Abstract
The 5S rRNAs from Bombyx mori and Dictyostelium discoideum were end-labeled with [32-P] at either the 5' or 3' end and sequenced using enzymatic digestion. The secondary structure of these molecules was studied using the single-strand specific S1 nuclease and the base-pair specific cobra venom ribonuclease. Computer analysis of these results was performed and was used to generate a consensus secondary structure for each molecule. A comparison of these results with those of other workers is presented.
Full text
PDF




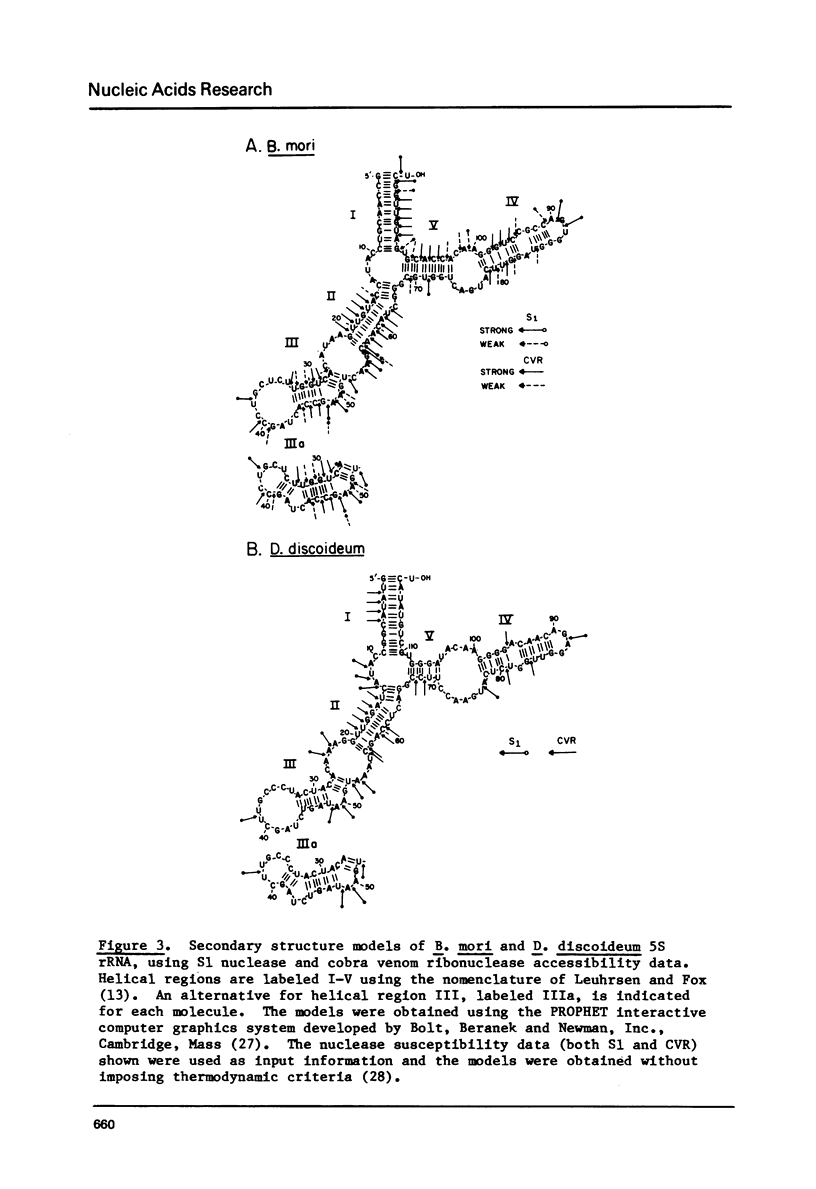
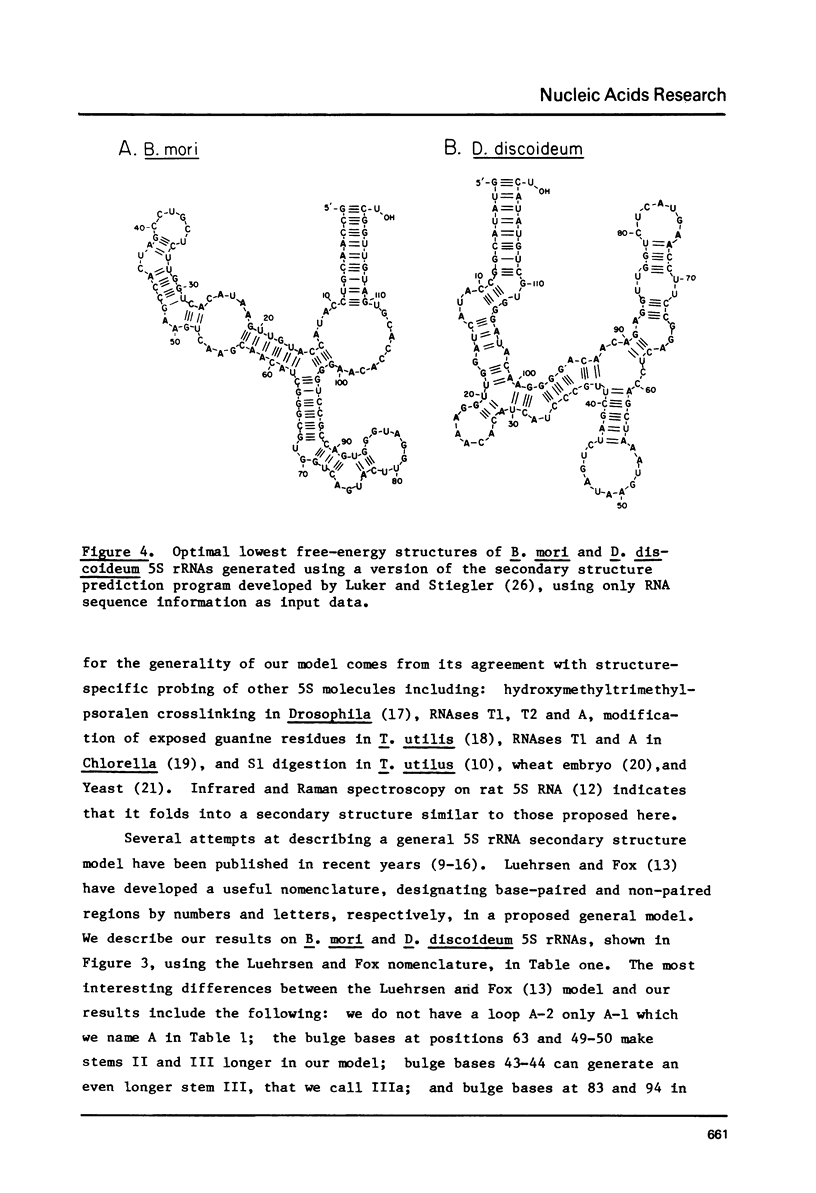
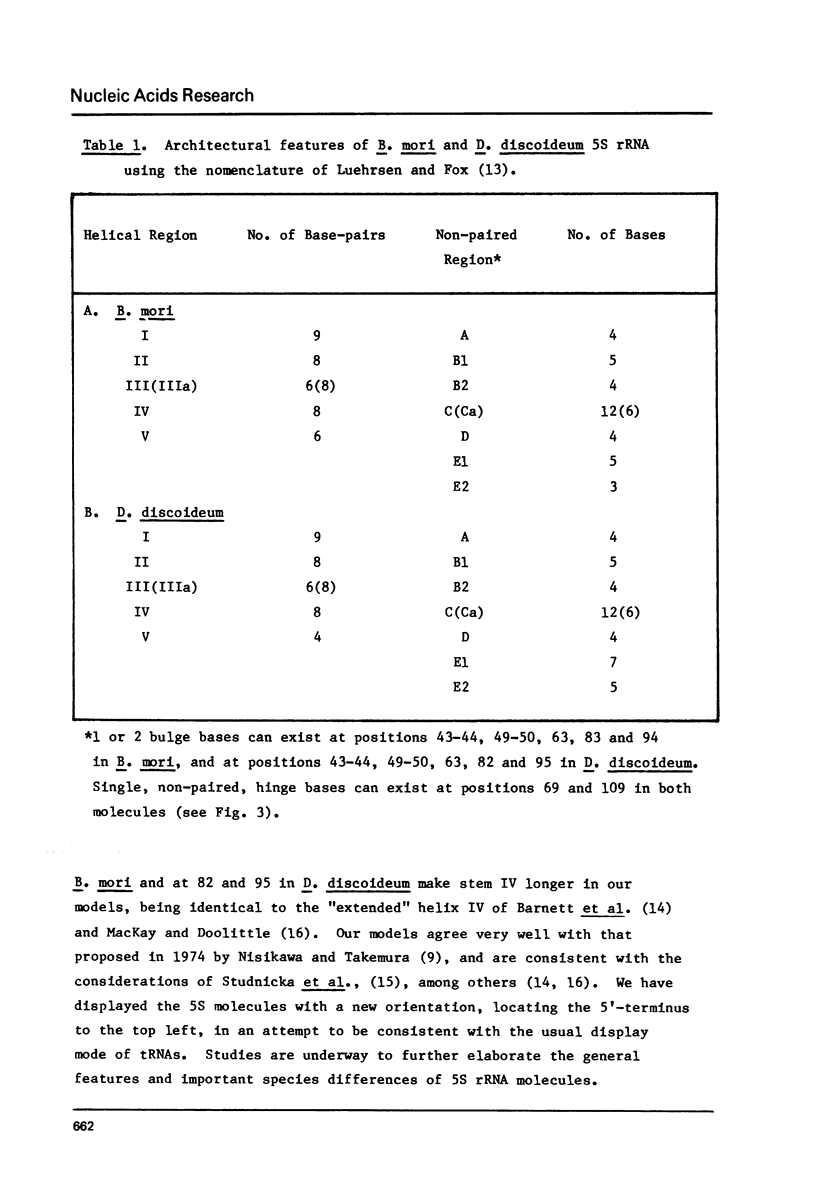


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Averner M. J., Pace N. R. The nucleotide sequence of marsupial 5 S ribosomal ribonucleic acid. J Biol Chem. 1972 Jul 25;247(14):4491–4493. [PubMed] [Google Scholar]
- Barber C., Nichols J. L. Conformational studies on wheat embryo 5S RNA using nuclease S1 as a probe. Can J Biochem. 1978 May;56(5):357–364. doi: 10.1139/o78-057. [DOI] [PubMed] [Google Scholar]
- Boguski M. S., Hieter P. A., Levy C. C. Identification of a cytidine-specific ribonuclease from chicken liver. J Biol Chem. 1980 Mar 10;255(5):2160–2163. [PubMed] [Google Scholar]
- Branlant C., Krol A., Ebel J. P., Gallinaro H., Lazar E., Jacob M. The conformation of chicken, rat and human U1A RNAs in solution. Nucleic Acids Res. 1981 Feb 25;9(4):841–858. doi: 10.1093/nar/9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Vournakis J. N., Donis-Keller H., Chaconas G., Dougall D. K., Kafatos F. C. End labeling of enzymatically decapped mRNA. Nucleic Acids Res. 1977 Dec;4(12):4165–4174. doi: 10.1093/nar/4.12.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashner M. S., Vournakis J. N. Specific hydrolysis of rabbit globin messenger RNA by S1 nuclease. Nucleic Acids Res. 1977 Jul;4(7):2307–2319. doi: 10.1093/nar/4.7.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget B. G., Weissman S. M. The nucleotide sequence of ribosomal 5 S ribonucleic acid from KB cells. J Biol Chem. 1969 Jun 25;244(12):3148–3165. [PubMed] [Google Scholar]
- Fresco J. R., Adams A., Ascione R., Henley D., Lindahl T. Tertiary structure in transfer ribonucleic acids. Cold Spring Harb Symp Quant Biol. 1966;31:527–537. doi: 10.1101/sqb.1966.031.01.068. [DOI] [PubMed] [Google Scholar]
- Gartland W. J., Sueoka N. Two interconvertible forms of tryptophanyl sRNA in E. coli. Proc Natl Acad Sci U S A. 1966 Apr;55(4):948–956. doi: 10.1073/pnas.55.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F., Dahlberg J. E. Specific cleavage of tRNA by nuclease S1. Nucleic Acids Res. 1975 Jun;2(6):865–871. doi: 10.1093/nar/2.6.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H. Molecular evolution of 5S RNA. Mol Gen Genet. 1976 May 7;145(2):119–123. doi: 10.1007/BF00269583. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S., Iwabuchi M. The nucleotide sequence of 5S rRNA from a cellular slime mold Dictyostelium discoideum. Nucleic Acids Res. 1980 Dec 11;8(23):5535–5539. doi: 10.1093/nar/8.23.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. R., Galling G., Jourdan R. Sequence and conformation of 5 S RNA from Chlorella cytoplasmic ribosomes: comparison with other 5 S RNA molecules. J Mol Biol. 1974 Aug 5;87(2):205–225. doi: 10.1016/0022-2836(74)90144-2. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Conformation of mammalian 5.8 S ribosomal RNA: S1 nuclease as a probe. FEBS Lett. 1976 Dec 15;72(1):105–110. doi: 10.1016/0014-5793(76)80823-x. [DOI] [PubMed] [Google Scholar]
- Komiya H., Kawakami M., Takemura S. Nucleotide sequence of 5S ribosomal RNA from the posterior silk glands of Bombyx mori. J Biochem. 1981 Mar;89(3):717–722. doi: 10.1093/oxfordjournals.jbchem.a133251. [DOI] [PubMed] [Google Scholar]
- Levy C. C., Karpetsky T. P. The purification and properties of chicken liver RNase: An enzyme which is useful in distinguishing between cytidylic and uridylic acid residues. J Biol Chem. 1980 Mar 10;255(5):2153–2159. [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Fox G. E. Secondary structure of eukaryotic cytoplasmic 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2150–2154. doi: 10.1073/pnas.78.4.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay R. M., Doolittle W. F. Nucleotide sequences of Acanthamoeba castellanii 5S and 5.8S ribosomal ribonucleic acids: phylogenetic and comparative structural analyses. Nucleic Acids Res. 1981 Jul 24;9(14):3321–3334. doi: 10.1093/nar/9.14.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J. L., Welder L. S1 nuclease as a probe of yeast ribosomal 5 S RNA conformation. Biochim Biophys Acta. 1979 Feb 27;561(2):445–451. doi: 10.1016/0005-2787(79)90152-7. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Structure and function of 5S ribosomal ribonucleic acid from Torulopsis utilis. II. Partial digestion with ribonucleases and derivation of the complete sequence. J Biochem. 1974 Nov;76(5):935–947. [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Structure and function of 5S ribosomal ribonucleic acid from Torulopsis utilis. III. Detection of single-stranded regions by digestion with nuclease S1. J Biochem. 1977 Apr;81(4):995–1003. doi: 10.1093/oxfordjournals.jbchem.a131566. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Structure and function of 5S ribosomal ribonucleic acid from Torulopsis utilis. IV. Detection of exposed guanine residues by chemical modification with kethoxal. J Biochem. 1978 Aug;84(2):259–266. doi: 10.1093/oxfordjournals.jbchem.a132126. [DOI] [PubMed] [Google Scholar]
- Pace N. R. Structure and synthesis of the ribosomal ribonucleic acid of prokaryotes. Bacteriol Rev. 1973 Dec;37(4):562–603. doi: 10.1128/br.37.4.562-603.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Pace B., Erikson R. L. The nucleotide sequence of chicken 5S ribosomal RNA. J Mol Evol. 1974;3(2):151–159. doi: 10.1007/BF01796560. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlakis G. N., Lockard R. E., Vamvakopoulos N., Rieser L., RajBhandary U. L., Vournakis J. N. Secondary structure of mouse and rabbit alpha- and beta-globin mRNAs: differential accessibility of alpha and beta initiator AUG codons towards nucleases. Cell. 1980 Jan;19(1):91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Pipas J. M., McMahon J. E. Method for predicting RNA secondary structure. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2017–2021. doi: 10.1073/pnas.72.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studnicka G. M., Eiserling F. A., Lake J. A. A unique secondary folding pattern for 5S RNA corresponds to the lowest energy homologous secondary structure in 17 different prokaryotes. Nucleic Acids Res. 1981 Apr 24;9(8):1885–1904. doi: 10.1093/nar/9.8.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Vournakis J. N., Celantano J., Finn M., Lockard R. E., Mitra T., Pavlakis G., Troutt A., van den Berg M., Wurst R. M. Sequence and structure analysis of end-labeled RNA with nucleases. Gene Amplif Anal. 1981;2:267–298. [PubMed] [Google Scholar]
- Walker T. A., Betz J. L., Olah J., Pace N. R. The nucleotide sequence of dolphin and bovine 5S ribosomal ribonucleic acid. FEBS Lett. 1975 Jun 15;54(2):241–244. doi: 10.1016/0014-5793(75)80083-4. [DOI] [PubMed] [Google Scholar]
- Wegnez M., Denis H., Mazabraud A., Clérot J. C. Biochemical research on oogenesis. RNA accumulation during oogenesis of the dogfish Scyliorhinus caniculus. Dev Biol. 1978 Jan;62(1):99–111. doi: 10.1016/0012-1606(78)90095-7. [DOI] [PubMed] [Google Scholar]
- Williamson R., Brownlee G. G. The sequence of 5S ribosomal RNA from two mouse cell lines. FEBS Lett. 1969 Jun;3(5):306–308. doi: 10.1016/0014-5793(69)80163-8. [DOI] [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]