Abstract
Hybrid organisms may fail to develop, be sterile or they may be more vigorous than either of the parents. Examples of hybrid vigour or hybrid necrosis in the F1 are often not inherited stably in subsequent generations if they are associated with overdominance. There can also be transgressive phenotypes that are inherited stably in these later generations, but the underlying mechanisms are not well understood. Here we have investigated the possibility that stable transgressive phenotypes in the progeny of crosses between cultivated tomato (Solanum lycopersicum cv. M82) and a wild relative (Solanum pennellii, accession LA716) are associated with micro or small interfering(si) RNAs. We identified loci from which these small(s)RNAs were more abundant in hybrids than in either parent and we show that accumulation of such transgressive sRNAs correlated with suppression of the corresponding target genes. In one instance this effect was associated with hypermethylation of the corresponding genomic DNA. Our results illustrate a potential role of transgressive sRNAs in plant breeding and in natural evolution with wild plants.
Keywords: hybrid phenotypes, miRNA, RNA silencing, siRNA, tomato
Introduction
The phenotypes of hybrid plants and animals are primarily a consequence of genetic effects at one or more loci. With a single-gene trait the phenotype in the hybrid reflects the activity of the dominant allele, whereas, with more complex quantitative traits, the phenotype will be affected by the composite action of the multiple loci and will normally be intermediate between the parents.
However, there are exceptions in which hybrids have more extreme phenotypes than the parents. The best-characterised exceptions are in the F1 hybrids of plants and animals, in which there is enhanced vigour (heterosis) or hybrid necrosis. These effects may be due to: complementation by dominant genes from one parent of defective or missing alleles in the other; epistasis between unlinked loci or heterozygote advantage (Bomblies and Weigel, 2007; Lippman and Zamir, 2007; Birchler et al, 2010; Chen, 2010). These F1 hybrid phenotypes are lost in later generations because they are caused by alleles or gene combinations that are inherited in only a proportion of the progeny (Baack and Rieseberg, 2007).
There are also heritable phenotypes in hybrids and hybrid lineages that are extreme to those of either parent. The heritability of these phenotypes indicates that they are distinct from those involved in heterosis or hybrid necrosis (Rieseberg et al, 1999), and the term transgressive segregation is often used to describe the niche divergence and phenotypic novelty of these hybrid lineages: the phenotypes transgress the parental range. Many eukaryotes exhibit transgressive segregation, but it is more frequent in plants than animals (Rieseberg et al, 2003).
The proposed mechanisms of transgressive segregation involve complementation and epistasis as in heterosis. Complementation would occur if defective genes from one parent are compensated by their functional homologues from the other parent. Epistasis would involve genes from one parent that interact in trans with non-homologous loci from the opposite parent (Rieseberg et al, 1999). Transgressive segregation is often associated with chromosome rearrangements (Rieseberg et al, 1996; Shaked et al, 2001; Lai et al, 2005), transposable element mobilization (Liu and Wendel, 2000; Michalak, 2009), DNA methylation (Shaked et al, 2001; Salmon et al, 2005) and changes to gene expression. Presumably, these changes are secondary to the initiating events associated with complementation or epistasis.
Any epistatic interactions associated with transgressive phenotypes could be mediated by proteins, DNA or RNA in various combinations affecting protein complexes or gene expression (Caicedo et al, 2004; Werner et al, 2005; Rowe et al, 2008). A previously unexplored possibility is based on RNA silencing at the chromatin or posttranscriptional level (Baulcombe, 2004), in which small(s)RNAs from one parent find novel targets in the genome or transcriptome of the opposite parent. Such interactions would establish patterns of gene expression at either the transcriptional or posttranscriptional level that would be specific to the hybrids.
To investigate the possible involvement of RNA silencing in hybrid phenotypes, we used high-throughput sequencing to characterise sRNAs in young seedlings of S. lycopersicum cv. M82 (hereafter abbreviated as M82) and S. pennellii (accession LA716) and in introgression lines (ILs), in which individual defined regions of the S. pennellii genome are present in the background of the M82 genome (Supplementary Figure S1A; Eshed and Zamir, 1995; Lippman et al, 2007). These lines have been studied extensively for their habit, leaf dissection, fruit morphology/yield and metabolic traits (Holtan and Hake, 2003; Schauer et al, 2006; Semel et al, 2006; Lippman and Zamir, 2007; Lippman et al, 2007).
The rationale for our approach was based on the ability of sRNAs in plants to trigger secondary small interfering RNAs (siRNAs; Baulcombe, 2004). We hypothesised that primary sRNA from one parent could initiate secondary siRNA on a target RNA from the opposite parent through an RNA-based mechanism. The initiator primary sRNA would associate with an Argonaute protein and stimulate an RNA-dependent RNA polymerase (RDR) to convert a target RNA into a double-stranded (ds) form. The dsRNA would then be processed into 21-nucleotide (nt) secondary siRNAs by a Dicer-like (DCL) protein. In this mechanism, there would be silencing of multiple RNAs including the primary target RNAs and any others that are complementary to the secondary siRNAs (Baulcombe, 2004).
At other loci the primary sRNA may direct methylation of a targeted DNA locus in the opposite parent. The methylated DNA would then become the template for secondary siRNA production via a mechanism that involves Polymerase IV, RDR2 and DCL3 (Wierzbicki et al, 2008; Matzke et al, 2009; Law and Jacobsen, 2010). Unlike the other secondary sRNA pathway this process would involve 24-nt siRNAs, and the silencing mechanism could be transcriptional or posttranscriptional. However, as in the posttranscriptional pathway, there would be silencing of both primary and secondary target loci and transgressive expression of sRNA.
Here we describe transgressive effects at many different sRNA loci in the hybrid lines of tomato and we investigated three of them in more detail. We describe how the expression of sRNA at these loci in hybrids affects gene expression and how they could be causal of transgressive phenotypes. Two of these examples involve siRNA loci and they could be explained by RNA-mediated epistasis. However, a third example is not easily reconciled with RNA-mediated epistasis because it involves a micro (mi)RNA whose biogenesis does not involve secondary sRNAs. These examples illustrate how different sRNA-based mechanisms could be involved in transgressive segregation.
Results
To find out whether miRNAs and siRNAs are transgressively expressed, we cloned and sequenced sRNA from 2-week-old seedlings of parental lines (S. lycopersicum cv. M82 and S. pennellii, accession LA716), their F1 and F2 hybrids and ILs. The sRNA sequences in the resulting data sets are predominantly 21- and 24-nt in length (Supplementary Figure S1B and Materials and methods section) with a 5′-nucleotide bias of U (21-nt) or A (24-nt), respectively (Supplementary Figure S1C). This bias is likely due to selective binding by Argonaute proteins (Baulcombe, 2004; Mi et al, 2008). Among all samples, 24-nt sRNAs accumulate to higher levels than those of 21-nt sRNAs.
Small RNA-enriched genomic regions (loci) were identified using SiLoCo (Moxon et al, 2008). The number of reads at each locus was normalised to the number of total reads as an indicator of the expression level (Materials and methods section). A transgressive index was calculated based on a comparison of locus representation in the different sRNA data sets. It was calculated as the ratio of the normalised counts for a genomic locus, for an IL or hybrid compared with the mean of the normalised counts of the most similar parent.
Transgressive expression of miR395 associated with salt stress
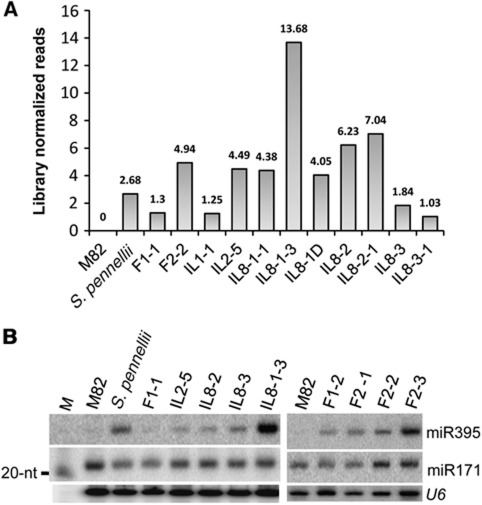
Among miRNAs the most striking transgressive effect was with miR395. This miRNA was abundant in S. pennellii but not in M82 seedlings (Figure 1A and B), and it was at a mid-parental level in the F1. However, in two of the three F2 samples and in a few of the ILs including IL8-1-3 (Figure 1A and B and Supplementary Figure S2A) this miR395 was hyper-abundant. This transgressive expression pattern was confirmed by northern blotting (Figure 1B and Supplementary Table SI).
Figure 1.
Transgressive expression of miR395. (A) Bar chart showing the abundance of miR395, normalised to the sizes of the sRNA data sets. Relative abundance was calculated using miRprof (UEA sRNA tool kit (Moxon et al, 2008)). (B) Accumulation of miR395 in parents and selected ILs. About 15 μg of total RNA from young seedlings were used for analysis of each sample. Total RNA was electrophoresed in 15% polyacrylamide gel, transferred to membrane and probed with corresponding DNA oligonucleotides (Supplementary Table SI) labelled with γ-32P-ATP. U6 serves as loading control. M, Size marker.
The miRNA 395 loci in the tomato genome are on chromosomes 2, 3 and 5, and not in the introgressed region of S. pennellii in IL8-1-3. The enhanced expression of miR395 is therefore likely to have been initiated by a component of the S. pennellii genome that acts either directly or indirectly on the M82 genome.
Expression of miR395 was induced by salt stress in S. pennellii (Supplementary Figure S3A), which is a naturally salt tolerant wild species (Frary et al, 2010). This miRNA is also induced by salt stress in tobacco (Frazier et al, 2011), maize (Ding et al, 2009) and poplar (Jia et al, 2009). In Arabidopsis, but not M82 or S. pennellii (Supplementary Figure S3B), miR395 is associated with sulphate stress (Jones-Rhoades and Bartel, 2004), and its target transcripts are ATP sulphurylase (APS; Supplementary Figure S3C) (Ding et al, 2009) and other mRNAs involved in both sulfur assimilation and allocation (Chiou, 2007; Kawashima et al, 2009).
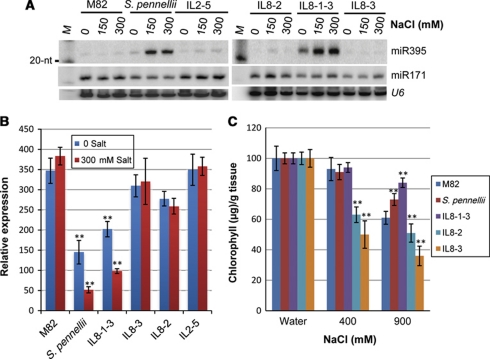
Similar to S. pennellii, miR395 was salt inducible in IL8-1-3, but not the other ILs tested (Figure 2A). The high-level expression of miR395 in S. pennellii and IL8-1-3 correlated with a low level of APS1 mRNA in the absence of salt. The APS1 mRNA level was further reduced in these two lines, but not in M82 or three other ILs under salt stress conditions (Figure 2B). There was a correspondingly greater tolerance to salt stress in S. pennellii and IL8-1-3 than in wild type, as indicated by reduced loss of chlorophyll in a leaf disc assay (Figure 2C) and in other assays including a measurement of leaf curling (Supplementary Figure S4). It is likely therefore that overexpression of miR395 in IL8-1-3 and S. pennellii contributes, at least in part, to their enhanced salt tolerance in comparison with M82. However, there are likely to be other determinants of salt tolerance because the miR395 target mRNA was at similar levels in IL8-1-3 and S. pennellii, although they differed in their salt tolerance. We also identified a second salt tolerant line (IL2-5) (Supplementary Figure S4A) in which miR395 levels were not elevated.
Figure 2.
miR395 expression contributes to a transgressive phenotype. (A) miR395 accumulation in salt treated (NaCl in mM) plants from parental lines (M82 and S. pennellii), IL8-1-3 and three other ILs. (B) Transcript analysis for APS1. RNA extraction and qRT–PCR was performed as described. GAPDH served as control. Data are mean±s.d. of three replicates. (C) Chlorophyll accumulation in salt-treated and control plants. Chlorophyll accumulation without salt treatment was normalised to 100%. Chlorophyll (μg/g fresh tissue weight) from three independent experiments (n>30) is shown. Data are mean±s.d. The double asterisks in B and C represent significance (one-way (B) or two-way (C) analysis of variance test (Holm-Sidak method; α=0.05)) at P<0.01.
Transgressive siRNA loci
When aligned with the available tomato genomic sequence (Materials and methods section), the M82 data set revealed 24 000 siRNA loci in which there were multiple matches to sRNA reads. The S. pennellii data set revealed 18 000 loci but, as the genome of this species is not available, the divergent sequence loci would not be identified and this number is an underestimate. More than 90% of the shared loci are represented at similar levels in the data sets from the two parental species, indicating that they are expressed at the same level in M82 and S. pennellii.
To identify differentially expressed siRNA loci, we restricted our analysis to regions of the genome with at least 100 reads in at least one of the data sets. We identified 153 transgressive loci that were expressed in F2 or IL lines at levels that are either higher than the high expressing parent or lower than the low expresser (Figure 3A and Supplementary Table SII). We used a stringent criterion of transgressivity and restricted our analysis to loci that were identified consistently using three different protocols. According to this criterion, there were no sRNA loci with transgressive expression in the F1 samples but, in the ILs and F2 plants, few siRNA loci were transgressively expressed (Supplementary Table SII). At present we cannot rule out that there are sRNA loci that are transgressively expressed in the F1. However, if such loci exist, their transgressive index would be substantially less than that with the loci identified here.
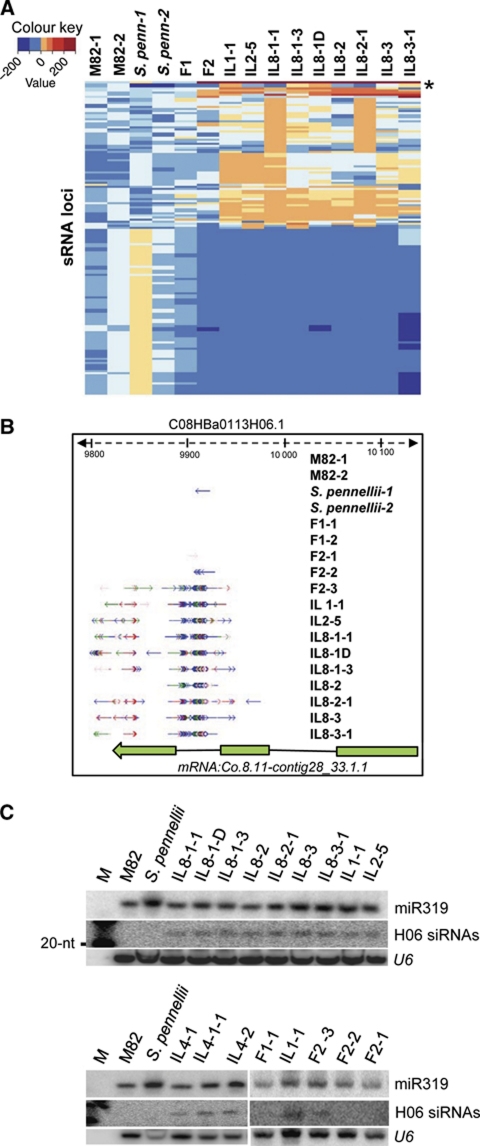
Figure 3.
Transgressive siRNA locus H06. (A) Heatmap showing changes in the level of sRNA from the most transgressive genomic loci in IL samples. The H06 locus is marked with *. Replicates of M82 (M82-1 and M82-2) and S. pennellii (S. penn-1 and S. penn-2) were used. Details of TI for each siRNA loci presented in the heatmap are given in Supplementary Table SII. (B) sRNAs from M82, S. pennellii, F1, F2 and introgression line samples aligned to the H06 genomic locus in M82 DNA. Arrows indicate direction of sRNA alignment, and the colour of the arrow indicates length: blue, 24-nt; red, 21-nt; and green, 22-nt. (C) Northern blot analysis of H06 siRNAs in parental lines, F1, F2 and in ILs in four different chromosomes. M, size marker. Oligonucleotide probe sequences (H06a and b) are given in Supplementary Table SI.
Two types of transgressive loci are present in these data sets: one type was less abundant in the ILs than in the parental lines (lower half, Figure 3A) and the second type was more abundant in ILs than in the parents (upper half, Figure 3A). The transgressive expression, depending on the locus, was evident either in a subset of ILs or in all of them.
Fewer than 1% of all loci were transgressive and they were distributed all over the genome similar to the total sRNA loci (Supplementary Figure S5A). Transgressive loci were also of similar length as any other sRNA loci (Supplementary Figure S5B). The transgressive loci overlap transposons, coding sequence and other genomic features (Supplementary Figure S6) but, with only 153 loci involved, we cannot determine whether the pattern is significantly different from total sRNA loci. The transgressive sRNAs were distinct from the total sRNAs in that they had a greater strand bias than the other sRNAs (Supplementary Figure S7A). They produce 21, 22 and 24-nt sRNAs, although the 24-nt sRNAs were more highly represented than in the total sRNA population (Supplementary Figure S7B). The relationship of these properties and mechanisms is not obvious, but they are consistent with the involvement of multiple biogenesis pathways in transgressive sRNA production.
A transgressive siRNA locus associated with hypermethylation of its target DNA
One of the 153 transgressive loci was overexpressed relative to the parents in all of the ILs (marked as * in Figure 3A). This locus is referred to as H06 and it overlaps a predicted protein coding gene on chromosome 8 (mRNA:Co.8.11-contig28_33.1.1; Figure 3B). There was only a single H06 siRNA read in the data sets from the S. pennellii parent and none from M82 tomato or the F1 hybrid, but there were hundreds of siRNA reads in each of nine IL data sets (Figure 3B and Supplementary Figure S8). Two F2 lines expressed H06 siRNAs, albeit at lower levels than the ILs (Figure 3B and Supplementary Figure S2B).
The activation of H06 siRNA could not be correlated with a particular region of S. pennellii DNA because the lines tested have introgressed DNA from four different chromosomes (1, 2, 8 and 4, Figure 3C). The initiation event was therefore likely to have been in a progenitor of the ILs. This event would have acted in trans to affect the H06 locus on chromosome 8 of the M82 genome. The transgressive F2 lines in this study (Figure 3B) would have recapitulated this initiation event.
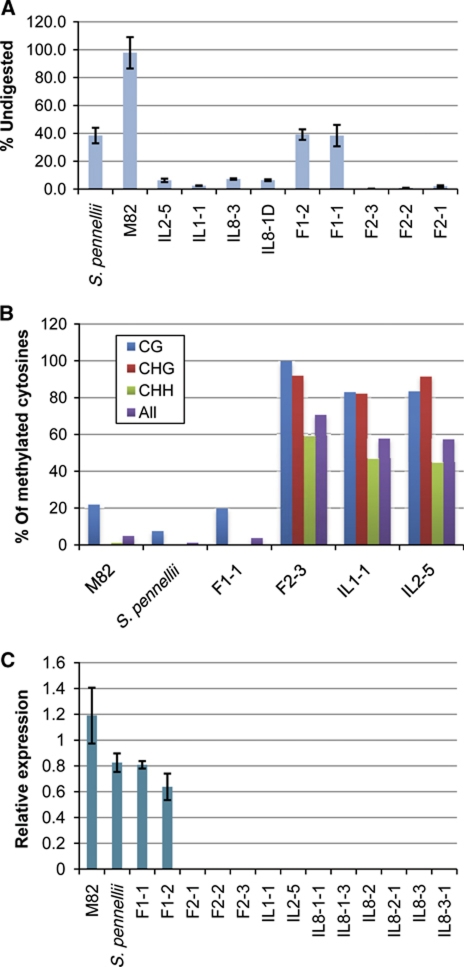
The H06 siRNAs were predominantly 24-nt in length and their overexpression in the ILs relative to the parental or F1 hybrid lines was confirmed by northern analysis (Figure 3B and C). The 24-nt size class of sRNA is associated with methylation of DNA with a similar sequence (Hamilton et al, 2002; Chan et al, 2004) and we therefore tested for methyl cytosine at the H06 locus using an McrBC methylation assay (Figure 4A). The H06 DNA from the parental lines and F1 was largely undigested in this assay indicating hypomethylation whereas, in the F2 and IL samples, the DNA was completely digested by McrBC indicative of hypermethylation. Using a sodium bisulphite analysis we confirmed that genomic DNA from parents and an F1 plant was largely unmethylated at this locus, but it was densely methylated in the F2-3 line and the two ILs that were analysed (Figure 4B and Supplementary Figure S9) at CG, CHG and CHH contexts.
Figure 4.
DNA methylation and expression analysis of H06 locus. (A) Methylation of H06 genomic DNA using restriction enzyme McrBC. Completely digested DNA indicates hypermethylation. Error bars indicate the 95% confidence intervals of three replicates. The assay was performed as described in Materials and methods section. (B) Sodium bisulphite analysis of H06 locus. Each sample had at least 10 clones analysed for cytosine methylation in replicate experiments. (C) Expression analysis of H06 mRNA using quantitative RT–PCR.
The H06 mRNA fragment detected by quantitative RT–PCR was less abundant in the ILs than in the parents or their F1 hybrid (Figure 4C), indicating that epigenetic modification of H06 DNA, siRNA production and silencing of H06 mRNA are interrelated phenomena.
Transgressive siRNA from the PAL loci are not associated with DNA methylation
In the absence of a fully sequenced and annotated tomato genome, we also searched for transgressive siRNA loci by aligning the siRNA sequence reads with expressed sequence tag cDNAs (ESTs). The transgressive expression of cDNA-specific siRNAs was more pronounced in ILs than in the F1 and F2 loci (Figure 5A and Supplementary Tables SIII and SIV).
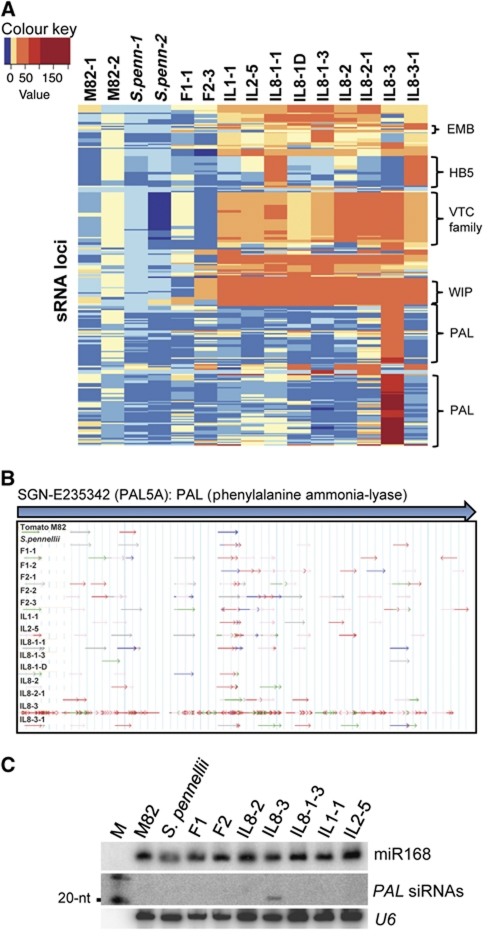
Figure 5.
Transgressive PAL siRNA accumulate in IL8-3. (A) Heatmap showing changes in the level of sRNA from the most transgressive cDNA loci in the IL samples. The major families of transgressive siRNAs correspond to the following genetic loci: PAL, phenylalanine ammonia–lyase; EMB, Embryo defective; HB5,Homeobox protein family; VTC, ‘Vitamin C deficient’ family; WIP, Wound inducible protein family. Transgressive siRNA loci are listed in Supplementary Table SIII. (B) sRNAs aligning to the PAL locus in M82, S. pennellii, F1, F2 and ILs. Arrows indicate direction of sRNA alignment, and the colour of the arrow indicates length: blue, 24-nt; red, 21-nt; and green, 22-nt. (C) Northern blot analysis of small RNAs hybridising to a PAL-specific probe.
The strongest transgressive effect of this type was in IL8-3, in which siRNAs corresponding to phenylalanine ammonia-lyase (PAL) mRNAs were increased by more than 100-fold relative to the parents or other ILs (Figure 5A and B and Supplementary Figure S2C). The PAL siRNAs in IL8-3 were predominantly 21-nt and their overexpression in IL8-3 was confirmed by northern analysis (Figure 5C). PAL siRNAs mostly aligned to the exons and matched predominantly to sense strands in the parents and most of the ILs. However, the transgressive siRNAs in IL8-3 matched to both sense and antisense strands (Supplementary Figure S10), indicating that they originated from a dsRNA molecule.
The PAL-derived sRNAs in IL8-3 matched numerous PAL ESTs (Supplementary Table SIII and SIV). A region in chromosome 9 (CO9SLe0130H12.3) has four PAL gene sequences (named as PAL5A, PAL5B, PAL5C and PAL5D) configured as a divergent inverted repeat (Supplementary Figure S10). These genes are 90–99% identical and also very similar (93–99%) to a PAL5 gene, previously identified from tomato that is expressed abundantly (Chang et al, 2008) (Supplementary Figure S11). There were unique siRNA sequence reads corresponding to each of the four PAL5 genes (Supplementary Table SV), indicating that the corresponding mRNAs are all templates for siRNA production. PAL5B had the highest proportion of unique sequence matches and it is likely to be the most abundant source of siRNAs.
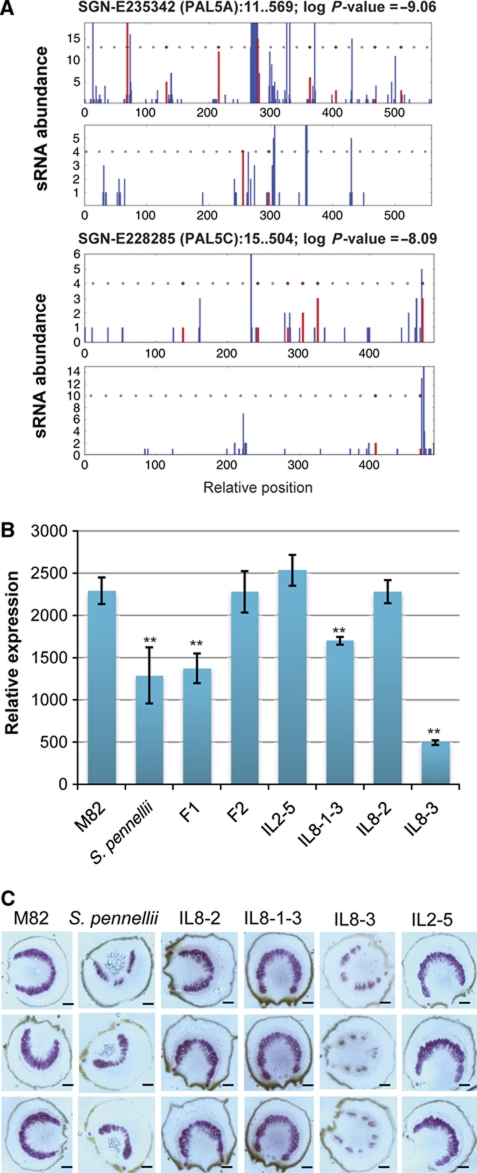
The alignment of PAL siRNAs with the PAL sequences was phased (Figure 6A) in a manner that is characteristic of trans-acting si (tasi)RNA in Arabidopsis and other plants (Peragine et al, 2004; Vazquez et al, 2004). An initiation event associated with these transgressive PAL siRNAs would have occurred in a progenitor of IL8-3 and it would have acted in trans to affect siRNA production at this chromosome 9 locus. A plausible scenario is that the initiator is an sRNA that is functionally equivalent to the miRNAs in tasiRNA biogenesis.
Figure 6.
PAL silencing results in reduced lignification. (A) Phasing of PAL-derived siRNAs along the PAL sequences. Two PAL ESTs with significant P-values (PAL5A, log P value=−9.06; PAL5C, log P-value=−8.09) for phasing are shown. Red bars indicate the start position of sRNAs in phase; and blue, the start position for those out of phase. Rectangles indicate expected phased positions and those in red indicate phased sequences. (B) PAL mRNA expression of parental and hybrid lines. See Materials and methods section for details. Data are mean±s.d. of three biological replicates. The double asterisks represent significance determined by one-way ANOVA at P<0.01 (α=0.05). (C) Phloroglucinol staining for total lignin in parental lines, IL8-3 and three other ILs. Violet stain indicates the presence and distribution of lignin. The micrographs are of sections derived from petioles of seedlings with staining concentrated in the vascular bundles. Size bar, 100 μM.
The level of PAL mRNA was assessed by RT–PCR using primers that would have detected all transcripts in the PAL5. It corresponded to the level of siRNAs in that it was lower in the IL8-3 line than in the parents, F1 or F2 hybrids or other ILs (Figure 6B). PAL initiates the phenylpropanoid pathway (Jones, 1984) that is associated with lignification of xylem vessels and, in line with the mRNA levels, the total lignin was lower in IL8-3 (and confined to fewer xylem vessels) than in parents or the other ILs tested (Figure 6C).
Discussion
Our analysis of tomato hybrids was prompted by the hypothesis that transgressive segregation would be affected by epistatic interactions between siRNAs and their targets from the opposite parent. Such interactions would be specific to the hybrids and the effect would persist in lines that inherit both interacting loci. In the most straightforward iteration of this hypothesis, the transgressive expression of sRNA would be fixed in lines in which the loci are both homozygous. The transgressive H06 and PAL siRNA loci are partly consistent with this hypothesis, because they are located outside the introgressed region of S. pennellii DNA, and their overexpression in the hybrids could have been induced by an epistatic interaction, involving primary sRNAs from S. pennellii DNA interacting with targets in the tomato genome.
However, simple epistasis can be ruled out because the transgressive expression of these loci was absent in the F1 when all the pairs of putatively interacting loci would have been present. It seems that there must be secondary effects following the initiating interaction. A formal possibility that the primary events involve mutation in male or female gametophytes seems unlikely, at least for H06, because the transgressive effect could be recapitulated in newly created F2 lines (Figures 1 and 3, and Supplementary Figures S2, S8). Genetic rearrangements are presumed to be rare and irregular and would be difficult to recreate.
An alternative explanation for H06 invokes epigenetic marks at H06 that are induced in the F1. These marks would then trigger sRNA expression in the F2 and in subsequent generations that inherit the epigenetically modified H06 DNA. To account for the absence of an effect in the F1, we propose that the epigenetic effects were initiated by interactions occurring during gametogenesis of the F1 and that they were subsequently reinforced by RNA-directed DNA methylation.
Consistent with that idea, the transgressive effect of H06 was associated with dense methylation of H06 DNA in CHH as well as CG and CHG (Figure 4A and B; Chan et al, 2004; Teixeira and Colot, 2010). The involvement of the 24-nt class of siRNA (Figure 3B and C) that is associated with RNA-directed DNA methylation and histone modification (Chan et al, 2004; Xie et al, 2004; Matzke and Birchler, 2005) is also consistent with RNA-directed mechanism at H06. An epigenetic effect could also explain the progressive increase of H06 over several generations that is implied by the comparison of F1, F2 and IL lines. Epigenetic effects at various transgene and endogenous siRNA loci similarly increase through several generations (Shiu et al, 2001; Volpe et al, 2002; Teixeira et al, 2009).
A clue as to the likely mechanism at the PAL locus is from the similarity of PAL siRNAs in IL8-3 to endogenous ta-siRNAs (Peragine et al, 2004; Vazquez et al, 2004). Both are predominantly 21-nt, derived from both strands of a dsRNA precursor, and their alignment with their target nucleotide sequence is phased (Figure 6A). In addition, the target DNA sequences of PAL siRNAs are not methylated in IL8-3, suggesting a posttranscriptional mechanism (Supplementary Figure S12).
The production of ta-siRNAs is initiated by miRNAs that target a non-coding ta-siRNA precursor (Allen et al, 2005). By analogy, in IL8-3, it is likely that there is an initiator sRNA of PAL siRNA biogenesis and that the PAL mRNA is equivalent to the ta-siRNA precursor. Presumably there was a genetic or an epigenetic change in one of the precursors of IL8-3 that activated production of this sRNA initiator of PAL silencing.
The transgressive expression of miR395 in IL8-1-3 is unlikely to be due to RNA-mediated epistasis, because miRNA biogenesis is not known to be initiated by sRNA interactions. An alternative explanation invokes a suppressor of miR395 encoded in the DNA of the IL8-1-3 introgression. The overexpression of miR395 in S. pennellii could be explained if the weak S. pennellii repressor is less effective with the tomato rather than the S. pennellii versions of miR395.
Similar transgressive expression of miRNAs and siRNAs has also been noted in natural and resynthesised Arabidopsis allopolyploids (Ha et al, 2009) and in Arabidopsis intraspecific F1 hybrids (Groszmann et al, 2011). However, these effects in Arabidopsis were evident in the F1, their heritability has not been tested and until now there has been no link with expression of protein coding genes. It remains possible that there are different mechanisms operating in tomato and Arabidopsis or, alternatively, that the more extensive genome coverage in Arabidopsis gives more sensitivity to the detection of transgressive sRNA loci.
Transgressive phenotypes have been frequently reported in tomato hybrids (Rick & Harrison, 1959; Devicente & Tanksley, 1993; Holtan and Hake, 2003), and their occurrence implies that phenotype of tomato parents is a poor predictor of their hidden potential to improve complex traits including that of yield (Tanksley & McCouch, 1997). On the basis of the findings reported here, we propose that miRNA and siRNAs that accumulate transgressively are manifestations of the hidden potential of parents that is released when hybrids are made. As with the examples described here these sRNAs would target protein coding genes affecting growth and responses to abiotic stimuli, and they would account, at least in part, for the transgressive phenotypes in hybrids. Further understanding of this mechanism should allow more efficient exploitation of transgressive segregation in plant breeding. It may also influence our understanding of variation and evolution in natural populations of outbreeding plants and animals.
Materials and methods
Plant growth
Tomato plants were raised from seeds in compost (Levington M3) and maintained in a growth room at 23°C with 16-h light and 8-h dark periods with 60% relative humidity and at light intensity of 300 μmol/m2/s. Two-week-old seedlings taken for analysis generally had two simple leaves and one compound leaf and a leaf bud.
Salt stress experiments were performed as outlined earlier (Verslues et al, 2006). Leaf disc assays and chlorophyll measurements were performed as described previously (Singla-Pareek et al, 2003).
RNA analysis and cloning
Total RNA was extracted from 500-mg plant tissue (2-week-old seedlings), grounded into fine powder in liquid nitrogen using TRIzol reagent (Invitrogen), according to the manufacturer's protocol. About 20- or 30-μg total RNA were resuspended in 10-μl loading buffer (0.10% bromophenol blue, 0.10% xylene cyanol in 100% de-ionised formamide), heated at 95°C for 1 min and loaded on 15% polyacrylamide denaturing gel (a 19:1 ratio of acrylamide to bisacrylamide, 8 M urea). The gel was run using electrophoresis apparatus (Bio-Rad) at 350 V for 4 h and then RNAs were transferred to a Hybond N+ membrane by electroblotting in 1 × TBE buffer at 10 V overnight. The hybridisation was performed at 35°C for 18–24 h in UltraHyb-oligo buffer (Ambion) using as a probe, short DNA oligos (Supplementary Table SI) end-labelled with 32P by polynucleotide kinase (New England Biolabs) and purified through MicroSpin™ G-25 columns (GE Healthcare), according to the manufacturers’ recommendations. The blot was washed twice with 2 × SSC, 0.5% SDS for 30 min at 35°C. The signal was detected after exposure to a phosphorimager screen using a Molecular Imager (Bio-Rad). For repeated hybridisation, the membrane was stripped with 0.5 × SSC, 0.5% SDS for 30 min at 80°C and then with 0.1 × SSC, 0.5% SDS for 30 min at 80°C.
Small RNA cloning was performed using an Illumina sRNA cloning kit. About 20 μg of total RNA was separated on a 15% 1 × TBE polyacrylamide gel, and small RNA region between 15 and 40-nt were excised and purified. This small RNA fraction was used for ligation of adapters and amplification of fragments. Sequencing was performed at Cancer Research UK, Cambridge.
Bioinformatics
Reference DNA sequence. The small RNA libraries were aligned to the tomato genome sequence (Mueller et al, 2005, 2009) downloaded from The International Tomato Genome Sequencing Consortium (http://solgenomics.net/), release bacs v340. This version was used in order to be compatible with small RNA locus definition tool ‘SiLoCo’ (see SiLoCo section).
Alignment. The small RNA high-throughput sequencing libraries were aligned to the reference sequence using the PatMaN (Prufer et al, 2008) alignment program. Only reads with 100% match to the genome were used in further analysis.
SiLoCo: defining the genomic small RNA loci. A number of pairwise comparisons were performed between parental and hybrid small RNA high-throughput sequencing libraries, using the University of East Anglia small RNA toolkit tool ‘SiLoCo’ (http://srna-tools.cmp.uea.ac.uk/plant). ‘SiLoCo’ generates ‘loci’ defined as regions of the genome, which produce overlapping small RNAs (Moxon et al, 2008). About 16 sets of loci were generated, one for each comparison and all loci from all comparisons were combined into a final set of loci; each final locus was defined by the minimum and maximum coordinates of loci from any of the subsets that overlapped.
Annotation reference data. The EST data were obtained from the SOL genomics FTP site (ftp://ftp.solgenomics.net), and the Unigene data mapping these ESTs to genes were obtained from ftp://ftp.sgn.cornell.edu/unigene builds/, using version Tomato unigene build 20070816.
Differential expression: baySeq, edger and DESeq. The bioconductor (Gentleman et al, 2004) package ‘baySeq’, version 1.3.4. (Hardcastle and Kelly, 2010) was used to test for differential expression between parental and hybrid samples at the loci defined by combining the SiLoCo loci. The small RNA counts for two parental replicates were compared in turn against the counts from each of the loci in each of the introgressed lines. Loci that were significantly different from both pairs of parents and that had ‘transgressive’ counts, that is, a count above or below both of the parental means, were selected for further study.
Two other bioconductor packages were also used to test differential expression and look for transgressive loci. These were edgeR and DESeq (Anders and Wolfgang, 2010). Only those loci identified as having differential expression by all three methods (baySeq, edger and DEseq) were used for further analysis.
Calculating and displaying the transgressive index. The transgressive index for the selected differentially expressing loci was calculated as the ratio of the normalised counts for a genomic locus, for an IL compared with the mean of the normalised counts of the most similar parent. Counts of zero were adjusted to 1 (to avoid infinite fold changes). When the count of the introgressed line was below that of nearest parental mean, the negative reciprocal was used in order to present the data as a negative-fold change. These fold changes were plotted as a heatmap using the R heatmap.2 function from the R ‘gplots’ library, using the default hclust hierarchical clustering algorithm, to cluster the rows.
Strand bias. The strand bias for each sRNA locus was calculated by dividing the number of sRNAs matching the most abundant strand to the opposite strand. To avoid extreme values due to the presence of very low abundant loci, only those loci with more than 50 counts matching to them were considered and counts of zero were adjusted to 1 to avoid infinite changes.
Annotation of sRNA loci. The bacs v340 sequence was mapped against the SL2.40 genomic sequence release using ‘blast 2.2.25’ (Camacho et al, 2009). All the results with the highest e-value and at least 75% of the alignment covering the original sequence were retrieved. To account for sequences matching more than one place in the genome, only the matches on the same chromosome where bacs aligned were retained. The redefined loci were then compared against the annotation from the ITAG2.3 annotation (ftp://ftp.solgenomics.net). To search for enrichment in the overlap between transgressive loci and each individual genomic feature when compared with the total loci, a Fisher test was performed using the function ‘fisher.test’ from the R statistical package.
Calculating phasing scores. The identification of phased segments in the genomes was performed by calculating the log P-value from the hypergeometric distribution, adapting the method used previously (Chen et al, 2007). The genome was divided into segments with each segment containing sRNAs that distribute within 42-nt from each other. The analysis was performed using one size-class at a time and all positions were considered to be possible start and end coordinates. The test was performed at all levels of abundance for each sRNA and only the lowest log P-value was reported. To estimate the level of false positives, the same method was applied to small RNA data sets from published data sets of Arabidopsis Col-0 dcl-2,3,4. For a log P-value smaller than −8.03, the false-positive rate was estimated to be smaller than 0.005.
RT–PCR and quantitative (q)PCR
cDNA synthesis was performed as per the manufacturer's protocol using random hexamers or oligo dT (Superscript II or III, Invitrogen). Quantitative PCR was performed using CFX96™ Real-Time System (Bio-Rad) using SYBR Green JumpStart Taq ReadyMix (Sigma Aldrich). Data were analysed as described (Schmittgen and Livak, 2008). Details of primers are given in Supplementary Methods.
DNA methylation assay and bisulphite sequencing
DNA was extracted using the CTAB method (Rogers and Bendich, 1985). The concentration of the purified DNA was estimated using a Nanodrop spectrophotometer. About 500 ng of DNA at 20 ng/μl was incubated with 20 U McrBC (New England Biolabs) or an equivalent volume of 50% glycerol for 2 h at 37°C followed by heat inactivation at 80°C for 20 min. Target regions were amplified by qPCR from 40-ng digested DNA using SYBR Green JumpStart Taq ReadyMix (Sigma S9194), using primers described in Supplementary Methods.
Total DNA (500 ng) from parents and hybrids was treated with sodium bisulphite using EZ-DNA methylation gold kit (Zymo Research) as per manufacturer's instructions. DNA was amplified using primer pair (5′-AAAYGAATTTAAAAGGAGAAGTATGAAATGATATATAGATTTGGG-3′ and 5′-CACRATTCATCTTAACTATATCATTRCRAATCTAATAAAAAAACATA-3′) and Go Taq DNA polymerase (Promega). Primers were designed as described previously (Henderson et al, 2010). Cycling parameters used are as follows: denaturation at 96°C 3 min, followed by 40 cycles of 96°C for 30 s, 50°C for 30 s, 62°C for 2 min and a final extension at 65°C for 2 min. PCR products were gel extracted (Qiagen) and cloned into pGEM-Teasy (Promega). Clones were sequenced using BigDye 3.1 (Applied Biosystems). Methylation analysis at various cytosine contexts were performed using online tools, Cymate (Hetzl et al, 2007) and Kismeth (Gruntman et al, 2008), with at least 10 individual clones per sample in replicate experiments.
Lignin staining
Phloroglucinol (Sigma-Aldrich) staining for total lignin was performed as described (Rohde et al, 2004). Photographs were taken under brightfield using a Leica DFC310 FX camera attached to Leica M165FC dissection microscope.
Supplementary Material
Acknowledgments
We thank Dani Zamir and Tomato Genetics Resources Centre, UC Davis for providing various tomato lines; Tamas Dalmay for discussions; and Jill Harrison, Attila Molnar and Donna Bond for comments on the manuscript. We thank the Genomics and Bioinformatics Core facilities (CRUK Cambridge Research Institute) for Illumina sequencing. We thank Solanaceous Genomics Network and the International Tomato Genome Sequencing Consortium for sequences. We thank K.A. Kelly and Kim Rutherford for processing the small RNA data and setting up genome browsers, and Jill Harrison for helping with phylogenetic analysis. We also thank Shuoya Tang for plant care. This work was supported by BBSRC (grant number BB/E006981/2), the Gatsby Charitable Foundation and a European Research Council Advanced Investigator grant 233325 REVOLUTION. DCB is a Royal Society Research Professor. RNA sequencing raw data are available from Gene Expression Omnibus GSE23562 (GSM577998 to GSM578015).
Author contributions: PVS and DCB designed the study and drafted the manuscript, PVS carried out the RNA and phenotypic analyses, RMD and BACMS carried out bioinformatic analysis and AB carried out DNA methylation analysis.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during Trans-Acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Anders S, Wolfgang H (2010) Differential expression analysis for sequence count data. Nat Precedings, http://hdl.handle.net/10101/npre.2010.4282.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baack EJ, Rieseberg LH (2007) A genomic view of introgression and hybrid speciation. Curr Opin Genet Dev 17: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA (2010) Heterosis. Plant Cell 22: 2105–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Weigel D (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8: 382–393 [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD (2004) Epistatic interaction between arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA 101: 15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST plus: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW-L, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE (2004) RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Chang A, Lim MH, Lee SW, Robb EJ, Nazar RN (2008) Tomato phenylalanine ammonia-lyase gene family, highly redundant but strongly underutilized. J Biol Chem 283: 33591–33601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Li YH, Wu SH (2007) Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA 104: 3318–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15: 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ (2007) The role of microRNAs in sensing nutrient stress. Plant Cell Environ 30: 323–332 [DOI] [PubMed] [Google Scholar]
- Devicente MC, Tanksley SD (1993) Qtl analysis of transgressive segregation in an interspecific tomato cross. Genetics 134: 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Zhang LF, Wang H, Liu ZJ, Zhang ZX, Zheng YL (2009) Differential expression of miRNAs in response to salt stress in maize roots. Ann Botany 103: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Gol D, Keles D, Okmen B, Pinar H, Sigva HO, Yemenicioglu A, Doganlar S (2010) Salt tolerance in Solanum pennellii: antioxidant response and related QTL. BMC Plant Biol 10: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier TP, Sun G, Burklew CE, Zhang B (2011) Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol Biotechnol 49: 159–165 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES (2011) Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA 108: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E, Qi YJ, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. Bmc Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen XM, Wang XJ, Chen ZJ (2009) Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA 106: 17835–17840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Voinnet O, Chappell L, Baulcombe DC (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle TJ, Kelly KA (2010) BaySeq: empirical bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Chan SR, Cao X, Johnson L, Jacobsen SE (2010) Accurate sodium bisulfite sequencing in plants. Epigenetics 5: 47–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O (2007) CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 51: 526–536 [DOI] [PubMed] [Google Scholar]
- Holtan HE, Hake S (2003) Quantitative trait locus analysis of leaf dissection in tomato using Lycopersicon pennellii segmental introgression lines. Genetics 165: 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia XY, Wang WX, Ren LG, Chen QJ, Mendu V, Willcut B, Dinkins R, Tang XQ, Tang GL (2009) Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol Biol 71: 51–59 [DOI] [PubMed] [Google Scholar]
- Jones DH (1984) Phenylalanine ammonia-lyase—regulation of its induction, and its role in plant development. Phytochemistry 23: 1349–1359 [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T (2009) Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J 57: 313–321 [DOI] [PubMed] [Google Scholar]
- Lai Z, Nakazato T, Salmaso M, Burke JM, Tang SX, Knapp SJ, Rieseberg LH (2005) Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics 171: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Semel Y, Zamir D (2007) An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr Opin Genet Dev 17: 545–552 [DOI] [PubMed] [Google Scholar]
- Lippman ZB, Zamir D (2007) Heterosis: revisiting the magic. Trends Genet 23: 60–66 [DOI] [PubMed] [Google Scholar]
- Liu B, Wendel JF (2000) Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43: 874–880 [PubMed] [Google Scholar]
- Matzke M, Kanno T, Claxinger L, Huettel B, Matzke AJM (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Mi SJ, Cai T, Hu YG, Chen Y, Hodges E, Ni FR, Wu L, Li S, Zhou H, Long CZ, Chen S, Hannon GJ, Qi YJ (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5‘ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P (2009) Epigenetic, transposon and small RNA determinants of hybrid dysfunctions. Heredity 102: 45–50 [DOI] [PubMed] [Google Scholar]
- Moxon S, Schwach F, Dalmay T, MacLean D, Studholme DJ, Moulton V (2008) A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24: 2252–2253 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Lankhorst RK, Tanksley SD, Giovannoni JJ, White R, Vrebalov J, Fei Z, van Eck J, Buels R, Mills AA, Menda N, Tecle IY, Bombarely A, Stack S, Royer SM, Chang S-B, Shearer LA, Kim BD, Jo S-H, Hur C-G et al. (2009) A snapshot of the emerging tomato genome sequence. The Plant Genome 2: 78–92 [Google Scholar]
- Mueller LA, Tanksley SD, Giovannoni JJ, van Eck J, Stack S, Choi D, Kim BD, Chen MS, Cheng ZK, Li CY, Ling HQ, Xue YB, Seymour G, Bishop G, Bryan G, Sharma R, Khurana J, Tyagi A, Chattopadhyay D, Singh NK et al. (2005) The tomato sequencing project, the first cornerstone of the International Solanaceae Project (SOL). Comparative and Functional Genomics 6: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Gen Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, Stenzel U, Dannemann M, Green RE, Lachmann M, Kelso J (2008) PatMaN: rapid alignment of short sequences to large databases. Bioinformatics 24: 1530–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick CM, Harrison AL (1959) Inheritance of five new tomato seedling characters. J Hered 50: 91–98 [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity 83: 363–372 [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Sinervo B, Linder CR, Ungerer MC, Arias DM (1996) Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272: 741–745 [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Widmer A, Arntz AM, Burke JM (2003) The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos Trans R Soc Lond B Biol Sci 358: 1141–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant-tissues. Plant Mol Biol 5: 69–76 [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, Van Driessche G, Van Beeumen J, Messens E, Boerjan W (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ (2008) Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon A, Ainouche ML, Wendel JF (2005) Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol Ecol 14: 1163–1175 [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, Willmitzer L, Zamir D, Fernie AR (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C-T method. Nat Protocols 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N, Pleban T, Lippman Z, Gur A, Zamir D (2006) Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci USA 103: 12981–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu PKT, Raju NB, Zickler D, Metzenberg RL (2001) Meiotic silencing by unpaired DNA. Cell 107: 905–916 [DOI] [PubMed] [Google Scholar]
- Singla-Pareek SL, Reddy MK, Sopory SK (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci USA 100: 14672–14677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277: 1063–1066 [DOI] [PubMed] [Google Scholar]
- Teixeira FK, Colot V (2010) Repeat elements and the Arabidopsis DNA methylation landscape. Heredity 105: 14–23 [DOI] [PubMed] [Google Scholar]
- Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, Voinnet O, Wincker P, Esteller M, Colot V (2009) A role for RNAi in the selective correction of DNA methylation defects. Science 323: 1600–1604 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert J-L, Bartel DP, Crete P (2004) Endogenous trans-acting siRNAs regulate the accumulation of arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu JH, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Volpe T, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen R (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D (2005) FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.