Abstract
Transcription regulation by steroid hormones and other metabolites is mediated by nuclear receptors (NRs) such as the vitamin D and retinoid X receptors (VDR and RXR). Here, we present the cryo electron microscopy (cryo-EM) structure of the heterodimeric complex of the liganded human RXR and VDR bound to a consensus DNA response element forming a direct repeat (DR3). The cryo-EM map of the 100-kDa complex allows positioning the individual crystal structures of ligand- and DNA-binding domains (LBDs and DBDs). The LBDs are arranged perpendicular to the DNA and are located asymmetrically at the DNA 5′-end of the response element. The structure reveals that the VDR N-terminal A/B domain is located close to the DNA. The hinges of both VDR and RXR are fully visible and hold the complex in an open conformation in which co-regulators can bind. The asymmetric topology of the complex provides the structural basis for RXR being an adaptive partner within NR heterodimers, while the specific helical structure of VDR's hinge connects the 3′-bound DBD with the 5′-bound LBD and thereby serves as a conserved linker of defined length sensitive to mutational deletion.
Keywords: cryo electron microscopy, DNA response element, nuclear receptor, retinoic acid, vitamin D
Introduction
The super-family of steroid/thyroid hormone/retinoid nuclear receptors (NRs) comprises two well-conserved core domains, the DNA-binding domain (DBD) and the ligand-binding domain (LBD). The N-terminal A/B domain is the most divergent among the NRs with respect to length and sequence conservation and contains an autonomous activation function, AF-1; however, the vitamin D receptor (VDR) contains only a short A/B region with no AF-1 function (Sone et al, 1991). DBDs and LBDs are connected through a hinge region, which is thought to be flexible in order to allow NR homodimers and heterodimers to bind to various DNA response element types (e.g., direct repeats, DRs, and inverted repeats, IRs). Response elements typically consist of two hexameric half-sites with 5′-AGGTCA-3′ as a consensus sequence in the case of non-steroidal receptors. For DR response elements, NRs bind as heterodimers with RXR as the common partner located on the upstream half-site (in the following, the first NR DBD named in a dimer defines the 5′ position within the promoter sequence upstream of the transcription start site, TSS), with the exception of RAR/RXR/DR1 and PPAR/RXR/DR1 complexes (retinoic acid receptor, RAR; peroxisome proliferator-activated receptor, PPAR) whose polarity is reversed (Kurokawa et al, 1994; Zechel et al, 1994a, 1994b; Rastinejad et al, 2000). The number of spacer nucleotides between the half-sites varies significantly, initially characterized in a simplified manner by the ‘1–5 rule’: RXR/RXR and RAR/RXR (DR1), RXR/RAR (DR2), RXR/VDR (DR3), RXR/TR (DR4) and RXR/RAR (DR5) (Umesono et al, 1991; Mangelsdorf and Evans, 1995) (thyroid hormone receptor, TR). However, recent data indicate a much more variable spacer usage (beyond five nucleotides) in different genes and cellular contexts, for example, in the case of the RAR (Delacroix et al, 2010). Several NRs also bind as homodimers to DR response elements (including VDR on DR3).
Since NRs are involved in the ligand-dependent regulation of gene expression they represent important drug targets directly linked to various severe diseases. 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3, or calcitriol) in particular regulates various biological functions, such as cell growth, differentiation, anti-proliferation, apoptosis, adaptive/innate immune responses, bone mineralization and calcium/phosphate homeostasis. 1α,25(OH)2D3 functions through the binding to VDR which is found in prostate, ovary, breast and skin, and also in brain, heart, pancreas, kidney, intestine and colon (Banerjee and Chatterjee, 2003). Consequently, deregulation of VDR function may lead to severe diseases such as cancers, psoriasis, rickets, renal osteodystrophy and autoimmunity (multiple sclerosis, rheumatoid arthritis, inflammatory bowel diseases, type I diabetes; Friedrich et al, 2002; Holick, 2003; Pinette et al, 2003; Bouillon et al, 2006; Fiévet and Staels, 2009). Because of the high pharmaceutical potential of VDR ligands, several crystal structures of the VDR LBD with ligands have been determined in the past (Rochel et al, 2000; Hourai et al, 2008; Kakuda et al, 2008; Nakabayashi et al, 2008; Antony et al, 2010; Sakamaki et al, 2010). The VDR LBD contains a dimerization interface with the RXR LBD, a 50 residue insertion of unknown function, and a ligand-dependent transcriptional activation region, AF-2. Agonist ligand binding induces a conformational change of the AF-2 helix (H12) that allows the recruitment of co-activators from the p160 family steroid receptor co-activator (SRC; Oñate et al, 1995) that remodel chromatin, or of the DRIP/TRAP mediator complexes that interact with the basal transcriptional machinery and help recruiting the RNA polymerase to the TSS (Lonard and O'Malley, 2006).
The regulation of gene activity through the binding of specific ligands involves the full NR (including DBDs and LBDs) through its DNA-binding and protein co-regulator recruitment and exchange abilities (Glass and Rosenfeld, 2000) that regulate chromatin modifications and remodelling. While the crystal structures of the isolated LBDs and also the DNA-bound DBDs of VDR and/or RXR are known, there is still no crystal structure available for the full RXR/VDR complex. In contrast, the crystal structure of the DNA complex of the RXR heterodimer with PPAR has been described (Chandra et al, 2008). This complex has an inverted polarity compared with most other RXR heterodimers as it is bound to a DR1 response element. In the crystal structure, the PPAR LBD was found in close contact with the RXR DBD resulting in a rather compact structure. The hinge areas between LBDs and DBDs are not visible for RXR and appeared rather disordered for PPAR (high temperature factors). The A/B domains were not visible. Using small angle X-ray scattering (SAXS), we recently characterized the architecture of several heterodimer complexes (RXR/VDR/DR3, RXR/RAR/DR5, RXR/RAR/DR1 and PPAR/RXR/DR1; Rochel et al, 2011). These solution structures show that the complexes exhibit an open asymmetric conformation and reveal how the DNA directs the positioning of co-activators. On the dynamic side, the existence of long-range allosteric communication between VDR and RXR has been documented by HDX experiments (Zhang et al, 2011). Besides a better resolution that allows a detailed interpretation of the molecular architecture of the DNA-bound RXR/VDR complex, the cryo-EM structure allows to unambiguously position the A/B domain of VDR and for the first time to visualize the hinge domains of both receptors. The well-defined hinges stabilize the complex in a precise conformation and provide the structural basis for understanding the specific role of each receptor within the RXR/VDR heterodimer. The structural similarity between VDR and TR hinges suggests that the conclusions of the present work can be extended also to the functional RXR/TR/DNA complex.
Results
The liganded RXR/VDR DNA complex has been formed from co-purified VDR (full-length, lacking the VDR-specific insertion sequence residues 166–216) and RXR (lacking the A/B domain which is the less conserved part of NRs) and incubated with a double-stranded DNA segment of 20 nucleotides containing a DR3 consensus response element (see Materials and methods and Supplementary Figure S1). The DNA comprises the sequence acAGGTCAcagAGGTCActc, the 5′ and 3′ half-sites being recognized by RXR and VDR, respectively (Mangelsdorf and Evans, 1995). The structures of the RXR/VDR DNA complexes have been determined by single-particle cryo-EM and 3D reconstruction (Figure 1) at a resolution of 10–12 Å (as estimated from Fourier-shell correlation and consistent with the features resolved in the map; see below and Supplementary Figures S2 and S3). The structure reveals an L-shaped organization with one side split into a V-shape module, similar to a slingshot frame (Figure 1). The shape and size of the V-shaped part (∼40 × 50 × 75 Å) can immediately be recognized as the LDB dimer, whereas the longer side of the L has proportions (∼75 × 35 × 32 Å) characteristic for a DBD-bound DNA segment. The structure reveals that the RXR/VDR complex forms an open architecture (Figure 1), with the LBD heterodimeric unit oriented almost perpendicular to the DBD/DNA part. Notably, a clear density attributable to the LBD–DBD hinge region connects the LBD part to the DNA-DBDs subcomplex.
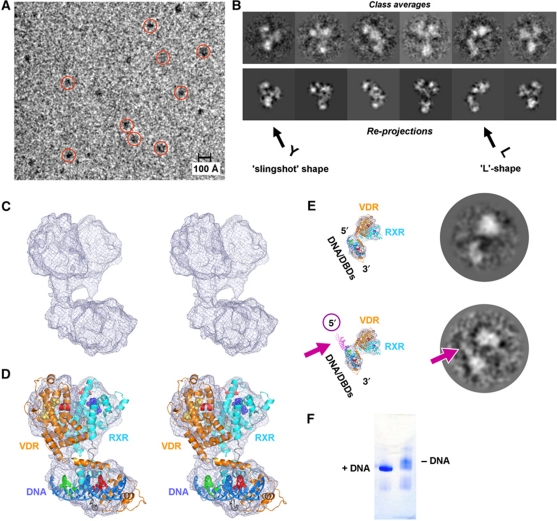
Figure 1.
Structure determination and overall description of the RXR/VDR/DNA complex. (A) CCD image of a flash-frozen hydrated sample of the RXR/VDR/DNA complex; scale bar is 100 Å; recorded on a transmission electron microscope (see Materials and methods) under cryo conditions at a defocus of ∼5 μm at 200 kV (for processing, data were collected at a defocus of −2.0 up to −4.0 μm). Some particles are marked with red circles. A similarly good contrast is obtained also closer to focus (−2.5 μm defocus) when images are recorded at 100 kV (see Supplementary Figures S8 and S9). (B) Comparison of class averages (first lane, obtained by multivariate statistical analysis and classification) with the corresponding re-projections of the 3D reconstruction (second lane). The characteristic ‘L’ and ‘slingshot’ shapes can be recognized in the class averages (compare these views with the side and front views in Figure 2A and B). A comparison with raw particle images can be found in Supplementary Figure S10. (C) Stereo representation of the cryo-EM map revealing the global architecture of the RXR/VDR/DNA complex. (D) Stereo representation of the cryo-EM map with the fitted crystal structures of the individual RXR and VDR LBDs and DBDs, resulting in a molecular model of the full RXR/VDR/DNA complex. (E) Assignment of the DNA polarity within the complex via a 5′-extended DNA oligomer. Comparison of the class averages of the RXR/VDR complex with the 20-bp DNA (top lane) and with the longer DNA extended by 15 nucleotides on the 5′-end (bottom lane); the corresponding views with the modelled DNA are displayed on the left. The pink arrow indicates the additional density in the class average, which is observed at the expected 5′ position. These data confirm the localization of the DNA within the complex and the annotation of the DNA 5′–3′ polarity in the complex. (F) Native gel analysis of RXR/VDR in the presence (left) and absence (right) of the DNA. The sharp band in the presence of DNA indicates a conformational stabilization of the complex.
In order to address the localization of the individual NR domains in more detail and provide a full interpretation, we fitted the available crystal structures of the LBDs and DNA-bound DBDs into the cryo-EM map (Figures 1 and 2). For the RXR/VDR LBD part, we used the RXR/RAR LDB crystal structure (Bourguet et al, 2000) and the VDR LBD crystal structure (Rochel et al, 2000) taking into account the high sequence conservation of the LDB dimer interface. Because there is no functional DNA-bound RXR/VDR DBD structure available for a DR3 element with RXR and VDR DBDs on the proper 5′ and 3′ DNA ends, respectively, we built a model using the VDR part of a DNA-bound 5′-VDR/RXR-3′ complex (Shaffer and Gewirth, 2004) and the DR4 RXR/TR DBD (Rastinejad et al, 1995) adapted to a DR3 element (see Materials and methods). Fitting the RXR/VDR LBD and DBD models allows unambiguously annotating the RXR and VDR parts in the map and assigns the handedness of the complex (Figure 1C and D), as validated also by calculating the cross-correlation coefficients between the maps and the different possible fittings (see Supplementary Figure S4). The cryo-EM structure is consistent with the small angle X-ray diffraction (SAXS) experimental data (Supplementary Figure S5; χ2=2.2 between the model derived from the cryo-EM structure and the SAXS curve).
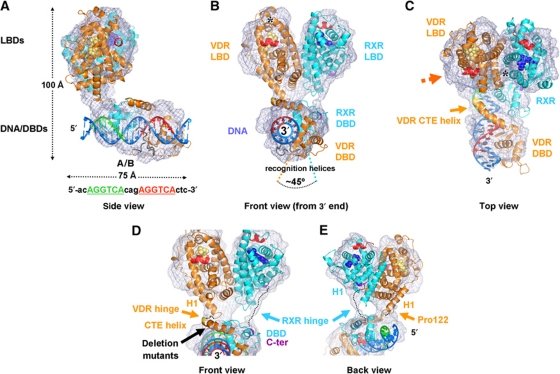
Figure 2.
Structure of the RXR/VDR/DR3 DNA nuclear receptor complex. (A) Side view of the structure, with the 5′ DNA end on the left. The cryo-EM map is shown in magenta, the fitted DNA/DBD and LBD heterodimer parts are shown with their backbone secondary structure (models are derived from available crystal structures, see main text). The DNA is shown in blue with the first half-site of the response element in green and the second half-site in red. The DBDs of RXR (cyan) and VDR (orange) are bound on the back, and a density close to the recognition helix of VDR is attributable to the A/B domain of VDR (17 residues). The LBDs are oriented perpendicular to the DNA, anchored through the 5′ side of the response element. The dimensions of the complex are indicated as well as the sequence of the response element. (B) Front view of the complex as seen from the 3′-end of the DNA (rotated 90° with respect to the view in A). The VDR LBD is on the left whereas the RXR LBD is on the right. The DBDs sit side-on to the DNA and are rotated by ∼45° with respect to each other because of the three nucleotide spacer (DR3) between the half-sites. The ligands 1α,25-dihydroxyvitamin D3 and 9-cis retinoic acid for VDR and RXR are shown in yellow and blue van der Waals spheres. The area comprising helices H2, H3n and the β-sheet of VDR is marked with a star. Since the RXR partner is specific for co-activator binding, the site on VDR that includes trans-activation helix H12 (in red) is available for the recruitment of chromatin-modifying co-regulator proteins on the side oriented opposite to the DBDs. (C) Top view of the complex as seen along the pseudo two-fold axis through the interface of the LBDs (rotated 70° downwards with respect to the view in B). The C-terminal extension helix of VDR protruding from the DBD and crossing the DNA minor groove is indicated, and the LBD interface contact comprising helix 4, loop 8/9 of VDR and helix H7 of RXR are marked with a star. (D, E) Description of the hinge regions between the LBDs and DBDs of RXR (cyan) and VDR (orange). The map resolves the individual hinges of the complex, revealing that these are well defined, thus providing the connectivity between the LBD and DBD core domains. The hinges adopt a parallel arrangement and thus neither cross nor interact with each other. RXR has a 25 residue linker (dotted line) between the C-terminal end (dark red) of the DBD to the N-terminal end of helix H1 (dark red). For VDR, there is a direct connection between the C-terminal end (yellow) of the CTE and the N-terminal end of helix H1. The conserved Pro 122 marks the kink between the VDR CTE helix and helix H1. The representations in (D) and (E) are seen from the front (rotated 10° around the horizontal axis with respect to the view in B to better see the CTE helix) and from the back (rotated 150° around the vertical axis compared with D).
The VDR and RXR LBDs can be identified unambiguously in the map thanks to two characteristic asymmetric features in the VDR LBD, the VDR segment comprising helix H4/loop 8/9 and RXR helix H7, and the area comprising helices H2, H3n and the ß-sheet (marked with a star in Figure 2B and C). Importantly, these features allow resolving the possible ambiguity residing in the pseudo-symmetry of the RXR and VDR LBD heterodimer (see view on the pseudo two-fold symmetry axis in Figure 2C). For the DNA-DBDs part, we fitted the DR3 RXR/VDR DBD model into the cryo-EM map (Figure 2A–C). The fit determines the 5′-3′ orientation of the DNA/DBD complex thanks to the precision of the DBD densities, and it is consistent with the connectivity constraint between the LBDs and DBDs of RXR and VDR, respectively. In addition, a complex with a DNA extended by 15 nucleotides on the 5′ side confirms the polarity assignment of the DNA (Figure 1E). The map clearly reveals that the DBDs lie ‘side-on’ with respect to the DNA (Figure 2B), with a part on the DNA side facing the LBDs and not opposite from them. The fitting also addresses the polarity of the complex with respect to the DNA, showing that the main anchor point to the DNA occurs on the 5′ side (Figure 2A) through the hinge region and the DBDs rather than for example centred on the response element. The RXR DBD is indeed found on the 5′-end as can be anticipated for a DR3 element.
The structure of the complex reveals that the LBD heterodimer is a rather rigid and stable unit of the complex also in the presence of DBDs and DNA. This is consistent with the high sequence conservation of the LBD interface (Brelivet et al, 2004) and the stability of LBD heterodimers which has facilitated the determination of several crystal structures (RXR/RAR, Bourguet et al, 2000; RXR/PPAR, Gampe et al, 2000; RXR/LXR, Svensson et al, 2003) but not yet for RXR/VDR. The present structure suggests that the RXR/VDR LBD adopts the same arrangement as RXR/RAR (Bourguet et al, 2000) or RXR/PPAR (Gampe et al, 2000) LBDs. Notably, the map indicates the position of the central helices of the LBD interface comprising helices H9 and H10/11 of VDR and RXR (Figure 2C; Supplementary Figure S2), and a nice piece of density is attributable to the LDB interface contact between helix H4, loop 8/9 (VDR) and helix H7 (RXR; Figure 2C). The orientation of the LBD dimer is approximately perpendicular to the DNA, with only a small tilt (the pseudo two-fold symmetry axis through the LBD heterodimer is tilted ∼20° towards the downstream side of the DNA, and the heterodimer is rotated by ∼5° on the VDR side). In the presence of an agonist ligand as in the complex here, the trans-activation helix H12 (marked in red in Figure 2) is known to close the occupied ligand-binding pocket and adopt an active conformation required for the recruitment of co-activators such as SRCs (McKenna et al, 1999). Notably, the structure of the RXR/VDR DNA complex shows that H12 and the co-activator binding site of VDR point side-on to the DNA (opposite to the DBDs; Figure 2B and C), suggesting that a co-activator which preferably binds to the partner of RXR could bind to the NR complex. This finding represents a key concept revealing that DNA-bound NRs maintain the trans-activation helix H12 area accessible for recruiting chromatin-modifying co-regulator proteins that can act on the nucleosome-bound region of the DNA.
The RXR and VDR DBDs are resolved as individual entities in the map, thus revealing their precise position on the DNA. They are rotated with respect to each other by ∼45° around the DNA axis, which is consistent with the 3-nucleotide spacer between the 6-nucleotide DRs (Figure 2B). The RXR DBD is positioned on the 5′-side of the DNA, while the VDR DBD is on the 3′-side, as can be recognized from the characteristic density observed for the VDR-specific C-terminal extension (CTE) helix expanding from the DBD core, while the RXR DBD has no such feature. From the proximity of RXR and VDR DBDs on the DNA it seems likely that they interact with each other (Figure 2C). An additional density is visible in the major groove next to the VDR recognition helix, which may correspond to the short A/B domain in VDR (17 residues at the N-terminus, Figure 2A). The A/B domain sequence MEAMAASTSLPDPGDFD shows prominent conserved residues within the VDR family, notably Ser/Thr, Glu/Asp and Phe which could provide additional base-specific interactions with the DNA major groove (notably the 5′ bases of the half-site and bases on the complementary strand). Interestingly, natural shortening of the N-terminus by three amino acids as it occurs in VDR polymorphism influences the repressive activity of VDR in the absence of ligand (Alimirah et al, 2010), while in the presence of ligand no significant effect is observed (Gross et al, 1998); this VDR polymorphism has been associated with several diseases. The VDR CTE connects directly to the N-terminal helix H1 of the LBD (Figure 2D; residue Leu 120 is the terminal residue seen in the individual LBD and DBD crystal structures). For RXR, the DBD C-terminal end extends into a discrete density connected to the RXR LBD, thus indicating the position of the hinge between the two core domains (Figure 2D and E). The CTE of RXR has not been modelled here since its structure is unknown and it has no sequence similarity with the CTE helix region of VDR or TR. However, the features in the map and the distance between the terminal residues of the RXR LBD and DBD (∼35 Å) suggest that the RXR linker contains a rather well-defined secondary structure (possibly two helical segments with a short extended loop; see dotted line in Figure 2D and E). While VDR has a straightforward connection between the DBD and LBD through the CTE helix, the RXR hinge needs to reach the LBD helix H1 at the back side of the LBD (oriented 5′ on the response element) and thereby crosses the tip of helix H9 which is part of the LBD interface.
The first part of the two hinges protruding from helix H1 to the DBDs is resolved as two individual segments (Figure 2D and E), and the hinges remain independent also when they kink into the DBD/DNA subcomplex (Figure 2A). This suggests that residues of the VDR and RXR hinge regions do not interact with each other, that is, RXR and VDR LBDs and DBDs adopt a parallel organization without crossover. However, the fact that the hinges provide two independent anchor points to the DNA clearly contributes to the stabilization of the complex in a precise conformation with respect to the DNA strand. The well-defined conformation of the complex, notably between LBDs and DBDs, is also suggested by a 3D re-sampling and classification (3D-SC; Simonetti et al, 2008) test that when applied to the data revealed no major variations in the structure. In this context, it is of particular interest that the VDR linker peptide carries a proline residue (Pro 122; Figure 2E) right at the junction of the two helical elements (CTE and H1, sequence KDSLRPKLSE), which favours the formation of the observed kink between the DBDs and LBDs thus leading to the characteristic L-shape conformation of the complex (Figure 2A). The proline residue is conserved in the VDR family and also in several other NRs (RAR, TR, PPAR, LXR, PXR and CAR) including their common heterodimeric partner RXR and therefore marks the N-terminal edge of the LBD domain. The conserved charged residues of the RPK motif (residues 121–123, tip of LBD helix H1) may contribute to an additional stabilization of the complex through interactions with the phosphate backbone of the central part of the 5′ half-site, in direct vicinity to the C-terminal tip of the RXR recognition helix (Figure 2E).
Discussion
The present study allows to better understand how RXR/VDR heterodimers are able to recognize specific DNA response elements in a cooperative manner, beyond the DBD part of the complex. A precise identification and localization of the RXR and VDR core domains with respect to the DNA are now available and provide a structural basis for numerous biochemical data previously analysed in the context of partial structural data. The structure exhibits an open conformation in which the RXR/VDR dimer binds to its target DNA in a highly asymmetric manner, resulting in the LBDs positioned at the 5′-end of the response element. For the first time, the VDR A/B domain and the VDR and RXR hinges can be localized in the RXR/VDR/DNA complex.
An important feature underlined by the present RXR/VDR/DNA complex is the organization of the hinge region, which has key implications for NR function. Within the topology of the complex, the long and partially extended CTE region of RXR is needed to reach helix H1 on the side opposite from the RXR DBD (i.e., further up in the 5′ direction; Figure 2D and E). This explains why RXR cannot have a rigid CTE helix, in contrast to that observed for VDR and TR (Figure 3); if RXR had a CTE helix such as VDR positioned at the 5′ side of the response element (Figure 3A and B) it would entirely change the topology of the complex. Furthermore, the flexible linker of RXR is required for the adaptation to different DR response elements (DR2, 3, 4, 5) with the 5′-bound heterodimerization partner RXR, or DR1's with a 3′-bound RXR. The RXR partner, however, being positioned with its DBD on the 3′ side of the DNA response element far away from the LBD (∼35 Å), requires a long straight segment to reach the LBD N-terminus. This is achieved through the VDR CTE helix (Figure 3B). The architecture of the complex thus rationalizes the existence of the CTE helix, which protrudes from the DBD core and crosses the DNA strand perpendicular to the minor groove in order to join the first α-helix of the LBD (H1; Figure 2C and D). The overall 5′ LBD topology of the complex probably also applies to TR, which has a CTE helix oriented in the same way over a DR4 element (Rastinejad et al, 1995; Figure 3C). The combination of a rigid geometry of the CTE in VDR, TR and possibly other related NRs, with a more flexible RXR CTE suggests that the adaptability of full NR heterodimers to different response elements resides largely in RXR. The structure of the entire RXR/VDR complex suggests that the CTE helix stabilizes the 5′ LBD position and conformation with respect to the DNA. Other NR complexes and response elements (particularly DR1's) may adopt a different organization. However, the stable conformation of RXR/VDR on the DNA suggests that binding to different response elements is based on the structural adaptability of RXR rather than an intrinsic flexibility or instability of the whole complex. Consistently, mutations in the hinge region of VDR and RXR show that the linker length has a critical role in VDR but not so in RXR (Shaffer et al, 2005). Deleting residues in the VDR CTE helix hinge (i.e., right before Pro 122 of the RPK motif which is located at the kink between DBDs and LBDs; Figure 2D and E) reduces transcriptional trans-activation, but shows no influence on DNA binding. However, replacing deleted residues at the C-terminal end of the helix by alanines restores activity (Shaffer et al, 2005), showing that a conserved length rather than a particular amino-acid sequence of the VDR hinge helix is required for full transcriptional activation. On the other hand, for the hinge region of RXR no helical structure is predicted and there is no sequence similarity with the CTE helix regions of VDR or TR. Consistently, deleting of up to 14 residues in the RXR hinge retains near wild-type activity. This is in agreement with the more flexible character of the hinge, which confers RXR plasticity and adaptability towards different response elements as a common partner in NR heterodimers. The structure thus rationalizes previous mutational data (Shaffer et al, 2005) by clarifying the role of the conserved long CTE helix which serves as a physical linker of defined, critical length and rather rigid character in the case of VDR, but not so for RXR. Changing the length of the CTE helix in VDR would clearly change the overall conformation of the complex and thereby affect co-activator recruitment, which explains the low trans-activation activity of the VDR deletion mutants.
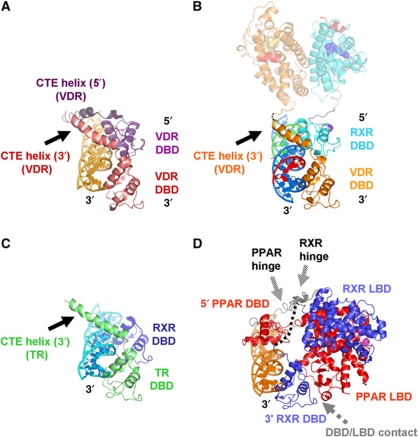
Figure 3.
Comparison of the topology of NR complexes on the DNA. (A, B) Comparison of the crystal structure of the VDR homodimer DBD on a DR3 response element (PDB ID 1KB4; Shaffer and Gewirth, 2002, A) super-positioned to the DBD part of the cryo-EM structure of the RXR/VDR/DR3 complex (B, viewing angle and colour code as in Figure 2D; the LBD part is shown in faded colours for simplicity). The 5′ VDR DBD (violet) corresponds precisely to the RXR DBD in the RXR/VDR/DR3 complex, with the exception of the CTE helix (dark violet) that is absent in RXR (compare also with Figure 2D). This similarity provides an independent validation of the model of the RXR/VDR/DNA subcomplex and suggests that in homodimers the 5′ VDR plays the role of the 5′ RXR. (C) The DNA-bound TR DBD shows the same organization as the VDR DBD (viewing angle as in A and B), notably the CTE helix that protrudes from the DBD in the same direction coming from the 3′-bound DBD, suggesting that an RXR/TR/DR4 complex has a similar overall topology to RXR/VDR/DR3. (D) Comparison with the crystal structure of the PPAR/RXR/DR1 complex (PDB ID 3DZU; Chandra et al, 2008) super-positioned through the DNA and the 3′ DBDs of the cryo-EM structure of the RXR/VDR/DR3 complex (viewing angle and colour code as in A–C; PPAR in blue, RXR in red, co-activator peptide in magenta; see also Supplementary Figure S6). The different response elements (DR3 versus DR1) lead to a reversal of the DNA polarity, but also to a transition from a side-on complex to an interaction from either sides of the DNA. The global architecture of the two complexes is also rather different with respect to the position of the LBDs which are perpendicular in the RXR/VDR complex while being in a closed conformation in the PPAR/RXR complex, resulting in an RXR DBD–PPAR LBD interaction (indicated in grey). In contrast, the RXR/VDR complex harbours only classical heterodimeric DBD–DBD and LBD–LBD interfaces. While the hinges are well defined in the cryo-EM structure of RXR/VDR (Figure 2), they are either not visible (black dotted line for the RXR hinge) or partially disordered (temperature factors of >90 Å2 in the PPAR hinge, the backbone is represented in grey).
The RXR/VDR DNA complex is a key representative of the class of RXR heterodimers that bind to DRs with 2, 3, 4 or 5 nucleotide spacers, resulting in response elements in which RXR occupies the 5′-end position: RXR/RAR (DR2), RXR/VDR (DR3), RXR/TR (DR4) and RXR/RAR (DR5; reviewed in Mangelsdorf and Evans, 1995). In contrast, DR1 elements bind RXR on the 3′ side with a reversed polarity on the DNA (e.g., PPAR/RXR, Chandra et al, 2008; RAR/RXR, Rastinejad et al, 2000; RXR/RXR, Zhao et al, 2000). Interestingly, the arrangement seen here for an RXR/VDR DR3 complex is fully consistent with that of the VDR DBD homodimer on a DR3 element (Shaffer and Gewirth, 2002; Figure 3A and B). In our structure, the RXR DBD core part takes the position of the 5′ VDR segment (without the distinct CTE part), suggesting a common architecture for DR3 NR complexes independently of whether these are heterodimers or homodimers. In other words, the 5′ DBD in VDR DBD homodimers appears to play the role of the RXR DBD (Figure 3A and B). In the homodimer structure, the tip of the 3′ VDR DBD CTE helix is bent towards the DNA axis (Figure 3A) and thus points to the connection point of helix H1, consistent with the VDR LBD position in the present structure. Note that the relative position of the CTE helix and the contacts with the DNA are maintained in the full VDR complex. The same relative orientation of the CTE helix was observed in the crystal structure of the TR DBD/DNA complex. This structural conservation in different crystalline and molecular environments is consistent with the key functional role of the CTE hinge as observed in the full RXR/VDR/DNA complex.
While the DBDs of VDR/VDR, RXR/VDR and RXR/TR show similar arrangements on DR3 and DR4 DNA response elements (Figure 3A–C), the organization is entirely different on a DR1, as shows the comparison of our RXR/VDR/DR3 structure with the crystal structure of PPAR/RXR/DR1 (Figure 3D; Supplementary Figure 6). The DNA in the PPAR/RXR complex contains an idealized DR1 element, with PPAR bound to the 5′ half-site, while RXR occupies the 3′ position. This results in a reversed polarity when compared with the RXR/VDR complex, because of the DR1 DNA as opposed to DR2, 3, 4, 5 elements. The DR1 leads to an arrangement in which the DBDs are rotated with respect to each other by ∼110°, with the DBDs on either side of the DNA (Figure 3D) rather than on the same side as in the RXR/VDR complex (Figure 3B). The two complexes also have a different overall architecture because (i) the 3′ RXR DBD has no CTE helix in order to reach the LBD and (ii) the PPAR DBD has its CTE region pointing into a completely different direction compared with the RXR CTE in the RXR/VDR complex because of an interactions with the 5′ DNA extension element, resulting in the LBDs facing the same side of the DNA as the DBDs (Figure 3D). The two hinge regions are rather disordered in the PPAR/RXR complex (either invisible or high temperature factors characteristic of a flexible area; Figure 3D), such that the role of the hinge region in stabilizing a particular conformation cannot be addressed here. Rather, the observed conformation is probably stabilized by an additional DBD–LBD contact between the PPAR LBD and the RXR DBD present in the crystal structure (Figure 3D; Supplementary Figure S6) but not seen in solution by SAXS (Rochel et al, 2011). In contrast, the RXR/VDR complex shows DBD–DBD and LBD–LBD contacts exclusively through the classical heterodimeric interfaces, the only DBD–LBD links being the well-structured hinge connectors (Figure 2D and E).
The cryo-EM structure of the DNA-bound RXR/VDR complex addresses the global architecture of a key representative of NR heterodimers. The topology of the complex has important functional implications. It identifies the relative positions of the DNA and DBDs with respect to LBDs where the natural ligands and pharmaceutical drugs bind. Rather than being located on the centre of the response element, the LBDs are positioned in an asymmetric manner on the 5′-end of the response element, shifted away from the centre of the two half-sites. This relieves potential ambiguity of response element recognition residing in the symmetrical arrangement of the two half-sites. As a result, the LBDs display a strong polarity with respect to the DNA. The asymmetric topology of the complex represents a key feature which clarifies the binding mode of the full complex to the DNA and explains the general role of RXR as an adaptive common partner within the class of heterodimeric NR complexes: RXR provides adaptability to different response elements, while the role of its variable partner is to stabilize the heterodimeric complex on a precise half-site DR (e.g., a DR3 for VDR and a DR4 for TR). An interesting aspect here is that the short A/B domain of VDR (17 residues at the N-terminus) interacts with the major groove next to the VDR recognition helix and may thus modulate DNA binding, possibly in a sequence-dependent manner; these findings will allow to design new experiments in order to analyse the role of the A/B domain in DNA binding. The structure also reveals the important role of the hinge regions between LBDs and DBDs that were thought until now to be disordered: being conformationally well defined they stabilize an overall open conformation of the whole complex without requiring any DBD/LBD inter-domain contacts. The thereby induced positioning of the LBDs makes the area of trans-activation helix H12 accessible. This is functionally relevant for the recruitment of co-regulators with chromatin-modifying activity and the subsequent transcription trans-activation.
The structure presented here suggests a general model for cooperative DNA binding of NR heterodimers. It highlights the importance of ligand and DNA binding, suggesting cooperative and allosteric effects between DBD and LBD parts in which ligands and DNA act together to fine-tune gene regulation. Ligand binding strengthens dimer formation (Cheskis and Freedman, 1996) and thus has a direct effect on DNA binding (i) because the recognition unit encompasses two DBDs and (ii) through helix H1 and the helical hinge domain of VDR, that is, the link between LBD and DBD may exert a distant allosteric action. Recent deuterium exchange experiments have suggested a relaxation of the DBD structure upon ligand binding (Zhang et al, 2011). The present structure excludes a direct contact between LBD and DBD playing a significant role in the allosteric control. Furthermore, ligand binding stabilizes the structure of the LBD in a conformation compatible with the binding of co-activator proteins. Allosteric effects between the LBDs of the heterodimers binding their individual ligands result in preferential binding of co-activators to the partner of RXR (e.g., RAR, VDR, TR and LXR; Son and Lee, 2010; Rochel et al, 2011). DNA binding promotes DBD–DBD interactions and stabilizes the whole complex in a precise global conformation (as illustrated by the native gel analysis in the presence and absence of DNA, Figure 1F). The latter aspect extends to the fact that natural DNA sequences can vary significantly from the canonical response elements based on AGGTCA half-sites and thus further fine-tune dynamic binding to promoter DNA and thereby regulate gene expression.
The analysis of a 100-kDa macromolecular complex by single-particle cryo-EM and 3D reconstruction to about 10–12 Å resolution has required some particular attention. The key parameters to overcome the technical difficulties related with the relatively small molecular weight of the complex resulting in a low contrast in the cryo-EM images have been: (i) rigorous screening of flash-frozen holey (rather than continuous) carbon grids for optimal ice thickness (Figure 1; Supplementary Figures S7–S9), (ii) defocus pairs to improve the image contrast and localize the objects (Supplementary Figure S7) and get a first 3D reconstruction through angular reconstitution and (iii) validation of particle selection and alignment through multivariate statistical analysis and classification of the images (Figure 1B; Supplementary Figure S10). Also, extra data were collected with an acceleration voltage of 100 kV at which the image contrast is notably better (Supplementary Figures S8 and S9), and used to produce an independent ab-initio reconstruction that is fully consistent with the structure refined using data collected at 200 kV (inter-map correlation of 0.87; Supplementary Figure S11). Moreover, the re-projections from the structure were cross-validated with the input class averages and with the raw particle images (Figure 1B; Supplementary Figure S10), and the cryo-EM map is also consistent with the size and features of the fitted LBD and DBD crystal structures (Figure 1; Supplementary Figure S12). Finally, extending the DNA by 15 nucleotides (Figure 1E) also validated the results. It should be noted too that an internal symmetry would be beneficial for processing such data, albeit not essential as illustrated by the present work on an entirely asymmetric object. Studies of this sort may in the future be facilitated by direct electron detectors, which are more sensitive than CCD cameras. The present work opens the way towards structural studies of many relatively small macromolecular objects such as protein/nucleic acid complexes, which represent important structural biology targets but may have been difficult to crystallize or judged too small for standard cryo-EM methods. Thus, the study of NR complexes with different response element types such as inverted, everted and direct repeats can now be envisaged.
Materials and methods
Purification
The HsVDRΔ (1–427Δ166–216) was expressed as a hexahistidine fusion protein. HsRXRαΔAB (130–462) was cloned into a pACYC plasmid encoding a non-tagged protein. Ligands (9-cis retinoic acid for RXR and 1α,25-dihydroxyvitamin D3 for VDR) were added in a two-fold excess to saturate the receptors. The annealed double-stranded DNA (sequence acAGGTCAcagAGGTCActc) was added in a 1.2 excess to the dimer. The complex was further purified on a gel-filtration S200 column. The final buffers were Tris 20 mM pH 7.5, NaCl 50 mM, KCl 50 mM, MgCl2 4 mM and DTT 5 mM. For the 5′-extended DNA, the sequence is agaggatggagtcag-cgAGGTCAcgaAGGTCAc.
Cryo electron microscopy
The RXR/VDR/DNA complexes were embedded in a thin layer of vitreous ice suspended across the holes of a holey carbon film as described in Klaholz et al (2004). The images have been collected at liquid-nitrogen temperature with the in-house FEI Tecnai20 field emission gun transmission cryo electron microscopy at 200 kV acceleration voltage with a dose of ∼20 electrons per Å2 at a magnification of 50 000 on photographic films (Kodak SO-163; Marzi et al, 2007; Simonetti et al, 2008). In order to determine the appropriate specimen concentration and best distribution of particles in vitreous ice, we screened numerous grids and grid areas best suitable for data collection. For this, CCD images were recorded at 50 K magnification in different areas in order to identify areas with best visibility of particles and best transparency of ice. All data were collected around these areas on previously unexposed zones. For starting the first structure determination, focal pairs of microphotographs were recorded (Supplementary Figure 7). The first close-to-focus microphotographs were taken at −2.5 to −4.5 μm underfocus. The second microphotographs were taken at −7.0 μm underfocus and were used for particle identification and selection purposes only. Once the particle selection and initial structure determination had been validated with the defocus pair data (6800 particles), further data were collected at a defocus of −2.0 up to −4.0 μm (19 938 particles from 19 micrographs with best power spectra from one single data collection session). Micrographs were digitized with a high-precision drum scanner (Heidelberger Druckmaschinen) resulting in a final step size of 1.0 Å per pixel (scanning step size 4.66 μm). The defocus values were estimated from the positioning of the contrast transfer function (CTF) rings from Fourier transforms of the scanned microphotographs by using the program CTFIT from the EMAN software package (Ludtke et al, 1999). To facilitate particle visibility and make the auto-boxing procedure more numerically stable, a smooth Gaussian real space filtering was applied, corresponding to a low-pass filter cutoff of 10 Å. The boxing of the initial data set has been done manually in BOXER (EMAN) from far-from-focus images and all coordinates have been transferred to the close-to-focus images. The subsequent data set has been boxed semi-automatically in BOXER with validation of all boxed particles by visual inspection. CTF correction was done by phase flipping using large particle boxes (512 pixels) using the CTFIT program of the EMAN suite. After particle selection and CTF correction, all boxed particles were coarsened twice resulting in a spatial resolution of 2.0 Å per pixel for further image processing. Data at 100 kV have been collected on the in-house FEI Polara electron microscope, which includes a field emission gun tunable between 80 and 300 kV and a more stable specimen holder system. CCD images were recorded at an underfocus of 2.0–3.0 μm and a magnification of 59K on a 4k × 4k Eagle CCD camera (FEI) after careful dark current and gain reference calibration (step size of 1.96 Å per pixel). About 2500 single-particle images were selected from 21 CCD images, CTF corrected (by examining the non-coherently averaged power spectra calculated from the selected particles) and processed as described above. The structure determination was done by angular reconstitution, as an ab-initio structure independent of the structure obtained from the 200-kV data set. The cross-correlation coefficient of the two structures is 0.87.
Three-dimensional reconstruction
The centering, determination of particles orientations and three-dimensional reconstruction has been done using the IMAGIC software package (van Heel et al, 1996). The initial reconstruction was obtained from 6800 particles using the common line approach applied to the results of the two-dimensional classification of particles centered to the total sum of the particle images. The initial class averages contained ∼25 particles per class. The original reconstruction was refined up to ∼20 Å resolution and validated by comparison of the re-projections with the original particle images (Figure 1; Supplementary Figure 10). Further refinements of the three-dimensional reconstruction were done using 19 938 particles collected without defocus pairs. According to the progress of the refinement procedure, the amount of particles per class was gradually reduced from 25 particles per class for the asymmetrical C1 start up procedure and angular reconstitution, to 5 particles per class for the final refinement steps. The angular distribution of particle views is rather homogeneous (Supplementary Figure 13). The resolution of the final three-dimensional structure was estimated with the Fourier-Shell Correlation (Saxton and Baumeister, 1982) according to the 0.5, 0.14 (Rosenthal and Henderson, 2003) and half-bit (van Heel and Schatz, 2005) criteria to 12.3/9.1/9.0 Å consistent with the features of the map (Supplementary Figure S2; e.g., the α-helix H9/H10 bundle at the LBD interface, the DBDs and the hinge region of RXR (Figures 1 and 2) can be resolved; the helical character of the DNA cannot be resolved because it is protein bound, that is, the DBDs and the VDR A/B domain fill the major groove of the DNA).
Map interpretation
The interpretation of the 3D density map has been done by fitting existing crystal structures taking into account the connectivity between the two major parts of the complex. For this, we built a model of the RXR/VDR LBD based on the RXR/RAR LDB crystal structure (PDB ID 1DKF; Bourguet et al, 2000) in which the RAR part was replaced by the VDR LBD crystal structure (PDB ID 1DB1; Rochel et al, 2000). For the DR3 RXR/VDR DBD complex, we built a model derived from the VDR part of a DNA-bound 5′-VDR/RXR-3′ DBD crystal structure (PDB ID 1YNW; Shaffer and Gewirth, 2004) and the DR4 RXR/TR DBD (PDB ID 2NLL; Rastinejad et al, 1995) adjusted to a DR3 response element by rotating and translating one DBD accordingly. The derived models were fitted into the density map using the PyMOL software (http://pymol.sourceforge.net) allowing to unambiguously annotate the RXR and VDR parts and assign the handedness of the complex. The quantitative estimation of the fitting has been done in IMAGIC by computation of the cross-correlations between the 3D density map and electron densities simulated from the fitted X-ray structures (Supplementary Figures S4 and S12). The fine scaling/magnification calibration of the map has been done according to the fitted crystal structure.
Supplementary Material
Acknowledgments
This work was supported by the Agence National pour la Recherche (ANR), the Fondation pour la Recherche Médicale (FRM), the Centre National pour la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the European Molecular Biology Organization (EMBO) Young Investigator Programme (YIP), the Institut du Développement et des Ressources en Informatique Scientifique (IDRIS, France), the European Commission as SPINE2-complexes (contract n_ LSHG-CT-2006-031220) and the IGBMC facilities. IO was supported by a PhD fellowship from the Alsace Region. The electron microscope facility is supported by the Alsace Region, FRM, INSERM, CNRS and the Association pour la Recherche sur le Cancer (ARC). The electron density map of the RXR/VDR/DR3 complex has been deposited to the EM Data bank with the accession number EMD-1985.
Author contributions: IO, NR and BPK performed the experiments and IO, NR, DM and BPK designed the experiments, analysed data, and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alimirah F, Vaishnav A, McCormick M, Echchgadda I, Chatterjee B, Mehta RG, Peng X (2010) Functionality of unliganded VDR in breast cancer cells: repressive action on CYP24 basal transcription. Mol Cell Biochem 342: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony P, Sigüeiro R, Huet T, Sato Y, Ramalanjaona N, Rodrigues LC, Mouriño A, Moras D, Rochel N. (2010) Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3. J Med Chem 53: 1159–1171 [DOI] [PubMed] [Google Scholar]
- Banerjee P, Chatterjee M (2003) Antiproliferative role of vitamin D and its analogs - a brief overview. Mol Cell Biochem 253: 247–254 [DOI] [PubMed] [Google Scholar]
- Bouillon R, Eelen G, Verlinden L, Mathieu C, Carmeliet G, Verstuyf A. (2006) Vitamin D and cancer. J Steroid Biochem Mol Biol 102: 156–162 [DOI] [PubMed] [Google Scholar]
- Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D (2000) Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell 5: 289–298 [DOI] [PubMed] [Google Scholar]
- Brelivet Y, Kammerer S, Rochel N, Poch O, Moras D. (2004) Signature of the oligomeric behaviour of nuclear receptors at the sequence and structural level. EMBO Rep 5: 423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. (2008) Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature 456: 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheskis B, Freedman LP (1996) Modulation of nuclear receptor interactions by ligands: kinetic analysis using surface plasmon resonance. Biochemistry 12: 3309–3318 [DOI] [PubMed] [Google Scholar]
- Delacroix L, Moutier E, Altobelli G, Legras S, Poch O, Choukrallah MA, Bertin I, Jost B, Davidson I. (2010) Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol 30: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiévet C, Staels B. (2009) Efficacy of peroxisome proliferator-activated receptor agonists in diabetes and coronary artery disease. Curr Atheroscler Rep 11: 281–288 [DOI] [PubMed] [Google Scholar]
- Friedrich M, Axt-Fliedner R, Villena-Heinsen C, Tilgen W, Schmidt W, Reichrath J. (2002) Analysis of vitamin D-receptor (VDR) and retinoid X-receptor alpha in breast cancer. Histochem J 34: 35–40 [DOI] [PubMed] [Google Scholar]
- Gampe RT Jr, Montana VG, Lambert MH, Miller AB, Bledsoe RK, Milburn MV, Kliewer SA, Willson TM, Xu HE (2000) Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell 5: 545–555 [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14: 121–141 [PubMed] [Google Scholar]
- Gross C, Krishnan AV, Malloy PJ, Eccleshall TR, Zhao XY, Feldman D. (1998) The vitamin D receptor gene start codon polymorphism: a functional analysis of FokI variants. J Bone Miner Res 13: 1691–1699 [DOI] [PubMed] [Google Scholar]
- Holick MF (2003) Vitamin D deficiency: what a pain it is. Mayo Clin Proc 78: 1457–1459 [DOI] [PubMed] [Google Scholar]
- Hourai S, Rodrigues LC, Antony P, Reina-San-Martin B, Ciesielski F, Magnier BC, Schoonjans K, Mouriño A, Rochel N, Moras D. (2008) Structure-based design of a superagonist ligand for the vitamin D nuclear receptor. Chem Biol 15: 383–392 [DOI] [PubMed] [Google Scholar]
- Kakuda S, Okada K, Eguchi H, Takenouchi K, Hakamata W, Kurihara M, Takimoto-Kamimura M (2008) Structure of the ligand-binding domain of rat VDR in complex with the nonsecosteroidal vitamin D3 analogue YR301. Acta Crystallogr Sect F Struct Biol Cryst Commun 64(Pt 11): 970–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaholz BP, Myasnikov AG, van Heel M (2004) Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 427: 862–865 [DOI] [PubMed] [Google Scholar]
- Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld MG, Heyman RA, Glass CK (1994) Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature 371: 528–531 [DOI] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW (2006) The expanding cosmos of nuclear receptor coactivators. Cell 125: 411–414 [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM (1995) The RXR heterodimers and orphan receptors. Cell 83: 841–850 [DOI] [PubMed] [Google Scholar]
- Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP (2007) Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell 130: 1019–1031 [DOI] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O'Malley BW. (1999) Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20: 321–344 [DOI] [PubMed] [Google Scholar]
- Nakabayashi M, Yamada S, Yoshimoto N, Tanaka T, Igarashi M, Ikura T, Ito N, Makishima M, Tokiwa H, DeLuca HF, Shimizu M (2008) Crystal structures of rat vitamin D receptor bound to adamantyl vitamin D analogs: structural basis for vitamin D receptor antagonism and partial agonism. J Med Chem 51: 5320–5329 [DOI] [PubMed] [Google Scholar]
- Oñate SA, Tsai SY, Tsai MJ, O'Malley BW (1995) Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270: 1354–1357 [DOI] [PubMed] [Google Scholar]
- Pinette KV, Yee YK, Amegadzie BY, Nagpal S (2003) Vitamin D receptor as a drug discovery target. Mini Rev Med Chem 3: 193–204 [DOI] [PubMed] [Google Scholar]
- Rastinejad F, Perlmann T, Evans RM, Sigler PB (1995) Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature 375: 190–191 [DOI] [PubMed] [Google Scholar]
- Rastinejad F, Wagner T, Zhao Q, Khorasanizadeh S (2000) Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J 19: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochel N, Ciesielski F, Godet J, Moman E, Roessle M, Peluso-Iltis C, Moulin M, Haertlein M, Callow P, Mély Y, Svergun DI, Moras D (2011) Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol 18: 564–570 [DOI] [PubMed] [Google Scholar]
- Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D (2000) The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell 5: 173–179 [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R (2003) Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333: 721–745 [DOI] [PubMed] [Google Scholar]
- Sakamaki Y, Inaba Y, Yoshimoto N, Yamamoto K (2010) Potent antagonist for the vitamin D receptor: vitamin D analogues with simple side chain structure. J Med Chem 53: 5813–5826 [DOI] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W (1982) The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc 127: 127–138 [DOI] [PubMed] [Google Scholar]
- Shaffer PL, Donald P, McDonnell P, Gewirth DT (2005) Characterization of transcriptional activation and DNA-binding functions in the hinge region of the vitamin D receptor. Biochemistry 44: 2678–2685 [DOI] [PubMed] [Google Scholar]
- Shaffer PL, Gewirth DT (2002) Structural basis of VDR-DNA interactions on direct repeat response elements. EMBO J 21: 2242–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer PL, Gewirth DT (2004) Structural analysis of RXR-VDR interactions on DR3 DNA. J Steroid Biochem Mol Biol 89–90: 215–219 [DOI] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP (2008) Structure of the 30S translation initiation complex. Nature 455: 416–420 [DOI] [PubMed] [Google Scholar]
- Son YL, Lee YC (2010) Molecular determinants of the interactions between SRC-1 and LXR/RXR heterodimers. FEBS Lett 584: 3862–3866 [DOI] [PubMed] [Google Scholar]
- Sone T, Kerner S, Pike JW (1991) Vitamin D receptor interaction with specific DNA. Association as a 1,25-dihydroxyvitamin D3-modulated heterodimer. J Biol Chem 266: 23296–23305 [PubMed] [Google Scholar]
- Svensson S, Ostberg T, Jacobsson M, Norström C, Stefansson K, Hallén D, Johansson IC, Zachrisson K, Ogg D, Jendeberg L (2003) Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation. EMBO J 22: 4625–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM (1991) Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65: 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- van Heel M, Schatz M (2005) Fourier shell correlation threshold criteria. J Struct Biol 151: 250–262 [DOI] [PubMed] [Google Scholar]
- Zechel C, Shen XQ, Chambon P, Gronemeyer H (1994b) Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J 13: 1414–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechel C, Shen XQ, Chen JY, Chen ZP, Chambon P, Gronemeyer H (1994a) The dimerization interfaces formed between the DNA binding domains of RXR, RAR and TR determine the binding specificity and polarity of the full-length receptors to direct repeats. EMBO J 13: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang Y, Busby SA, Pascal BD, Garcia-Ordonez RD, Bruning JB, Istrate MA, Kojetin DJ, Dodge JA, Burris TP, Griffin PR (2011) DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol 18: 556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Chasse SA, Devarakonda S, Sierk ML, Ahvazi B, Rastinejad F (2000) Structural basis of RXR-DNA interactions. J Mol Biol 296: 509–520 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.