Abstract
EMBO J 31 2, 257–266 (2012); published online December 16 2011
Hybrid seeds have been a key component of greatly increasing the yield of many important crops, foremost of maize. If the parents are properly chosen, non-additive interactions between diverged genomes can lead to strongly superior performance of the F1 progeny, known as heterosis. While many different explanations have been advanced, a consensus for the causes of genome-wide positive epistasis in hybrids has not emerged. In this issue of The EMBO Journal, Shivaprasad and colleagues describe a new mechanism that can account for heterosis often being a genome-wide phenomenon. These authors show that small RNA (sRNA) loci of tomato can exhibit transgressive activity, which can in turn lead to epigenetic and gene expression changes within hybrid progeny. This is particularly exciting because many sRNAs are produced from non-coding regions or transposable elements (TEs), which diverge more quickly than protein-coding genes and thus provide more opportunity for unexpected genetic interactions.
Epistasis is defined as a non-additive genetic interaction, where the interaction may be described as transgressive if the hybrid progeny is in some way either superior to the better or inferior to the worse parent. Transgression has been previously suggested to facilitate hybrid niche specialization and is particularly important in crop breeding (i.e., when hybrid yields are higher than those of either parent).
sRNAs play an important role in gene and genome regulation. MicroRNAs (miRNAs) and trans-acting small interfering RNAs (tasiRNAs) regulate coding transcript levels, while small interfering RNAs (siRNAs) guide DNA methylation and stable chromatin modifications predominantly at TEs and other repeat sequences. These epigenetic marks keep TEs repressed, thereby limiting potentially detrimental transposition events. Epigenetic and sRNA differences between and within species are relatively poorly described compared with genetic and transcriptome variation. Nonetheless, since genomic differences are overrepresented within TEs and repeat elements, which are controlled by siRNAs, one might expect that divergent epigenetic modifications could make major contributions to hybrid phenotypes. In agreement, TEs can be activated in interspecific hybrids, accompanied by changes in DNA methylation (Michalak, 2009), and TEs have been proposed to contribute to transgressive phenotypes through several mechanisms (Tenaillon et al, 2010).
In Arabidopsis thaliana, TEs and siRNAs (which are often associated with TEs) appear to have a stabilizing influence on DNA methylation patterns across generations and between different lines (Vaughn et al, 2007; Becker et al, 2011). TEs, however, can vary substantially between strains of the same species, and much of the intraspecific variation in plant siRNAs is associated with structurally variant TEs (Zhai et al, 2008). Both in Arabidopsis genomes, which have few TEs (He et al, 2011), and in maize genomes, which are enriched in TEs (Eichten et al, 2011), differences in siRNA-dependent TE silencing are predominantly controlled in cis. Yet, like miRNAs and tasiRNAs, siRNAs may in principle also act in trans.
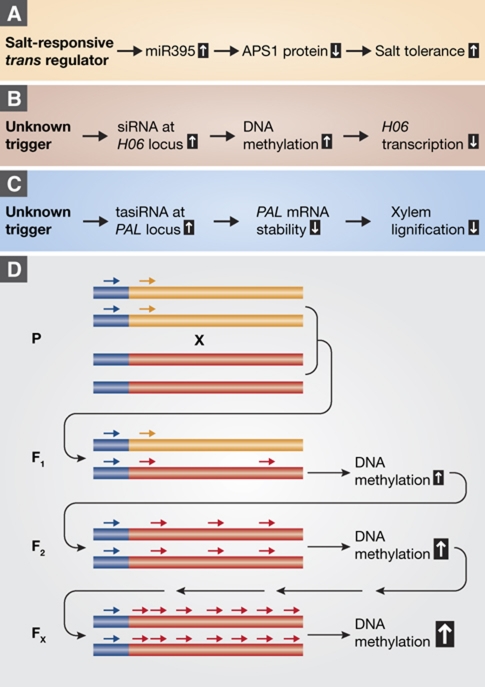
The mixing of genomes in hybrids is vital for the generation of new, favourable genetic combinations, both in natural and in artificial evolution, known as breeding. Thus, understanding non-additive interactions between genomes is of both fundamental interest and very practical utility. In this issue of The EMBO Journal, Shivaprasad et al (2012) report on a genome-wide survey for transgressive segregation at sRNA loci in hybrids and introgression lines between domesticated tomato (Solanum lycopersicum) and a wild relative (S. pennellii). Since the S. pennellii genome is not yet available, the reported number of sRNA loci and hence variation in sRNA expression are certainly underestimates. Still, the authors convincingly demonstrate transgressive effects for both miRNAs and siRNAs (Figure 1). The miRNA miR395 was much more highly expressed in some hybrid progeny, suggesting that one of the parents contributes an allele at a trans-regulatory locus that can specifically increase the abundance of the miRNA generated from the miR395 allele contributed by the other parent. A possible explanation may be a transcription factor that regulates expression of the miR395 precursor.
Figure 1.
(A–C) Three examples of transgressive effects of sRNAs (Shivaprasad et al, 2012). (A) Induction of miR395 increases salt tolerance. (B) Induction of siRNAs at the H06 locus induces local DNA methylation, leading to transcriptional gene silencing. (C) Induction of siRNAs at the PAL locus leads to post-transcriptional gene silencing. (D) Scenario for transgressive sRNA expression, through interaction between allelic or non-allelic loci that share only limited sequence identity (blue line). Small RNAs (small arrows) acting in trans. Specific features of the trans locus lead to spreading of small RNAs through a phenomenon called transitivity, and gradual amplification of sRNA production and DNA methylation over several generations.
For siRNAs, a small fraction of loci, 153 or about 1%, showed transgressive behaviour in the F2 generation or in introgression lines, but not in the F1 plants. In one particularly exciting example, the authors describe in detail an introgression line where siRNA production is apparently amplified in a secondary step, in a mechanism analogous to tasiRNAs. The siRNAs in this case target a protein-coding gene and reduce its mRNA expression.
This innovative paper suggests many new research directions for the field. One question to be answered is why transgression at sRNA loci appears often only in the F2 or even later generations (although changes in the F1 have been reported in intraspecific and interspecific Arabidopsis crosses; Ha et al, 2009; Groszmann et al, 2011). As suggested by the authors (Shivaprasad et al, 2012), interactions between the two genomes during meiosis and gametogenesis may be required to initiate transgressive patterns, or the extent of transgressive expression may be below detection in the F1 generation. Identification of the trans-acting loci will be key to understanding the underlying mechanism(s). Similarly interesting is the question whether cis interactions can also lead to transgression. Finally, why are transgressive interactions only established in a minority of lines?
Epigenetics has been hailed as a potential answer to the question of missing heredity in genome-wide-association studies, at least in humans. The paper by Shivaprasad et al (2012) provides some of the first concrete evidence for epigenetic phenomena that generate entirely new allelic states not easily explained by Mendelian laws.
Acknowledgments
LMS was supported by an European Community FP7 Marie Curie Fellowship (PIEF-GA-2008-221553) and an EMBO Long-Term fellowship. Small RNA studies in the Weigel laboratory are supported by European Community FP7 Collaborative Project AENEAS Contract KBBE-2009-226477 and the Max Planck Society.
Footnotes
The authors declare that they have no conflict of interest.
References
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D (2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249 [DOI] [PubMed] [Google Scholar]
- Eichten SR, Swanson-Wagner RA, Schnable JC, Waters AJ, Hermanson PJ, Liu S, Yeh C-T, Jia Y, Gendler K, Freeling M, Schnable PS, Vaughn MW, Springer NM (2011) Heritable epigenetic variation among maize inbreds. PLoS Genet 7: e1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES (2011) Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA 108: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang XJ, Chen ZJ (2009) Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA 106: 17835–17840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Zhang X, Hu J, Turck F, Dong X, Goebel U, Borevitz J, de Meaux J (2011) Widespread interspecific divergence in cis-regulation of transposable elements in the Arabidopsis genus. Mol Biol Evol (advance online publication, 15 November 2011; doi:10.1093/molbev/msr281) [DOI] [PubMed] [Google Scholar]
- Michalak P (2009) Epigenetic, transposon and small RNA determinants of hybrid dysfunctions. Heredity 102: 45–50 [DOI] [PubMed] [Google Scholar]
- Shivaprasad PV, Dunn RM, Santos BACM, Bassett A, Baulcombe DC (2012) Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J 31: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Hollister JD, Gaut BS (2010) A triptych of the evolution of plant transposable elements. Trends Plant Sci 15: 471–478 [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Tanurdzic M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD, Dedhia N, McCombie WR, Agier N, Bulski A, Colot V, Doerge RW, Martienssen RA (2007) Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol 5: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Liu J, Liu B, Li P, Meyers BC, Chen X, Cao X (2008) Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet 4: e1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]