4184–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capili AD, Lima CD (2007) Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol 17: 726–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, Holsboer F, Arzt E (2007) RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell 131: 309–323 [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, Varelas X, Koszela J, Wasney GA, Vedadi M, Dhe-Paganon S, Cox S, Xu S, Lopez-Girona A, Mercurio F, Wrana J et al. (2011) An allosteric inhibitor of the human cdc34 ubiquitin-conjugating enzyme. Cell 145: 1075–1087 [DOI] [PubMed] [Google Scholar]
- Chen V, Arendall W, Headd J, Keedy D, Immormino R, Kapral G, Murray L, Richardson J, Richardson D (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Collins CS, Kalish JE, Morrell JC, McCaffery JM, Gould SJ (2000) The peroxisome biogenesis factors pex4p, pex22p, pex1p, and pex6p act in the terminal steps of peroxisomal matrix protein import. Mol Cell Biol 20: 7516–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook WJ, Martin PD, Edwards BF, Yamazaki RK, Chau V (1997) Crystal structure of a class I ubiquitin-conjugating enzyme (Ubc7) from Saccharomyces cerevisiae at 2.9 angstroms resolution. Biochemistry 36: 1621–1627 [DOI] [PubMed] [Google Scholar]
- Crane DI, Kalish JE, Gould SJ (1994) The Pichia pastoris PAS4 gene encodes a ubiquitin-conjugating enzyme required for peroxisome assembly. J Biol Chem 269: 21835–21844 [PubMed] [Google Scholar]
- Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, McFeeters RL, Altieri AS, Ji X, Byrd RA, Weissman AM (2009) Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell 34: 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct 36: 131–150 [DOI] [PubMed] [Google Scholar]
- Eckert JH, Johnsson N (2003) Pex10p links the ubiquitin-conjugating enzyme Pex4p to the protein import machinery of the peroxisome. J Cell Sci 116: 3623–3634 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evans PR (1993) In Proceedings of the CCP4 Study Weekend, Sawyer L, Isaacs N, Bailey S (eds) pp 114–122. Warrington: Daresbury Laboratory [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y et al. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grou CP, Carvalho AF, Pinto MP, Huybrechts SJ, Sá-Miranda C, Fransen M, Azevedo JE (2009) Properties of the ubiquitin-pex5p thiol ester conjugate. J Biol Chem 284: 10504–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald M, Bono F (2010) Structure of Importin13–Ubc9 complex: nuclear import and release of a key regulator of sumoylation. EMBO J 30: 427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Girzalsky W, van Den Berg M, Erdmann R, Distel B (2000) Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J 19: 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1988) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst 21: 916–924 [Google Scholar]
- Kiel JA, Otzen M, Veenhuis M, van der Klei IJ (2005) Obstruction of polyubiquitination affects PTS1 peroxisomal matrix protein import. Biochim Biophys Acta 1745: 176–186 [DOI] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139: 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK (2007) Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J 26: 2797–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller A, Snyder WB, Faber KN, Wenzel TJ, Rangell L, Keller GA, Subramani S (1999) Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J Cell Biol 146: 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova Z, Mariano J, Scholz S, Koenig C, Weissman AM (2009) A Ubc7p-binding domain in Cue1p activates ER-associated protein degradation. J Cell Sci 122: 1374–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragt A, Voorn-Brouwer T, van den Berg M, Distel B (2005) The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J Biol Chem 280: 7867–7874 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (PDBeFold), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Merkley N, Shaw GS (2004) Solution structure of the flexible class II ubiquitin-conjugating enzyme Ubc1 provides insights for polyubiquitin chain assembly. J Biol Chem 279: 47139–47147 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA (2005) Auto-Rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr D Biol Crystallogr 61: 449–457 [DOI] [PubMed] [Google Scholar]
- Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F (2004) The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat Struct Mol Biol 11: 984–991 [DOI] [PubMed] [Google Scholar]
- Platta HW, Debelyy MO, El Magraoui F, Erdmann R (2008) The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p. Biochem Soc Trans 36: 99–104 [DOI] [PubMed] [Google Scholar]
- Platta HW, El Magraoui F, Baumer BE, Schlee D, Girzalsky W, Erdmann R (2009) Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol 29: 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R (2007) Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol 177: 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, Girzalsky W, Erdmann R (2004) Ubiquitination of the peroxisomal import receptor Pex5p. Biochem J 177: 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao ST, Rossmann MG (1973) Comparison of super-secondary structures in proteins. J Mol Biol 76: 241–256 [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk SJ, de Vries SJ, Kemmeren P, Huang A, Boelens R, Bonvin AM, Timmers HT (2009) A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol Syst Biol 5: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk SJ, Timmers HT (2010) The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J 24: 981–993 [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C (2001) Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105: 711–720 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G (2007) Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Wenzel DM, Stoll KE, Klevit RE (2011) E2s: structurally economical and functionally replete. Biochem J 433: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebel FF, Kunau WH (1992) The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature 359: 73–76 [DOI] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Distel B (2005) Saccharomyces cerevisiae Pex14p contains two independent Pex5p binding sites, which are both essential for PTS1 protein import. FEBS Lett 579: 3416–3420 [DOI] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Geers E, Distel B (2008) Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun 374: 620–624 [DOI] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Sprenger RR, Distel B (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem 282: 22534–22543 [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
- Worthylake DK, Prakash S, Prakash L, Hill CP (1998) Crystal structure of the Saccharomyces cerevisiae ubiquitin-conjugating enzyme Rad6 at 2.6 A resolution. J Biol Chem 273: 6271–6276 [DOI] [PubMed] [Google Scholar]
- Ye Y, Rape M (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang SE, Hayward GS (2005) The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22: 59–70 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B (2005) Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.