Abstract
Background:
There is clinical evidence that very low and safe levels of amplitude-modulated electromagnetic fields administered via an intrabuccal spoon-shaped probe may elicit therapeutic responses in patients with cancer. However, there is no known mechanism explaining the anti-proliferative effect of very low intensity electromagnetic fields.
Methods:
To understand the mechanism of this novel approach, hepatocellular carcinoma (HCC) cells were exposed to 27.12 MHz radiofrequency electromagnetic fields using in vitro exposure systems designed to replicate in vivo conditions. Cancer cells were exposed to tumour-specific modulation frequencies, previously identified by biofeedback methods in patients with a diagnosis of cancer. Control modulation frequencies consisted of randomly chosen modulation frequencies within the same 100 Hz–21 kHz range as cancer-specific frequencies.
Results:
The growth of HCC and breast cancer cells was significantly decreased by HCC-specific and breast cancer-specific modulation frequencies, respectively. However, the same frequencies did not affect proliferation of nonmalignant hepatocytes or breast epithelial cells. Inhibition of HCC cell proliferation was associated with downregulation of XCL2 and PLP2. Furthermore, HCC-specific modulation frequencies disrupted the mitotic spindle.
Conclusion:
These findings uncover a novel mechanism controlling the growth of cancer cells at specific modulation frequencies without affecting normal tissues, which may have broad implications in oncology.
Keywords: hepatocelullar carcinoma, electromagnetic fields, mitotic spindle, PLP2, XCL2
Treatment of hepatocellular carcinoma (HCC) is a major challenge given the limited number of therapeutic options available (Thomas and Zhu, 2005). We have developed a novel approach to treat advanced HCC, consisting of intrabuccal administration of very low levels of radiofrequency electromagnetic fields (RF EMF), amplitude-modulated at specific frequencies, and identified using biofeedback methods in patients with cancer (Barbault et al, 2009). The encouraging findings from a feasibility study (Barbault et al, 2009) led to the design of a phase I/II trial in patients with advanced HCC, and objective responses assessed by CT-scan and changes in alpha-fetoprotein levels were observed in several patients with biopsy-proven HCC (Costa et al, 2011). These findings prompted us to initiate reverse translational experiments to investigate the mechanism of action of amplitude-modulated electromagnetic fields. Two different in vitro exposure systems operating at 27.12 MHz were used to expose cells in culture, replicating patient-treatment conditions.
Proliferation of both HepG2 and Huh7 HCC cells was significantly decreased upon exposure to radiofrequency electromagnetic fields, which were modulated at HCC-specific modulation frequencies. To determine how such frequencies modulate cancer cell growth, we assessed differential gene expression with RNA-seq and found that the expression of several genes was significantly downregulated by HCC-specific modulation frequencies. Previous reports have shown that low intensity, intermediate frequency electric fields are capable of inhibiting cancer growth by interfering with the proper formation of the mitotic spindle (Kirson et al, 2004; Kirson et al, 2007). Similarly, we found that electromagnetic fields that are amplitude-modulated at HCC-specific frequencies disrupt the mitotic spindle of HCC cells. Thus, we provide novel evidence that very low level of amplitude-modulated electromagnetic fields block the growth of HCC cells in a tumour- and tissue-specific fashion.
Materials and methods
In vitro exposure devices
The design and construction of the two in vivo exposure devices (Figure 1) used to conduct these experiments is described in the Supplementary Information.
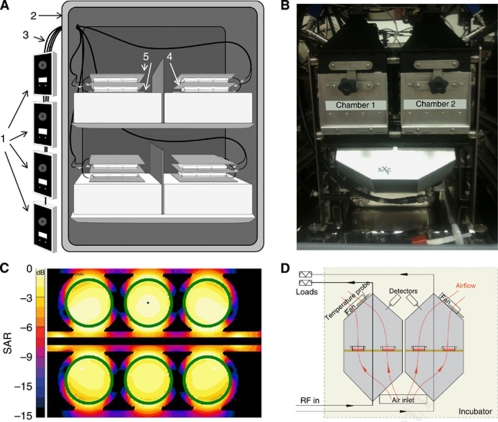
Figure 1.
In vitro exposure experimental setups. (A) Parallel plate capacitor. Emitting devices (1) are placed outside the incubator (2). Each device is connected to a coaxial cable (3), which is connected to a set of brass plates inside the incubator. The centre brass plate (4) is connected to the inner conductor of the emitting device coaxial cable. The outer two brass plates (5) are connected to the outer conductor of the emitting device coaxial cable. Plates containing cells are placed in between the brass plates. (B) TEM cell. The system contains two identical TEM cells placed in an incubator. (C) Distribution of the specific absorption rate (SAR) of cell monolayer in the TEM cell (1 dB per contour), (D) Schematic representation showing the air flow through the TEM cell.
Cell lines
HepG2 and Huh7 cells, both of Biosafety Level 1, were used as representative HCC cell lines. HepG2 cells were obtained from ATCC (Manassas, VA, USA), and Huh7 cells were a gift from Dr Nareej Saxena (Emory University). Normal hepatocytes, THLE-2 cells, were also obtained from ATCC. The breast adenocarcinoma cell line MCF-7 was used as a representative non-HCC malignant cell line (ATCC). The breast epithelial cell line MCF-10A (ATCC) was used to represent normal breast cells. Lymphoblastoid cell lines from healthy individuals enrolled in IRB-approved protocols were provided by Dr Jeff Edberg (UAB).
[3H]thymidine incorporation assay
Growth inhibition (GI) was assessed in HCC cells exposed to HCC-specific modulation frequencies as previously described (Rosman et al, 2008).
Luminescent cell viability assay
Cell proliferation was quantitated using the Promega CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA), a method to determine the number of viable cells in culture based on ATP quantitation.
RNA-seq
We performed RNA-seq as previously described (Reddy et al, 2009). We used HepG2 cells exposed to either HCC-specific modulation frequencies or to randomly chosen frequencies. We double-selected polyA-containing mRNA from 3 μg of total RNA by using oligo-dT magnetic beads. We fragmented the mRNA with RNA fragmentation buffer and removed free-ions with a G-50 Sepharose spin column. Fragmented mRNA was used as a template to synthesise single-stranded cDNA with SuperScript II reverse transcriptase with random hexamer primers in the presence of RNAseOUT (Invitrogen by Life Technologies Corporation, Carlsbad, CA, USA). We synthesised double-stranded DNA (dsDNA) for sequencing by ligating Illumina (Illumina, San Diego, CA, USA) sequencing adaptors to blunted and dA-extended dsDNA, and size-selected fragments of 200–300 bp from a 2% Invitrogen gel and purified with a Qiagen Gel Extraction kit (Qiagen, Valencia, CA, USA). Lastly, we amplified the dsDNA library with 15 rounds of PCR with Illumina sequencing primers. Sequencing was performed on an Illumina GenomeAnalyzer IIx and the paired 36 bp reads were mapped to the hg18 reference genome by using ELAND (Illumina), allowing up to two mismatches per read and 10 or fewer map locations. By using the ERANGE software package (http://woldlab.caltech.edu/rnaseq), we placed uniquely mapped reads against 29 673 transcripts from NCBI build 36.1 of the human genome. After placing unique reads, ERANGE assigned multiple mapping reads and reads mapping to splice junctions according to the number of unique reads in potential transcripts. Once all reads were mapped, ERANGE reported gene expression in units of reads per kilobase of exon and per million tags sequenced (RPKM).
Quantitative PCR
At the conclusion of the AM-EMF exposure portion of the experiment, RNA extraction (Qiagen) and reverse transcription (TaqMan, Applied Biosystems by Life Technologies Corporation) were performed to generate cDNA. Experiments comparing gene expression in HCC cells receiving HCC-specific AM-EMF with gene expression in HCC cells not receiving any exposure were conducted using Applied Biosystems pre-designed TaqMan Gene Expression Assays (PLP2, cat#Hs01099969_g1; XCL2, cat#Hs00237019_m1; Applied Biosystems by Life Technologies Corporation). Real-time quantitation was completed in quadruplicate according to the manufacturer's instructions using an ABI 7900HT Real-Time PCR System (ABI by Life Technologies Corporation), with analysis performed using ABI SDS2.2 software. Quantitative values of gene expression were determined by comparing PCR amplification curves to a known standard curve generated in tandem with the experimental samples. Each sample was individually normalised to the average corresponding to endogenous expression of GAPDH (GAPDH, cat#Hs99999905_m1, TaqMan, Applied Biosystems by Life Technologies Corporation). Averages of the normalised values from each condition were then used to compare the relative gene expression between the experimental groups. The s.e.m. was determined for each experimental condition.
Confocal laser scanning microscopy
Cells undergoing mitosis were imaged using the Zeiss LSM 710 Confocal Laser Scanning Microscope (Carl Zeiss, Inc., Thornwood, NY, USA). For imaging experiments, 22 mm square microscope cover glass (Corning Life Sciences, Lowell, MA, USA, cat#2865–22) were flame-sterilised with 200-proof ethanol and placed in 6-well or 35 mm Falcon tissue culture plates (BD Biosciences, Franklin Lakes, NJ, USA). Approximately 300 μl of cell suspension/growth media was added directly to the top of the cover slips, and cells were plated at varying concentrations (4 × 105–5 × 105 cells per ml) on separate cover slips for each assay to control for variability in antibody affinity between different experiments. Once the cells were given 8–18 h to attach to the cover slips, 3 ml of complete growth media was added to each well containing a cover slip. Following RF EMF exposure, indirect immunofluorescent microscopy compared the cells receiving HCC-specific modulation frequencies with cells not receiving any exposure (Microtubule Marker (AE-8) sc-73551, Fluorescent Secondary Alexa Fluor 488 goat anti-mouse IgG (H+L): A-11001; Santa Cruz Biotechnologies, Santa Cruz, CA, USA).
Karyotype analysis
To determine whether these changes were associated with karyotypic changes, HepG2 cells exposed to HCC-specific modulation frequencies or unexposed were harvested, slides prepared, and metaphase chromosomes G-banded using standard methods. The chromosomes were analysed and the karyotype described according to the International System for Cytogenetic Nomenclature (Brothman et al, 2009).
Statistical analyses
One sample two-sided t-test was performed to test the significance of cell proliferation exposed to RF EMF amplitude-modulated at tumour-specific or randomly chosen frequencies. ANCOVA analysis: For the long-term (7 weeks) GI analysis and the GI analysis for varying SAR values (0.05, 0.1, 0.4, and 1.0 W kg−1), data were fit to a linear model, and time point and dosage level were considered as covariates in determining significance.
Results
Assessment of cell proliferation in the presence of RF EMF
Cell proliferation assays were conducted after 7 days, that is, 21 h of exposure to amplitude-modulated RF EMF. Treatment with HCC-specific modulation frequencies (Supplementary Table 1) significantly reduced the proliferation of HepG2 and Huh7 cells using both the parallel plate capacitor and the transverse electromagnetic (TEM) setups (Figure 1). The observed growth-inhibitory effect on HepG2 cells was of the same magnitude when using a tritium incorporation assay and a bioluminescence assay based on ATP consumption (Figure 2A). Having shown similar results with two different assays, the remainder of the cell proliferation experiments were conducted with the more commonly used tritium incorporation assay. Cell proliferation of HepG2 and Huh7 cells exposed to HCC-specific modulation frequencies was significantly lower than the proliferation of cells exposed either to randomly chosen frequencies (Supplementary Table 2) or not exposed to RF EMF (Figure 2A, columns 1–3). When HepG2 cells were exposed for only 1 h daily, we did not observe any significant inhibition of cell proliferation (Figure 2B). Daily exposure for 6 h instead of 3 h resulted in the same level of cell-proliferation inhibition (Figure 2B). To determine when HCC-specific modulation frequencies begin to exert anti-proliferative effects on HepG2 cells, we assessed cell proliferation following 3 days (9 h) of exposure and did not find any significant difference between cells exposed to HCC-specific modulation frequencies and unexposed cells (Figure 2B).
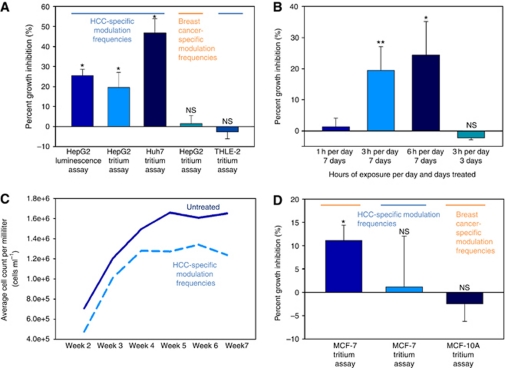
Figure 2.
Cell proliferation assays of cell lines exposed to HCC-specific or breast cancer-specific modulation frequencies. (A) Cells were not split after initial seeding; medium was exchanged every 48 h. Experiments were performed with both equipment setups. Left to right columns: (1) HepG2 cells exposed to HCC-specific modulation frequencies with GI evaluated with a luminescence assay, 25.46±3.22% GI (P=0.0009). (2) HepG2 cells exposed to HCC-specific modulation frequencies with GI evaluated using tritium incorporation, 19.44±7.60% GI (P=0.00993). (3) Huh7 cells exposed to HCC-specific modulation frequencies, 47.73±7.14% GI (P=0.018). (4) HepG2 cells are not significantly inhibited when exposed to breast cancer-specific modulation frequencies, 1.49±3.99% GI (P=0.8815). (5) THLE-2 cells are not affected by HCC-specific modulation frequencies, −2.54±3.54% GI (P=0.6550). Values represent average percent GI (n=6)±%STERR. (B) Cell proliferation assays exposing cells for varying hours per day. Left to right: 1 h per day 1.36±2.77% (P=0.8508); 3 h per day 19.44±7.60% (P=0.0099); 6 h per day 24.46±10.75% (P=0.0301); 3 h per day for 3 days −2.12±0.66% (P=0.4067). Values represent average percent GI (n=6)±%STERR. (C) Cumulative decrease in cell counts over time when HepG2 cells are exposed to HCC-specific modulation frequencies. Samples were subcultured by volume every 7 days (1 : 20 split by volume). Average total cells mL−1 per week: week 2: 7.07 × 105, 4.75 × 105; week 3: 1.20 × 106, 1.01 × 105; week 4: 1.50 × 105, 1.28 × 105; week 5: 1.66 × 105, 1.22 × 105; week 6: 1.61 × 106, 1.34 × 106; week 7: 1.65 × 106, 1.24 × 106 for untreated and treated samples, respectively. For the duration of the 7-week experiment with time considered as a covariate: P=0.005751. (D) Left to right columns: (1) MCF-7 cells exposed to breast tumour-specific modulation frequencies, 11.08±3.30% GI (P=0.0230). (2) MCF-7 cells are not significantly inhibited when exposed to HCC-specific modulation frequencies, 1.49±3.99% (P=0.8815) GI, respectively. (3) MCF-10A cells are not affected by breast tumour-specific modulation frequencies, −2.46±3.75% GI (P=0.8579). Values represent average percent GI (n=6)±%STERR.
Further, to determine whether the growth-inhibitory effect of HCC-specific modulation frequencies persists over time and results in a decrease in the total number of tumour cells, we counted the number of HepG2 cells following treatment with HCC-specific modulation frequencies and that of untreated HepG2 cells weekly for up to 7 weeks. Cells that were either exposed to HCC-specific modulation frequencies or not exposed were split weekly at the same ratio over a period of 7 weeks. As shown in Figure 2C, when compared with unexposed HepG2 cells, the number of HepG2 cells following exposure to HCC-specific modulation frequencies decreased steadily over 7 weeks, resulting in a cumulative loss of 1.71 × 106 cells per ml at week 7.
The average specific absorption rate (SAR) for cells exposed in the parallel capacitor plate system is 0.03 W kg−1 (Supplementary Information). All initial experiments conducted with the TEM system were conducted at a SAR of 0.4 W kg−1. To determine the range of SARs within which significant GI was observed, additional cell proliferation experiments were performed at 0.05, 0.1 and 1.0 W kg−1. A significant anti-proliferative effect was observed at all SARs ranging from 0.05 to 1.0 W kg−1 (P=0.0354). All subsequent assays with the TEM system were conducted at an SAR of 0.4 W kg−1.
Inhibition of cell proliferation is tumour and tissue specific
Our previous clinical observations revealed that patients with HCC had biofeedback responses to specific modulation frequencies that were different from those identified in patients with other types of cancer, such as breast cancer (Barbault et al, 2009). To experimentally assess the relevance of these findings on the proliferation of tumour cells, we determined the specificity of frequencies identified in patients with these two tumour types given the documented objective clinical responses that included one complete and one partial response in two patients with metastatic breast cancer (Barbault et al, 2009) and three partial and one near-complete responses in four patients with HCC (Costa et al, 2011). A total of 194 breast cancer-specific modulation frequencies ranging in the same modulation frequency band from 100 Hz to 21 kHz have been identified in patients with a diagnosis of breast cancer (Supplementary Table 3). In all 9 (4.6%) of the HCC-specific modulation frequencies are identical to breast cancer-specific modulation frequencies.
The two patients with metastatic breast cancer who had experienced an objective response to breast cancer-specific modulation frequencies had tumours that over-expressed oestrogen receptor (ER+) and progesterone receptor (PR+), but did not over-express ERBB2 (ERBB2−) (Barbault et al, 2009). We therefore chose the MCF-7 cell line as it represents the same tumour phenotype, that is, ER+, PR+, ERBB2−. Although the growth of MCF-10A breast cells was unaffected by exposure to breast cancer-specific modulation frequencies, exposure of MCF-7 breast cancer cells to breast cancer-specific modulation frequencies significantly inhibited cell proliferation (Figure 2D). However, exposure of HepG2 cells to the same breast cancer-specific modulation frequencies did not affect cell proliferation (Figure 2A). Similarly, the proliferation of MCF-7 cells was not affected by exposure to HCC-specific modulation frequencies (Figure 2D). Consequently, the observed anti-proliferative effect on HCC and breast cancer cells was observed only upon exposure to tumour-specific modulation frequencies previously identified in patients with a diagnosis of HCC and breast cancer, respectively, despite the fact that more than 57% of the modulation frequencies only differed by <1% (Supplementary Tables 1 and 3).
Having demonstrated that the anti-proliferative effect of amplitude-modulated frequencies was tumour specific, we sought to determine whether the HCC-specific modulation frequencies have an effect on the proliferation of THLE-2 normal hepatocytes. As shown in Figure 2A, exposure of THLE-2 cells to HCC-specific modulation frequencies did not have any measurable effect on cell proliferation. These findings provide strong support for the novel notion that a combination of narrowly defined, specific modulation frequencies identified in a group of patients with the same type of cancer is capable of inhibiting cell proliferation in a tumour- and tissue-specific fashion.
Tumour-specific modulation frequencies and gene regulation
To study the mechanism by which tumour-specific modulation frequencies inhibit cell proliferation, we assessed the gene expression profile of HepG2 cells exposed to HCC-specific modulation frequencies using RNA-seq, as it provides a more comprehensive assessment of differential gene expression across a broader range of expression levels than microarray-based analysis (Wang et al, 2009). Overall, we did not observe statistically significant differences in transcript levels when comparing two HepG2 cultures exposed for 1 week, 3 h a day to HCC-specific modulation frequencies with two HepG2 cultures exposed to randomly chosen modulation frequencies (Supplementary Figure 1). However, we did observe a small number of genes with an absolute fold-change >1.5 and a minimum mean RPKM of 1.5 following exposure to HCC-specific modulation frequencies. Two genes with an absolute fold-change >1.8 appeared to be downregulated in HepG2 cells exposed to HCC-specific modulation frequencies, PLP2 and XCL2, and were considered to be candidates worthy of further experiments. We validated the downregulation of PLP2 and XCL2 with quantitative PCR in both HepG2 as well as Huh7 cells exposed to HCC-specific modulation frequencies (Figures 3A and B). There was no significant downregulation of PLP2 and XCL2 in MCF-7 breast cancer cells (Figure 3C). Similarly, there was no downregulation of PLP2 and XCL2 in nonmalignant cells, that is, in THLE-2 normal hepatocytes (Figure 3D), or in lymphoblastoid cell lines from healthy individuals (Figures 3E and F). These findings support the novel notion that the demodulation effects of RF EMF amplitude-modulated at specific frequencies inhibit cell proliferation and affect the expression of several genes in a tumour- and tissue-specific fashion.
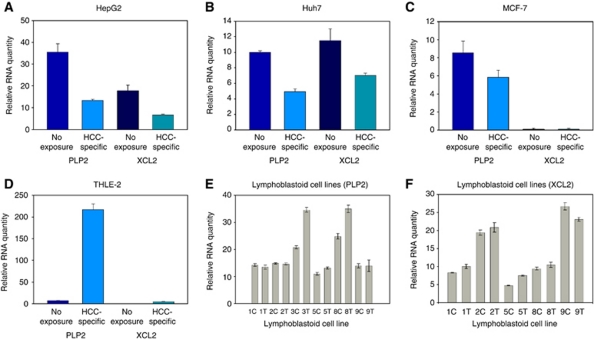
Figure 3.
Expression of XCL2 and PLP2 receiving HCC-specific RF EMF compared with cells not receiving exposure. (A) HepG2: PLP2 (35.46±3.85; 13.17±0.70) and XCL2 (17.87±2.49; 6.52±0.48) (P=9.0371e-3 and P=0.0179, respectively). (B) Huh7: PLP2 (10.02±0.19; 4.95±0.35) and XCL2 (11.52±1.49; 7.02±0.29) (P=9.4981e-5 and P=0.0536, respectively). (C) MCF-7: PLP2 (8.52±1.30; 5.84±0.77) and XCL2 (levels not detectable). (D) THLE-2: PLP2 (7.11±0.14; 216.89±13.18) and XCL2 (0.03±0.01; 4.55±1.04) in THLE-2 cells exposed to HCC-specific modulation frequencies (P=5.5108e-4 and P=0.0221, respectively). (E) Expression levels of PLP2 in lymphoblastoid cell lines (C=unexposed; T=HCC-specific exposure) (for all cell lines compiled P=0.418), LCL 3 expression was significant P=0.0021 as was LCL8 P=0.0159 (F) Expression levels of XCL2 in lymphoblastoid cell lines (for all cell lines compiled (P=0.899), LCL 1 expression difference was significant P=0.0002. Values represent average relative RNA expression (n=4)±s.e.m. Levels were normalised to levels of GAPDH.
Tumour-specific modulation frequencies and disruption of the mitotic spindle
There is evidence that the proliferation of several rodent and human cancer cell lines is arrested by exposure to sinusoidal electric fields of 100–200 V m−1 at a frequency of 100–300 kHz (Kirson et al, 2004). This approach has also shown efficacy in animal and human tumour models as well as promising results in the treatment of patients with cancer (Kirson et al, 2004; Kirson et al, 2007; Salzberg et al, 2008; Kirson et al, 2009). The anti-tumour effect of this therapeutic approach appears to be caused by disruption of the mitotic spindle mediated by interference of spindle tubulin orientation and induction of dielectrophoresis (Kirson et al, 2004; Kirson et al, 2007). In contrast to the sinusoidal signals (Kirson et al, 2004), the carrier frequency of the signal applied in our experiments is more than 100 times higher; the peak E-field amplitude of the carrier at 0.4 W kg−1 corresponds to approximately 35 V m−1 inside the cell medium when the signal is sinusoidally amplitude-modulated at specific frequencies with 85% modulation depth (Kirson et al, 2004).
Despite these significant differences, confocal laser scanning microscopy revealed pronounced disruption of the mitotic spindle in more than 60% of HepG2 cells exposed for 1 week, 3 h per day to HCC-specific modulation frequencies whereas there was no disruption of the mitotic spindle in unexposed HepG2 cells (Figures 4A and B). Specifically, the observed cytoskeletal disruption in cells exposed to HCC-specific modulation frequencies was apparent in cells in mitosis, in which we saw centrosomal distortion and poor chromosomal separation at anaphase (Figure 4D). We found no evidence of karyotypic differences between HepG2 cells exposed to HCC-specific modulation frequencies and unexposed HepG2 cells.
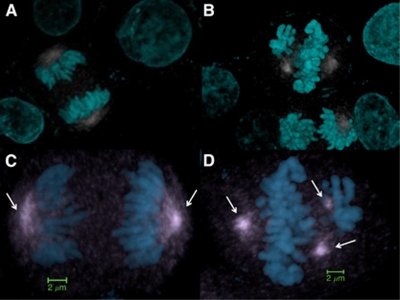
Figure 4.
Mitotic spindle disruption in cells receiving HCC-specific RF EMF compared with cells not receiving exposure. (A) HepG2 efficiently assembles a bipolar mitotic spindle, allowing cells to pass through the mitotic assembly checkpoint and successfully progress from metaphase to anaphase, (B) >60% of dividing HepG2 cells exposed to HCC-specific modulation frequencies exhibit microtubule-associated anomalies, (C) high magnification of unexposed HepG2 cells in mitosis (D) high magnification of HepG2 cells exposed to HCC-specific modulation frequencies shows errors such as tripolar spindle formation (Cyan=DAPI; Gray=Microtubules; Arrows=mitotic spindle).
Discussion
By exposing HCC cells to 27.12 MHz RF EMF sinusoidally amplitude-modulated at specific frequencies, which were previously identified in patients with a diagnosis of HCC (Barbault et al, 2009) and result in therapeutic responses in patients with HCC (Costa et al, 2011), we demonstrate a robust and sustained anti-proliferative effect. This effect was seen within SARs ranging from 0.03 to 1.0 W kg−1, that is, within the range of exposure in humans receiving treatment administered intrabuccally (Barbault et al, 2009; Costa et al, 2011). HCC-specific modulation frequencies began to hinder cell proliferation after 7 days of exposure and the anti-proliferative effect increased over a 7-week period. The anti-proliferative effect HCC-specific modulation frequencies were observed only in HCC cells, but not in breast cancer cells or normal hepatocytes.
The specificity of modulation frequencies is exemplified by the fact that two sets of similar modulation frequencies (breast cancer-specific and randomly chosen) within the same range, that is, from 100 Hz to 21 kHz, did not affect the proliferation of HCC cells. Similarly, the proliferation of breast cancer cells was affected only by breast cancer-specific modulation frequencies, but neither by HCC-specific nor by randomly chosen modulation frequencies. The fact that >50% of the modulation frequencies from these three programs differed by <1%, provides strong experimental evidence that the biological effects are only mediated by a combination of narrowly defined, tumour-specific modulation frequencies.
The modulation-frequency specific laboratory findings are consistent with the clinical observation of a complete response in a patient with breast cancer metastatis to the adrenal gland and the bone while a primary malignancy of the uterus continued to grow (Barbault et al, 2009). This suggests that a combination of precise tumour-specific modulation frequencies is needed to block cancer growth in vitro and in patients with a diagnosis of cancer. The clinical results reported by Barbault et al, (2009) and Costa et al, (2011) as well as laboratory evidence included in this report provide support for the novel and transformational concept that the growth of human tumours arising from the same primary tissue may be effectively blocked by identical modulation frequencies. While receiving treatment with HCC-specific modulation frequencies, one black and three white patients with advanced carcinoma had partial responses (Costa et al, 2011). Furthermore, proliferation of the Huh7 HCC cell line, which is derived from a Japanese patient's tumour (Nakabayashi et al, 1982), exhibited the most pronounced response to HCC-specific modulation frequencies (Figure 2A). This indicates that the frequency signature and biological effects of HCC-specific modulation frequencies are likely independent of ethnic status.
There is no known biophysical mechanism accounting for the effect observed in these experiments; however, other modulation-frequency dependent effects have been observed in biological systems at similarly low exposure levels. Documented effects have occurred in cellular processes controlling cell growth, proliferation, and differentiation (Blackman, 2009). Further, modulation of the signal appears to be a critical factor in the response of biological systems to electromagnetic fields (Blackman, 2009). The amount of electromagnetic energy delivered is far too low to break chemical bonds or cause thermal effects, necessitating alternative mechanistic explanations for observed biological outcomes. Several theories have been put forth to explain biological responses to electromagnetic fields. Some reports have shown that low levels of electromagnetic fields can alter gene expression and subsequent protein synthesis by interaction of the electromagnetic field with specific DNA sequences within the promoter region of genes (Blank and Goodman, 2008; Blank and Goodman, 2009). Such changes have been demonstrated in the family of ‘heat shock’ proteins that function in the cell stress response (Blank and Goodman, 2009).
To thoroughly interrogate gene expression changes in cells exhibiting decreased cell proliferation, we used high-throughput sequencing technologies to sequence the cells' cDNA, a technique that has become invaluable in the study of cancer (Maher et al, 2009). Tumour cell GI was associated with downregulation of PLP2 and XCL2 as well as with disruption of the mitotic spindle. PLP2 encodes an integral membrane protein that localises to the endoplasmic reticulum in epithelial cells. The encoded protein can multimerise and may function as an ion channel (Breitwieser et al, 1997). PLP2 enhances chemotaxis of human osteogenic sarcoma cells (Lee et al, 2004) and PLP2 downregulation is associated with decreased metastasis in a mouse model of cancer (Sonoda et al, 2010). XCL2 encodes for a protein that enhances chemotactic activity for lymphocytes and downregulation of XCL2 has been shown to be associated with good prognosis in patients with breast cancer (Teschendorff et al, 2007; Teschendorff and Caldas, 2008). The pronounced disruption of the mitotic spindle seen in the majority of HepG2 cells exposed to HCC-specific modulation frequencies undergoing mitosis is not associated with karyotypic changes, but may be a major determinant of the anti-tumour effects of HCC-specific modulation frequencies accounting for the therapeutic responses seen in patients receiving the same modulation frequencies (Costa et al, 2011).
Exposure of HCC cells to the same RF EMF modulated at slightly different modulation frequencies did not result in changes in gene expression, which demonstrates that inhibition of cell proliferation is associated with changes in gene expression levels.
In conclusion, we show that very low levels of 27.12 MHz radiofrequency electromagnetic fields, which are comparable to the levels administered to patients, inhibit tumour cell growth when modulated at specific frequencies. The exciting findings presented in this report suggest that the anti-proliferative effect of modulation frequencies is both tumour- and tissue-specific, and is mediated by changes in gene expression as well as disruption of the mitotic spindle. These findings uncover a new alley to control tumour growth and may have broad implications for the treatment of cancer.
Acknowledgments
We thank Dr Carl F. Blackman, 3413 Horton Street, Raleigh, NC, Dr Richard B. Marchase, UAB, and Dr Bernard Veyret, University of Bordeaux for reviewing the manuscript. We also thank Dr Andrew Carroll, Cytogenetics Laboratory, UAB Department of Genetics, for performing the karyotype analysis. We would also like to thank Dr Jeff Edberg for the lymphoblastoid cell lines from the UAB Control Population. Finally, we would like to thank Dr Nareej Saxena, Emory University for his expertise and the gift of Huh7 cells.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Barbault A, Costa F, Bottger B, Munden R, Bomholt F, Kuster N, Pasche B (2009) Amplitude-modulated electromagnetic fields for the treatment of cancer: discovery of tumor-specific frequencies and assessment of a novel therapeutic approach. J Exp Clin Cancer Res 28(1): 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman C (2009) Cell phone radiation: evidence from ELF and RF studies supporting more inclusive risk identification and assessment. Pathophysiology 16: 205–216 [DOI] [PubMed] [Google Scholar]
- Blank M, Goodman R (2008) A mechanism for stimulation of biosynthesis by electromagnetic fields: charge transfer in DNA and base pair separation. J Cell Physiol 214: 20–26 [DOI] [PubMed] [Google Scholar]
- Blank M, Goodman R (2009) Electromagnetic fields stress living cells. Pathophysiology 16: 71–78 [DOI] [PubMed] [Google Scholar]
- Breitwieser GE, McLenithan JC, Cortese JF, Shields JM, Oliva MM, Majewski JL, Machamer CE, Yang VW (1997) Colonic epithelium-enriched protein A4 is a proteolipid that exhibits ion channel characteristcs. Am J Physiol 272(3 Pt 1): C957–C965 [DOI] [PubMed] [Google Scholar]
- Brothman AR, Persons DL, Shaffer LG (2009) Nomenclature evolution: changes in the ISCN from the 2005 to the 2009 edition. Cytogenet Genome Res 127: 1–4 [DOI] [PubMed] [Google Scholar]
- Costa FP, de Oliveira AC, Meirelles R, Machado MCC, Zanesco T, Surjan R, Chammas MC, de Souza Rocha M, Morgan D, Cantor A, Zimmerman J, Brezovich I, Kuster N, Barbault A, Pasche B (2011) Treatment of advanced hepatocellular carcinoma with very low levels of amplitude-modulated electromagnetic fields. Br J Cancer 105(5): 640–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirson ED, Dbaly V, Tovarys F, Vymazal J, Soustiel JF, Itzhaki A, Mordechovich D, Steinberg-Shapira S, Gurvich Z, Schneiderman R, Wasserman Y, Salzberg M, Ryffel B, Goldsher D, Dekel E, Palti Y (2007) Alternating electric fields arrest cell proliferation in animal tumor models and human brain tumors. PNAS 104(24): 10152–10157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirson ED, Giladi M, Gurvich Z, Itzhaki A, Mordechovich D, Schneiderman RS, Wasserman Y, Ryffel B, Goldsher D, Palti Y (2009) Alternating electric fields (TTFields) inhibit metastatic spread of solid tumors to the lungs. Clin Exp Metastasis 26(7): 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirson ED, Gurvich Z, Schneiderman R, Dekel E, Itzhaki A, Wasserman Y, Schatzberger R, Palti Y (2004) Disruption of cancer cell replication by alternating electric fields. Cancer Res 64(9): 3288–3295 [DOI] [PubMed] [Google Scholar]
- Lee SM, Shin H, Jang SW, Shim JJ, Song IS, Son KN, Hwang J, Shin YH, Kim HH, Lee CK, Ko J, Na DS, Kwon BS, Kim J (2004) PLP2/A4 interacts with CCR1 and stimulates migration of CCR1-expressing HOS cells. Biochem Biophys Res Commun 324: 768–772 [DOI] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM (2009) Transcriptome sequencing to detect gene fusions in cancer. Nature 458(7234): 97–101, doi:10.1038/nature07638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J (1982) Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res 42: 3858–3863 [PubMed] [Google Scholar]
- Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM (2009) Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res 19(12): 2163–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosman DS, Phukan S, Huang CC, Pasche B (2008) TGFBR1*6A enhances the migration and invasion of MCF-7 breast cancer cells through RhoA activation. Cancer Res 68(5): 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg M, Kirson E, Palti Y, Rochlitz C (2008) A pilot study with very low-intensity, intermediate-frequency electric fields in patients with locally advanced and/or metastatic solid tumors. Onkologie 31(7): 362–365. [DOI] [PubMed] [Google Scholar]
- Sonoda Y, Warita M, Suzuki T, Ozawa H, Fukuda Y, Funakoshi-Tago M, Kasahara T (2010) Proteolipid protein 2 is associated with melanoma metastasis. Oncol Rep 23: 371–376 [PubMed] [Google Scholar]
- Teschendorff AE, Caldas C (2008) A robust classifier of high predictive value to identify good prognosis patients in ER-negative breast cancer. Breast Cancer Res 10(4): R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C (2007) An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol 8(8): R157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MB, Zhu AX (2005) Hepatocellular carcinoma: the need for progress. J Clin Oncol 23(13): 2892–2899 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.