Abstract
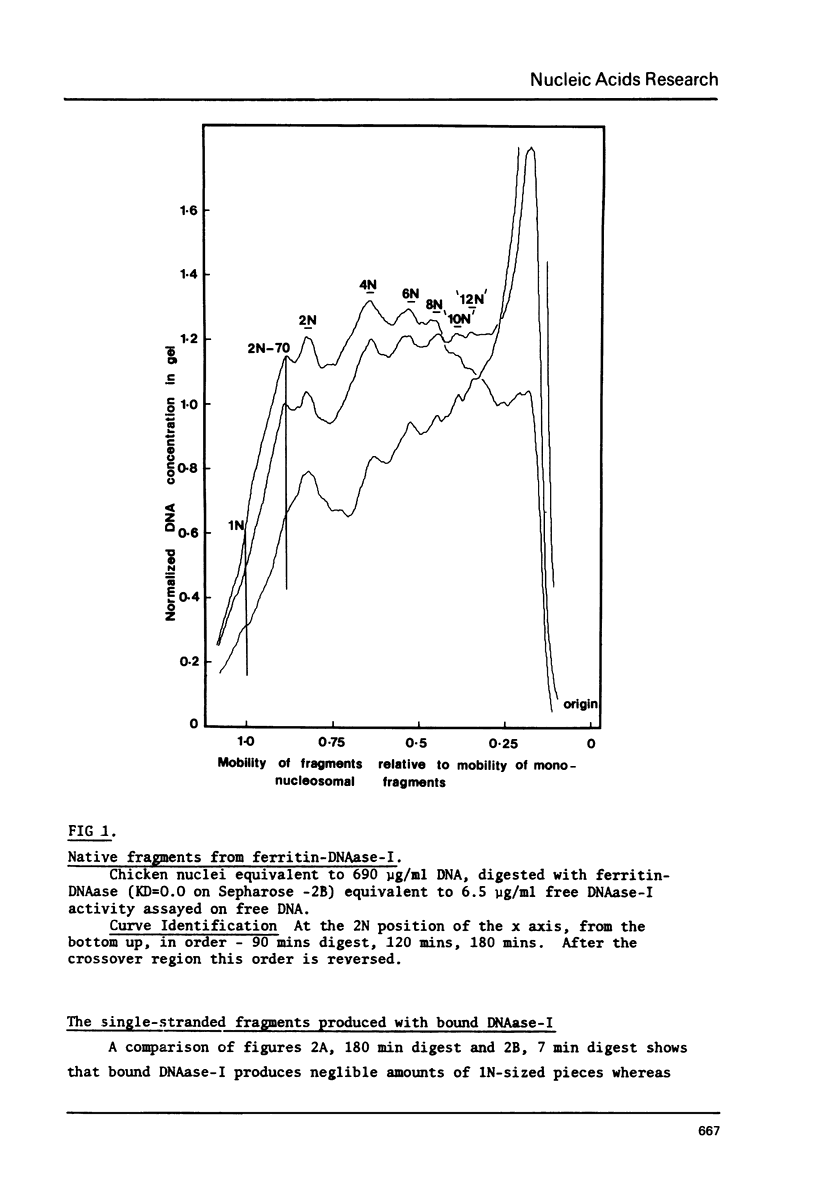
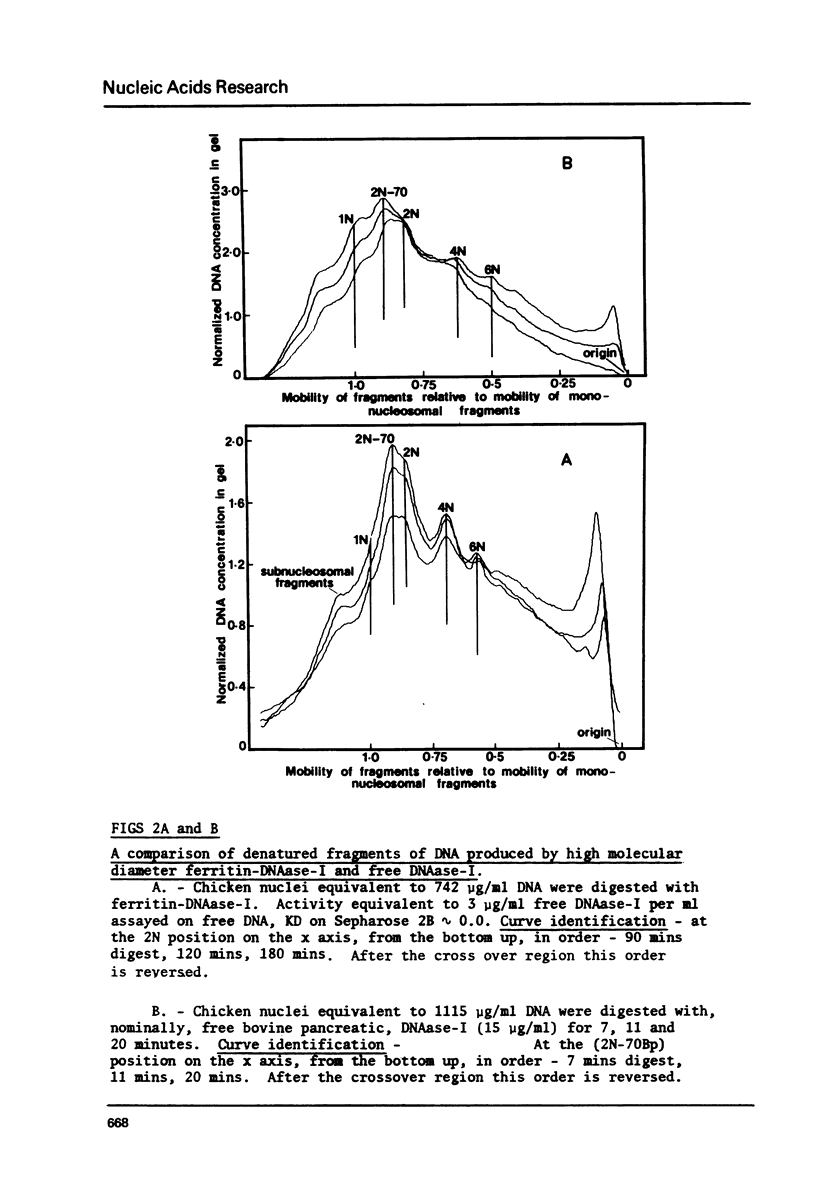
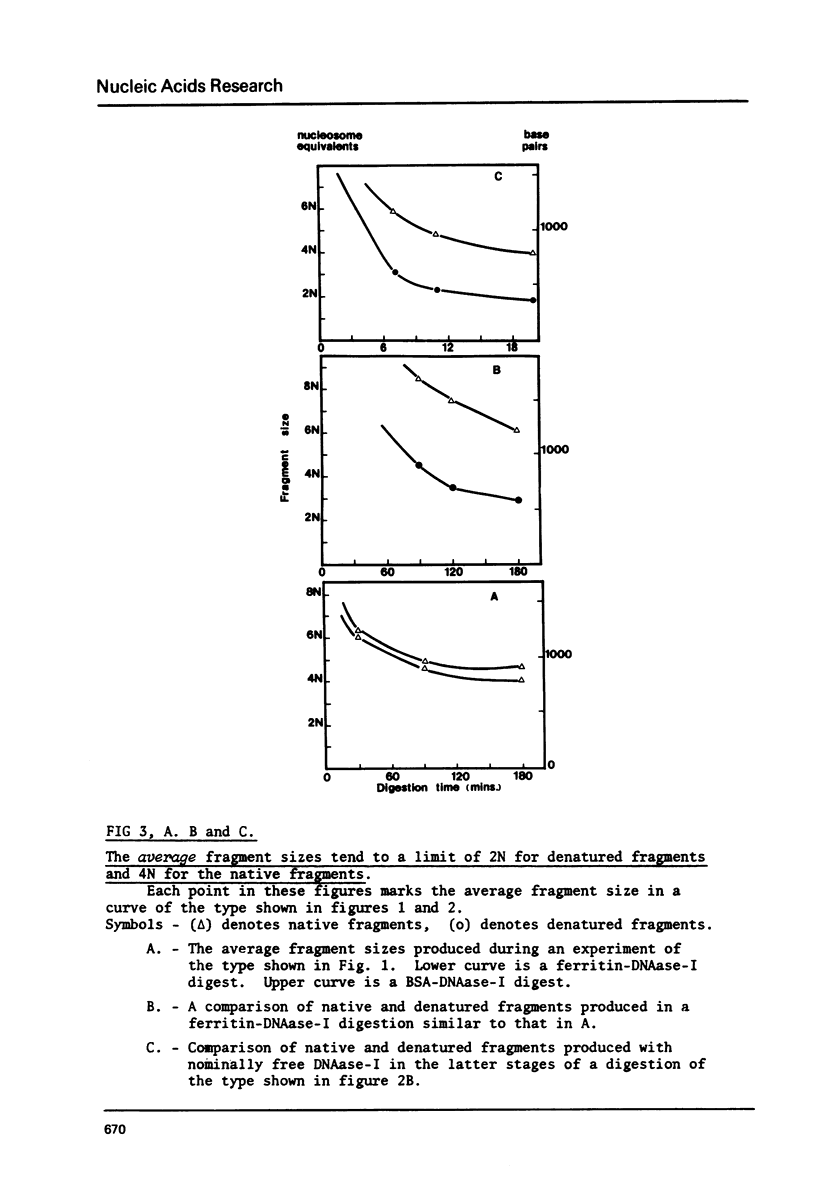
Bulky probes composed of glutaraldehyde-cross-linked-complexes of ferritin or bovine serum albumen armed with bovine pancreatic DNAase-I have been used to generate the typical dinucleosome-based series of native or denatured DNA fragments from chicken erythrocytes. Double stranded DNA fragments have shown that the 2N series extends to at least 10N without any clearly defined end-point. The susceptible nucleosomes were observed to be nucleolysed by two major, different methods of attack that generated multiple peaks in the dinucleosomal size class and some characteristics of the digestion patterns have been taken as indications of the presence of another level of structure above that of the alternating structure. The chromatin structure becomes resistant to the probes when the average fragment length falls to approximately 4N but this 'core' is not a single integral multiple of nucleosomes.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arceci R. J., Gross P. R. Sea urchin sperm chromatin structure as probed by pancreatic DNase I: evidence for a noval cutting periodicity. Dev Biol. 1980 Nov;80(1):210–224. doi: 10.1016/0012-1606(80)90509-6. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Skinner J. D. Chromatin superstructure: the next level of structure above the nucleosome has an alternating character. A two-nucleosome based series is generated by probes armed with DNAase-I acting on isolated nuclei. Biochem Biophys Res Commun. 1981 Apr 15;99(3):893–899. doi: 10.1016/0006-291x(81)91247-x. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Skinner J. D., Marshall A. Analysis of the penetrable space within the nucleus. J Cell Sci. 1978 Jun;31:1–11. doi: 10.1242/jcs.31.1.1. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Skinner J. D. Probing the free space within rat and chicken chromatin with active and passive probes. J Cell Sci. 1979 Jun;37:85–96. doi: 10.1242/jcs.37.1.85. [DOI] [PubMed] [Google Scholar]
- Khachatrian A. T., Pospelov V. A., Svetlikova S. B., Vorob'ev V. I. Nucleodisome - a new repeat unit of chromatin revealed in nuclei of pigeon erythrocytes by DNase I digestion. FEBS Lett. 1981 Jun 1;128(1):90–92. doi: 10.1016/0014-5793(81)81087-3. [DOI] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6326–6330. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannherz H. G., Goody R. S., Konrad M., Nowak E. The interaction of bovine pancreatic deoxyribonuclease I and skeletal muscle actin. Eur J Biochem. 1980 Mar;104(2):367–379. doi: 10.1111/j.1432-1033.1980.tb04437.x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Strogatz S., Riley D. Structure of chromatin and the linking number of DNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]



