Abstract
A quality control method is described for verifying that the coordinate systems of a PET and MR image from the same subject have the same relative left–right orientation. Such ambiguities can arise, for example, at the coordinating center receiving data from many institutions in large multisite studies. In this study, image registration was performed on the PET/MR image pair. The MR image was then reflected (left/right reversed) and the image registration was performed again. A comparison of the values of the cost function describing the accuracy of the registration was used to identify the correct relative orientation. In 122 studies using data with known orientations, 100% accuracy of the method was observed.
Introduction
A common method in image-based neuro research is to acquire separate functional and anatomical images, in particular, using PET and MRI. Typically, the MR image is used to define anatomically based regions of interest (ROIs); to produce a segmentation of the brain into its tissue components; or to provide a means of warping the image set to fit a predefined “standard” brain anatomy. Each of these tasks requires that the two images be brought into spatial alignment, and a number of accurate registration techniques exist for this purpose (Gholipour et al., 2007).
Usually, the PET and MR images will have known geometric properties such as voxel dimensions and coordinate system orientation. However, the number of large multisite trials is increasing in which several types of scanners are employed along with a myriad of processing software, software that may change during the course of a project. Personnel at an analysis site, remote from the initial acquisition site, can be faced with uncertainties, in particular, about the relative left–right orientation of the coordinate systems of PET and MR images. Unlike other issues of coordinate system orientation, because of the left–right symmetry of the human brain, it is, when possible, difficult and time consuming to visually distinguish between a correctly oriented image and a mirror image.
We report on a quality control method for deducing the correct relative left–right orientation of a PET/MR image pair. The procedure is based on the hypothesis that the small degree of left–right asymmetry in the human brain (Toga and Thompson, 2003; Volkau et al., 2006) is sufficient and that the asymmetry persists sufficiently between functional and anatomical imaging modalities such that a registration procedure operating on consistently oriented PET and MR images will yield a better alignment, as measured by the registration-algorithm cost function (i.e., the measure of the goodness of fit), than when operating on images with an incorrect relative left–right orientation. Under this condition, the proper relative left–right orientation of an image pair can be determined by performing the PET/MR image registration twice, once with each of the possible MR left-right orientations, and comparing the two resulting cost-function values.
Materials and methods
Subjects
The data used in the evaluation were obtained from our existing database. All studies were performed using protocols approved by the University of Pittsburgh Institutional Review Board (IRB). The studies presented include five different radiotracers and are summarized in Table 1. Our primary interest is in establishing a quality control procedure for imaging using [11C]6-OH-BTA-1 [Pittsburgh Compound B (PIB)] as well as [18F]Fluorodeoxyglucose (FDG) as radiotracers. All available in-house PIB studies of control subjects were used in the evaluation. The FDG studies included in this work comprise all available control subjects acquired as part of a particular set of projects. No other selection criteria were applied. Control subjects were chosen because they are less likely to have pathology-related anatomical or metabolic asymmetries that could simplify the discrimination task.
Table 1. Study demographics.
| Tracer | Number of subjects (number female) | Age range (years) | Mean age ± standard deviation (years) | Fractional average cost-function difference ± standard deviation |
|---|---|---|---|---|
| [11C]PIB | 44(24) | 39–88 | 71±11 | 0.13 ±0.04 |
| [18F]FDG | 37(18) | 23–90 | 57±24 | 0.14±0.04 |
| [11C]raclopride | 10(10) | 20–38 | 26±6 | 0.17±0.02 |
| [15O]water | 10(8) | 60–94 | 74±9 | 0.16±0.03 |
| [11C]WAY | 11(9) | 60–82 | 74±6 | 0.15±0.02 |
The final column summarizes the study outcome as described in the Results and discussion section.
These data were supplemented with sets of studies using [11C]raclopride, [15O]water, and [carbonyl-11C]WAY100635 (WAY) and include normal controls as well as non-controls. In each case, all available studies acquired for particular project sets were used. We note that the water and WAY data came from the same project and there is an overlap of 8 subjects in the two studies.
Data acquisition and processing
All PET data were acquired using an HR+ (Siemens) scanner. Subjects were placed in the scanner and a 10 min transmission scan was performed. Dynamic emission studies were acquired with the scanner operating in 3D mode. For all studies, except those with FDG, scanning was initiated simultaneously with the intravenous injection of tracer. In the FDG studies, scanning was initiated 40 to 60 min after injection. Each PET frame was reconstructed, using the manufacturer's software, by filtered back projection into a 128 × 128 × 63 voxel array with voxel sizes 0.21 cm × 0.21 cm × 0.24 cm (axial). Reconstruction included corrections for attenuation, scatter, and radioactive decay.
Following our standard procedure, a PET image, for use in registration, was formed by summing frames. The summed image typically spanned the full 30 min for FDG, the first 5 to 8 min for 11C compounds, and the full 180 s for [15O]water studies. In some cases, an internal frame-to-frame registration to correct for patient motion was used. The details of the production of this registration image vary slightly on a case-by-case basis, depending on findings made during the data analysis. However, in each case, the PET registration image used for the current evaluation was produced prior to this study as part of the original data analysis.
The MR images were acquired on a 1.5 T Sigma (General Electric) scanner using a spoiled gradient recalled (SPGR) pulse sequence (TE = 5, TR = 25, flip angle = 40° NEX = 1, slice thickness = 1.5 mm/0 mm interslice).
All processing was performed using in-house protocols resulting in images with known orientations. A second set of MR images was produced by reflecting each original image through a sagittal plane. Thus, each patient study has associated three images, PET, MR with the correct left–right orientation (MR +), and MR with the wrong left–right orientation (MR−). For each patient study, two registrations, PET/MR+ and PET/MR−, were performed using the procedure described below.
Registration method
While a variety of registration algorithms are available (Gholipour et al., 2007), we conducted tests using only a single method that our laboratory routinely employs. This registration procedure uses the Ratio Image Uniformity (RIU) cost function (Woods et al., 1992) as implemented in the AIR 3 package (Woods et al., 1998). In the procedure, a voxel-by-voxel ratio between the two images is calculated and the registration parameters are adjusted with the goal of achieving a smooth ratio image as measured by the ratio standard deviation (the RIU cost function). For our purposes, the procedure was conducted as a PET-to-MRI alignment. The registration was carried out using a 4-step sampling technique (Woods et al., 1992) in which 1/27 of the image voxels were used in the initial registration followed by refinements using 1/9, 1/3, and finally, all voxels.
Prior to registration, MR images were manually cropped such that pixels belonging to the skull were set to zero. In each case, the MRI image was centered (Minoshima et al., 1992). Using AIR 3 routines, it was then resampled to match the PET voxel size and a thresholding procedure applied to eliminate pixels outside of the brain.
Evaluation
Summed PET data
For each subject, the values of the RIU cost function resulting from the PET/MR+ and PET/MR− registration using the summed PET images were compared. Method accuracy over all subjects was computed as the fraction of cases in which the PET/MR+ registration resulted in a lower final value of the cost function than did the PET/MR− registration.
For illustrative purposes, for two subjects, aligned PET/MR ratio images were produced for both the MR+ and MR− registrations. For each registration, variance images were formed in which each pixel takes the value Vi = Σi(Ri − R̄)2 where Ri is the ratio value of pixel i and R̄ is the average ratio value. In this procedure, pixels in which the MR value was less than 10% of the maximum MR value were excluded. The variance image is related to the pixel-by-pixel registration quality as measured by the RIU cost function. To exhibit the pixel-by-pixel differences in registration quality, the MR+ variance image was subtracted from the MR− variance image.
Single frame PET data
An additional study to test the robustness of the method in the case of high PET noise levels was performed using the [15O]water data. The evaluation procedure outlined above was repeated, except that the final frame of each of the 10 PET studies was used instead of the summed image. For these studies, the final frame acquisition was of 20 s duration commencing 160 s following injection of, nominally, 4.4×108 Bq of tracer.
Results and discussion
Summed PET data
Fig. 1 is a scatter plot comparing the cost-function values resulting from PET/MR+ registration with those from the PET/MR− registration in which the summed PET data were used. In all tests of the method, the post-registration cost-function value associated with the PET/MR+ image pair was found to be less than the value associated with the PET/MR− pair. The last column of Table 1 shows the fractional average difference r̄ in the PET/MR+ and PET/MR− cost function for each tracer, computed as where is the PET/MR± cost function for subject i. The sums are taken over all subjects in a particular tracer group. The standard deviation of the terms in the calculation is also shown.
Fig. 1.
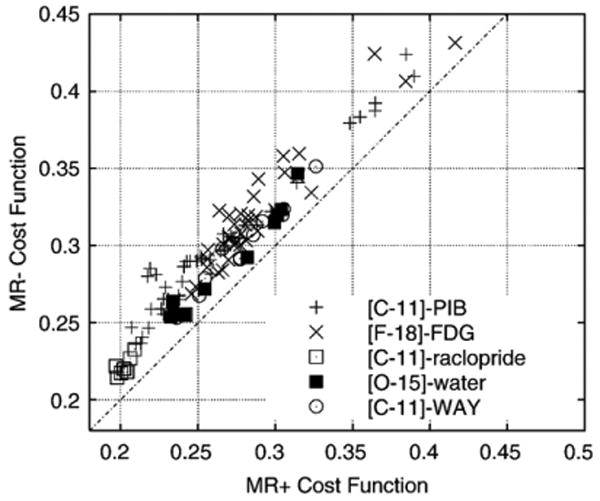
Each subject is represented by a point on the scatter plot. The coordinates of each point are the RIU cost-function value at the termination of the MR−/PET registration procedure (vertical axis), and the corresponding MR+/PET value (horizontal axis). Equivalence between the two values is indicted by the dashed line. Plot symbols represent the tracer used in the study. All points fall above the equivalence line, indicating a better registration for the case of the MR+/PET registration.
Fig. 2 shows a transaxial and coronal slice through a ratio variance-difference image (color scale) superposed on the MR+ image (gray scale) at the mid-brain level for an FDG control subject. The transparent to increasingly red pixels show regions in which the PET/MR+ registration yielded a better fit than did the PET/MR− registration as measured by the image ratio variance. The transparent to dark blue pixels show regions in which the PET/MR− registration yielded a better fit. For these illustrative slices, it can be seen that regions of greatest difference in fit quality between the PET/MR+ and PET/MR− registration correspond to regions of known gray matter asymmetry (Sowell et al., 2002), specifically, around the temporal lobes and the Sylvian fissure region. Differences can also be seen in individual gyri patterns and the CSF/gray matter border of the occipital lobes as well as the interhemispheric fissure regions of the frontal lobe.
Fig. 2.
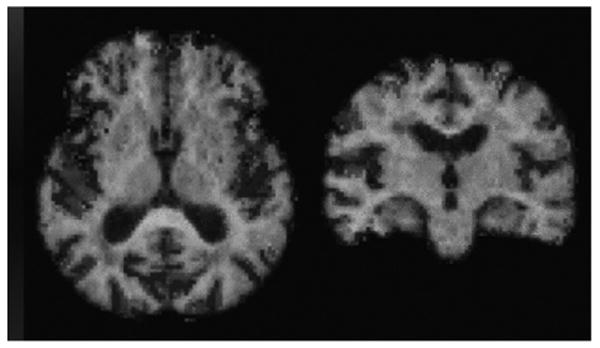
Transverse (left) and coronal (right) slices from a superposed MR image (gray scale) and PET/MR ratio variance-difference image (color scale) for an FDG control subject. Zero difference corresponds to transparency at the center of the color scale. Positive values (toward red) indicate regions in which the PET/MR+ registration produced a better fit, as measured by the ratio variance, than did the PET/MR− registration. Negative values (toward blue) indicate regions of better fit with the PET/MR− registration.
The described procedure can be used with specific receptor binding agents because the early images, which trace blood flow, are included and there is a significant cortical contribution to the asymmetry. This point is illustrated by Fig. 3, equivalent to Fig. 2 but for a raclopride subject. Regions of differences in quality between the PET/MR+ and PET/MR− registrations are similar to those seen in the FDG case (Fig. 2).
Fig. 3.
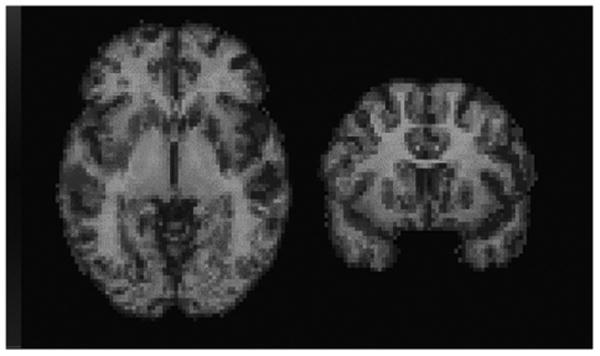
Transverse (left) and coronal (right) slices from a superposed MR image (gray scale) and PET/MR ratio variance-difference image (color scale) for a raclopride subject. Zero difference corresponds to transparency at the center of the color scale. Positive values (toward red) indicate regions in which the PET/MR+ registration produced a better fit, as measured by the ratio variance, than did the PET/MR− registration. Negative values (toward blue) indicate regions of better fit with the PET/MR− registration.
Single frame PET data
The robustness study in which the final frame of each of the 10 [15O]water studies was used in place of the summed-frame images also yielded 100% method accuracy.
The concept of performing two alignments with different relative left–right orientations could, alternatively, be considered a single registration procedure in which an additional discrete registration parameter, i.e., a relative image mirroring, has been added to the traditional continuous rotational and translational parameters. In this view, the question of method robustness becomes a question of parameter robustness. In the single-frame, high-noise studies, we observe that each PET image still contains sufficient information to obtain a reasonable traditional registration. Thus, it is not unreasonable that the information content is also enough to correctly obtain a discrete mirroring parameter as well. Nevertheless, the robustness of the method for different imaging parameters or high image noise levels may require additional validation.
Conclusion
Based on this observed accuracy of 100%, we conclude that the use of a registration procedure constitutes a rational approach to verifying proper PET/MR left–right image orientation. The processing used in this study was, by intent, a minor extension of the methods followed by our laboratory for PET/MR image registration. Other laboratories that wish to implement such a procedure using different registration algorithms or with different tracers should verify these findings for their particular situation.
Acknowledgments
We thank these principle investigators for the data used in this project: S.T. Dekosky, W.H. Kaye, W.E. Klunk, C.A. Mathis, C.C. Meltzer, R.D. Nebes, and E.A. Nofzinger. Support for projects from which data were used was provided by NIH grants AG020677, AG025516, MH01410, MH061566, MH070729, MH42984; by the Pittsburgh Mind-Body Center; and by the Universiy of Pittsburgh Pilot Funding Program. We also thank Ms. Kathi Antonetti for assistance in the preparation of this manuscript.
Footnotes
Supported by the Dana Foundation.
References
- Gholipour A, Kehtarnavaz N, Briggs R, Devous M, Gopinath K. IEEE Trans Med Imag. 2007;26:427–451. doi: 10.1109/TMI.2007.892508. [DOI] [PubMed] [Google Scholar]
- Minoshima S, Berger KL, Lee KS, Mintun MA. J Nucl Med. 1992;33:1579–1585. [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Rex D, Kornsand D, Tessner KD, Jernigan TL, Toga AW. Cereb Cortex. 2002;12:17–26. doi: 10.1093/cercor/12.1.17. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Nat Rev, Neurosci. 2003;4:37–48. doi: 10.1038/nrn1009. [DOI] [PubMed] [Google Scholar]
- Volkau I, Prakash B, Ananthasubramaniam A, Gupta V, Aziz A, Nowinski WL. Acad Radiol. 2006;13:752–758. doi: 10.1016/j.acra.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Woods RP, Cherry SR, Mazziotta JC. J Comput Assist Tomogr. 1992;16:620–633. doi: 10.1097/00004728-199207000-00024. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. J Comput Assist Tomogr. 1998;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]


