Abstract
EMBO J 31 2, 291–300 (2012); published online December 16 2011
Nuclear receptors (NRs) can be conceptualized as highly dynamic scaffold proteins, where binding of ligand, DNA or transcriptional coregulator proteins can allosterically change the scaffold structure and direct changes in subsequent binding events. In this issue of The EMBO Journal, Orlov et al present the first cryo-EM structure of a NR complex, a technically challenging feat for the 100-kDa complex of the heterodimer of the vitamin D receptor (VDR), Retinoid X receptor (RXR) and their cognate DNA response element. VDR is one of the few NRs for which the hinge between the ligand-binding domain (LBD) and DNA-binding domain is an extended helix, which enforced a bend in VDR/RXR to an L-shaped architecture. The hinge domain is thus a key regulator of the relative orientation of the LBDs to the DNA, which will impact how transcriptional coregulator complexes are oriented towards the chromatin. It further suggests that the positioning of the hinge may serve as a conduit of structural information, determining how specific DNA sequences can modulate activity in the LBDs.
Nuclear receptors (NRs) are a superfamily of transcription factors that regulate key aspects of development, metabolism and disease (Huang et al, 2010). NRs typically form transcriptionally active homodimers or heterodimers with another NR, that is, Retinoid X receptor (RXR). The classic modular domain organization of NRs includes a variable N-terminal domain (NTD) that may contain a ligand-independent transcriptional activation function (AF-1); a centrally located zinc-finger DNA-binding domain (DBD); and a C-terminal ligand-binding domain (LBD) that contains ligand-dependent AF-2 (Chandra et al, 2008). The DBD and LBD are well structured and connected via a flexible linker or ‘hinge’ region.
NR-interacting proteins include transcriptional coregulators that enzymatically regulate post-translational modification (PTM) of histones and associated proteins (Metivier et al, 2008). However, NRs also interact with other types of enzymes, for example acting as scaffolding proteins for DNA repair enzymes or kinases, or modulating the activity of metabolic and apoptotic proteins (Schultz-Norton et al, 2011). A critical feature of these interactions is that they are highly dynamic, where the NR may transiently interact with DNA, and coregulator complexes that remodel the chromatin with PTMs, which in turn define binding sites for different coregulators (Metivier et al, 2008). Unlike the ligand- and DNA-binding functions, which are restricted to distinct specialized domains, multiple surfaces including AF-1, AF-2 and the hinge contain sites for protein–protein interaction and PTMs. We propose that NRs should be viewed as multi-modal scaffold proteins, where a binding event (e.g., ligand, DNA, protein, PTM) can alter the nature of the scaffold and determine subsequent molecular interactions and activity profiles.
The work by Orlov et al (2012) of The EMBO Journal represents the first cryo-EM structure of a NR complex. There are a number of factors that make this an ideal candidate for pushing the resolution limits of cryo-EM. There are crystal structures of all three components (LBD, DBD and DNA). Each of these components has a unique size and shape. The components are rigid. Further, vitamin D receptor (VDR) is one of the few NRs with the hinge domain characterized by X-ray crystallography, as discussed further below. While there is symmetry between dimers, there are also significant differences that allowed for the unambiguous fitting of the crystal structures into the EM map. The DNA also adds to the likelihood of success in two ways: it has greater scattering due to the phosphates, and provided an opportunity to establish polarity and validate the results by comparing structures with different lengths of oligo. Lastly, the work is consistent with the published small angle X-ray scattering (SAXS) structure (Rochel et al, 2011), further validating it.
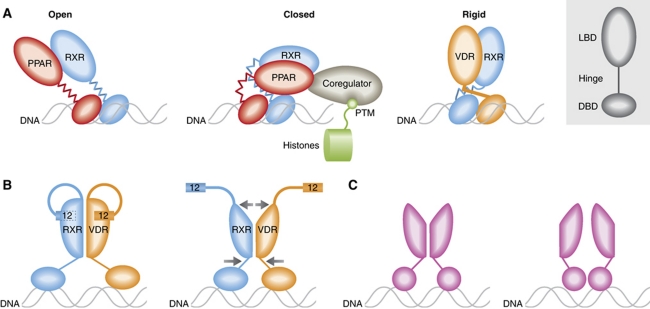
The VDR/RXR structure cryo-EM structure is significantly different from the full-length crystal structure of PPARγ/RXR (Chandra et al, 2008). Both VDR and the thyroid receptor display a structured helix in the hinge domain, termed the C-terminal extension (CTE). Here, it is shown to maintain VDR in an extended, L-shaped conformation, which is also evident in the RXR dimer partner (Figure 1A). A proline at the juncture between hinge CTE and helix 1 of the LBD induces a kink, causing the overall L-shape of the complex. In contrast, PPARγ/RXR assumed a compact architecture, where the PPARγ LBD directly contacts the RXR DBD. Importantly, SAXS experiments showed that PPARγ/RXR adopts an open extended conformation in solution, as does RAR/RXR as well (Rochel et al, 2011).
Figure 1.
(A) Architecture of full-length NRs. VDR has an extended helix in the hinge domain that positions the LBD, and may limit inter-domain flexibility. (B) A model for regulation of the LBD by DNA binding. Changes in the positioning of the hinge could modulate the LBD dimer interface, and in turn change the conformation of helix 12, which forms part of the coactivator binding site. (C) For the steroid receptors AR, GR, MR and PR, interaction with DNA (or other proteins) could stabilize different low affinity dimer interfaces in the LBD.
The concept of NR as scaffold protein is important in evaluating the relevance of different inter-domain architectures. While PPARγ/RXR may only adopt the compact architecture a small percent of the time in solution, doing so in cells is almost certainly going to be determined by the ensemble of interacting proteins (Figure 1A). The important point is that the hinge domains of PPARγ and RXR allow for conformational flexibility. In contrast, the single long helix in the hinge of VDR would interfere with adoption of the compact architecture, and likely renders the entire complex more rigid than PPARγ/RXR and other NRs.
As discussed by Orlov et al and Rochel et al (2011), the inter-domain structure of the NR impacts how histone-modifying enzymes are spatially directed relative to the chromatin. However, it is important to remember that signalling between the chromatin and NR is bidirectional. Thus, many of these enzymes have independent interactions with the chromatin, and may thus reorient the receptor as well (Figure 1A).
The importance of the hinge in determining receptor conformation suggests a model to explain how DNA sequence can alter coregulator binding to the LBD (Figure 1B). A recently published study of full-length VDR/RXR showed that DNA binding alters hydrogen/deuterium exchange rates in the dimer interface of the LBD (Zhang et al, 2011). We and others have shown that the dimer interface can also allosterically control the coactivator binding site in the LBD (Nettles et al, 2008). This suggests a model, where DNA-induced changes in the position of hinges can modulate the LBD dimer interface, and in doing so modulate the coactivator binding site.
This model likely does not apply to the subset of NRs that do not contain the helix 11 dimer interface, including GR, PR, MR and AR. These receptors have much lower LBD dimer affinity, and dimerization likely only occurs with DNA binding. Several different putative LBD dimer interfaces have been identified from crystal structures. This suggests that for these receptors, different DNA sequences (or interacting proteins) might actually induce different LBD dimer conformations (Figure 1C).
While full-length NRs have proven very difficult to crystallize, the ability to apply cryo-EM to complexes of this size will enable the studies needed to unravel structural features of the NR signalling scaffold, and the critical role of inter-domain communication.
Footnotes
The authors declare that they have no conflict of interest.
References
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F (2008) Structure of the intact PPAR-gamma-RXR-nuclear receptor complex on DNA. Nature 456: 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Chandra V, Rastinejad F (2010) Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Ann Rev Physiol 72: 247–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Huet G, Gallais R, Finot L, Petit F, Tiffoche C, Merot Y, LePeron C, Reid G, Penot G, Demay F, Gannon F, Flouriot G, Salbert G (2008) Dynamics of estrogen receptor-mediated transcriptional activation of responsive genes in vivo: apprehending transcription in four dimensions. Adv Exp Med Biol 617: 129–138 [DOI] [PubMed] [Google Scholar]
- Nettles KW, Bruning JB, Gil G, Nowak J, Sharma SK, Hahm JB, Kulp K, Hochberg RB, Zhou H, Katzenellenbogen JA, Katzenellenbogen BS, Kim Y, Joachmiak A, Greene GL (2008) NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Nat Chem Biol 4: 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlov I, Rochel N, Moras D, Klaholz BP (2012) Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J 31: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochel N, Ciesielski F, Godet J, Moman E, Roessle M, Peluso-Iltis C, Moulin M, Haertlein M, Callow P, Mely Y, Svergun DI, Moras D (2011) Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol 18: 564–570 [DOI] [PubMed] [Google Scholar]
- Schultz-Norton JR, Ziegler YS, Nardulli AM (2011) ERalpha-associated protein networks. Trends Endocrinol Metab 22: 124–129 [DOI] [PubMed] [Google Scholar]
- Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang Y, Busby SA, Pascal BD, Garcia-Ordonez RD, Bruning JB, Istrate MA, Kojetin DJ, Dodge JA, Burris TP, Griffin PR (2011) DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol 18: 556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]