Abstract
Background:
It remains important to understand the biology and identify biomarkers for less studied cancers like testicular cancer. The purpose of this study was to determine the methylation frequency of several cancer-related genes in different histological types of testicular cancer and normal testis tissues (NT).
Methods:
DNA was isolated from 43 seminomas (SEs), 14 non-SEs (NSEs) and 23 NT, and was assayed for promoter methylation status of 15 genes by quantitative methylation-specific PCR. The methylation status was evaluated for an association with cancer, and between SEs and NSEs.
Results:
We found differential methylation pattern in SEs and NSEs. MGMT, VGF, ER-β and FKBP4 were predominately methylated in NSEs compared with SEs. APC and hMLH1 are shown to be significantly more methylated in both subtypes in comparison with NT. When combining APC, hMLH1, ER-β and FKBP4, it is possible to identify 86% of the NSEs, whereas only 7% of the SEs.
Conclusions:
Our results indicate that the methylation profile of cancer-associated genes in testicular cancer correlates with histological types and show cancer-specific pattern for certain genes. Further methylation analysis, in a larger cohort is needed to elucidate their role in testicular cancer development and potential for therapy, early detection and disease monitoring.
Keywords: testicular cancer, DNA methylation, epigenetics, biomarker, seminoma, non-seminoma
Testicular cancer is the most commonly diagnosed malignancy among young men aged 15 to 40 years, and its incidence has doubled in the past 40 years (Chia et al, 2010). An annual increase of 3–6% is reported for Caucasian populations. In the United States, approximately 8500 newly diagnosed testicular cancer cases and 350 deaths were expected in 2010 (Jemal et al, 2010). Testicular germ cell tumours (TGCTs) represent over 95% of the testicular cancers and histopathologically are classified into two major groups of seminomas (SE) or non-SEs (NSE), frequently occurring as mixed tumours (Holmes et al, 2008). Histologically, SE resembles primordial germ cells/gonocytes, whereas NSE shows somatic, primitive embryonal or extra-embryonal differentiation (Horwich et al, 2006). Carcinoma in situ or intra-tubular germ cell neoplasia unclassified are believed to be the origin of both SE and NSE (Rajpert-De Meyts, 2006). They have a primordial germ cell/gonocyte origin, and it is important to emphasise that epigenetic reprogramming is known to occur during germ cells development.
The treatment of testicular cancer includes orchiectomy, and according to the metastatic condition surveillance, or adjuvant retroperitoneal surgery, radiation and chemotherapy are offered. The decision whether to choose the adjuvant treatment regimen is based only on clinical parameters, leading to around 25% development of metastasis in patients on surveillance or unnecessary adjuvant treatments in 20% of the patients. Genetic understanding of each tumour will enable to better tailor the treatment.
The importance of epigenetic alterations has long been demonstrated in carcinogenesis. Several studies have shown that methylation-associated silencing inactivates certain tumour suppressor genes (TSGs) as effectively as mutations and is one of the cancer-predisposing hits described in Knudson's two hit hypothesis. Promoter methylation, the most studied epigenetic alteration, is also increasingly recognised as a major mechanism of gene inactivation during TGCT progression (Manton et al, 2005; Ellinger et al, 2009). CG dinucleotide-rich regions, also known as CpG islands, in or near the proximal promoter regions of genes are targets for DNA methylation, leading to effective transcriptional silencing (Herman, 1999). In normal cells, CpG methylation is an important mechanism for regulating gene expression, whereas in cancer cells, aberrant promoter methylation (hypermethylation) can lead to abnormal gene silencing, including repression of TSGs. Another relevant aspect is that environmental and endogenous conditions can influence the epigenetic processes. The exposure to certain environmental risk factors may be related to the onset of cancer or to participate in carcinogenesis (Brait et al, 2009). There are many evidences that endogenous factors (such as oestrogens or androgens inhibitors) exposure may lead to cancer (Godmann et al, 2009), and endogenous factors (like hormonal stimuli) induce methylation of promoter region of certain genes (Kutanzi et al, 2010). Even though TGCT has high survival rates due to good responses to therapy, significant consequences of multimodality therapies exist with regard to general health, secondary late malignancies, reproduction and economic productivity (Sokoloff et al, 2007). A great need for understanding TGCT biology still exists to help curb its increasing incidence and potentially adopt effective prevention strategies.
Several genetic alterations have been shown in TGCTs. For example, abnormalities in the short arm of chromosome 12 (Mostert et al, 1998), as well as loss on chromosomes 1, 3, 5, 9, 11, 12q, 13q, 17p and 18q (Mathew et al, 1994; Lothe et al, 1995; al-Jehani et al, 1995; Honorio et al, 2003; Oosterhuis and Looijenga, 2005) have been reported. But major TSGs having a role in TGCTs are yet to be identified (Honorio et al, 2003). One of the most intriguing questions in the biology of TGCTs is how such distinct histological tumour subtypes (SE and NSE) can arise from the same cell type (cell of origin). Both subtypes exhibit similar cytogenetic abnormalities (Lutzker and Barnard, 1998). So far, molecular alterations that could distinct SEs from NSEs have not been clear yet. A pioneer study, performed in 1991, has shown that hypermethylation was abundant in NSEs, but not in SEs (Peltomaki, 1991). In 2002, Smiraglia et al (2002) demonstrated significant epigenetic differences between SEs and NSEs by a global methylation approach. In addition, studies of the X chromosome have demonstrated little or no methylation in SEs, and increased methylation in NSEs, particularly in more highly differentiated NSEs (Looijenga et al, 1997). Netto et al (2008) analysed global methylation status by immunohistochemistry and concluded that SEs cells generally retain the lack of methylation that occurs due to normal developmental erasure of methylation marks, whereas NSEs do not, thus showing more methylation. This was independently shown as well (Wermann et al, 2010).
In the present study, we used a candidate gene approach to investigate the methylation profile of 57 primary TGCT (of which 43 were SEs and 14 were NSEs) and 23 normal testis (NT) by quantitative methylation-specific PCR (QMSP). QMSP has been successfully used in other tumour models and has the benefit of providing accurate and precise data regarding the level of methylation in the various tumours. Six of the genes we evaluated, including ARF, APC, MGMT, RAR-β2, CCNA1 and hMLH1, were previously shown to be aberrantly methylated in TGCT (Muller et al, 2000; Koul et al, 2002; Honorio et al, 2003; Olasz et al, 2005; Lind et al, 2007). The remaining nine genes we studied has been found to be methylated in other cancer types, but not yet tested in TGCT; these genes include AIM1, PGP9.5, S100A2, ER-α, ER-β, MCAM, VGF, FKBP4 and SSBP2. Among the later genes, four genes (MCAM, VGF, FKBP4 and SSBP2) were recently discovered by our group, using a pharmacological unmasking strategy in other cancer types (Hoque et al, 2008). We compared the promoter methylation profiles of SEs and NSEs along with NT to better understand the role of epigenetic silencing in testis tumourigenesis. Relationships between methylation values and clinicopathological parameters were further assessed.
Materials and methods
Study cohort
The TGCT and NT samples were retrospectively collected from the Johns Hopkins Medical Institutions tissue archive. To be included in the cohort, an eligible patient had to have a confirmed diagnosis of TGCT (or normal tissue) and a sufficient amount of archived tumour (or normal) material to allow for DNA extraction (tissue preserved in sectioned blocks; >50% of tumour cells). A total of 75% (43 out of 57) of our TGCT samples were SEs and 25% (14 out of 57) were NSEs. We also analysed 23 NT from archived samples (testis removed on castration procedures). Thus, a total of 80 samples (57 tumours and 23 normals) were tested for methylation pattern by QMSP. Demographic and clinical information was obtained from the computerised tumour registry at the Johns Hopkins Healthcare System. The tumours were classified according to the WHO Classification of Tumours, Pathology and Genetics of the Urinary System and Male Genital Organs (Looijenga, 2009). Patient characteristics included in this study are summarised in Table 1. Approval for research on human subjects was obtained from the Johns Hopkins University Institutional Review Boards. This study qualified for exemption under the US Department of Health and Human Services policy for protection of human subjects (45 CFR 46.101(b)) (IRB 03-11-12-06e).
Table 1. Demographic and clinical characteristics of testicular cancer patients (N=57, seminomas=43, non-seminomas=14) and normals (N=24).
| Characteristic |
Number of patients (%)
|
||
|---|---|---|---|
| Hystologic diagnosis | Seminoma | Non-seminoma | Normal |
| Total samples | 43 (100) | 14 (100) | 23 (100) |
| Site | |||
| Right | 25 (58.1) | 10 (71.4) | 5 (21.7) |
| Left | 18 (41.9) | 4 (28.6) | 3 (13.1) |
| Unknown | 0 (0) | 0 (0) | 15 (65.2) |
| Age (years) | |||
| Median age (range) | 34 (22–62) | 27.5 (23–38) | 55 (20–81) |
| Race | |||
| Caucasian | 35 (81.4) | 10 (71.4) | 18 (78.3) |
| African American | 4 (9.3) | 2 (14.2) | 5 (21.7) |
| Others | 4 (9.3) | 3 (21.4) | 0 (0) |
| Stage | |||
| I | 40 (93.0) | 6 (42.9) | |
| II | 3 (7.0) | 5 (35.7) | N/Aa |
| III | 0 (0) | 3 (21.4) | |
N/A=Not applicable.
Gene selection
A total of 15 genes were selected for promoter methylation status analysis. Among these genes, six were previously assessed on testicular tissues (ARF, CCNA1, MGMT, MLH1, APC and RAR-β2; Muller et al, 2000; Koul et al, 2002; Honorio et al, 2003; Olasz et al, 2005; Lind et al, 2007), five were not yet tested in this tissue type (AIM1, PGP9.5, S100, ER-α and ER-β; Fiegl et al, 2004; Brait et al, 2008; Carvalho et al, 2008), and four were recently identified genes by our pharmacological unmasking strategy in different cancer types (MCAM, VGF, FKBP4 and SSBP2) (Hoque et al, 2008).
DNA extraction
After initial patient de-identification, all original histological slides from the TGCT or normal specimens were reviewed by a pathologist to reconfirm the diagnosis. A representative block was retrieved for DNA extraction. Microdissected NT samples from autopsy material were used for controls. Histological slides from the formalin-fixed, paraffin-embedded tissue were prepared. Slides were microdissected to obtain >50% neoplastic cells. DNA was extracted using standard protocols as previously described (Brait et al, 2008). Briefly, DNA was obtained by digestion with 50 μg ml−1 proteinase K (Roche, Indianapolis, IN, USA) in the presence of 1% SDS at 48 °C for 2 days, followed by phenol/chloroform extraction and ethanol precipitation, and finally dissolved in 20 μl of LoTE (2.5 mmol l−1 EDTA and 10 mmol l−1 Tris-HCL) and stored at −20 °C until used.
Sodium bisulphite treatment
DNA extracted from tumour or normal tissue was subjected to bisulphite treatment with the EpiTect Bisulfite kit (QIAGEN, Valencia, CA, USA), according to the manufacturer's instructions. Treated DNA was stored at −80 °C until used.
Methylation analysis
Bisulphite-modified DNA was used as a template for fluorescence-based real-time PCR, as previously described (Brait et al, 2009). Amplification reactions were carried out in triplicate in a final volume of 20 μl containing 1 μl of bisulfite-modified DNA, 600 nM concentrations of forward and reverse primers, 200 nM probe, 0.6 U of platinum Taq polymerase (Invitrogen, Frederick, MD, USA), 200 μM concentrations each of dATP, dCTP, dGTP and dTTP, and 6.7 mM MgCl2. Primers and probes were designed to specifically amplify the promoters of the 15 genes of interest and the promoter of a reference gene, β-actin; primer and probe sequences, and annealing temperatures are provided in Supplementary Table 1. Amplifications were carried out using the following profile: 95 °C for 3 min, followed by 50 cycles at 95 °C for 15 s and 60 °C for 1 min. Amplification reactions were carried out in 384-well plates in a 7900HT sequence detector (Applied Biosystems, Foster City, CA, USA) and were analysed by a sequence detector system (SDS 2.3; Applied Biosystems). Each plate included studied DNA samples, positive (in-vitro methylated leukocyte DNA) and negative (normal leukocyte DNA or DNA from a known unmethylated cell line) controls and multiple water blanks. Leukocyte DNA from a healthy individual was methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc., Beverly, MA, USA) to generate completely methylated DNA, and serial dilutions (90–0.009 ng) of this DNA were used to construct a calibration curve for each plate. All samples were within the assay's range of sensitivity and reproducibility, based on the amplification of the internal reference standard (threshold cycle (CT) value for β-actin of 40). The relative level of methylated DNA for each gene in each sample was determined as a ratio of methylation-specific PCR-amplified gene to β-actin (reference gene), and then multiplied by 1000 for easier tabulation (average value of triplicates of the gene of interest divided by the average value of triplicates of β-actin × 1000).
This measure represents the relative level of methylation in a particular sample and was used for direct comparison of samples. For a tumour sample to be considered methylated at a specific gene, it had to meet two specific criteria. Amplification must have been present in at least two of the three reaction wells in the triplicate run, and the β-actin-normalised mean methylation value must have fallen within the range of the serial standard curve dilutions. The lack of DNA contamination was verified by the absence of amplification of a distilled water negative control for each QMSP run.
Statistical analysis
The primary objective in this study was to describe the methylation patterns of 15 well-established or putative TSGs in NT and two TGCT types, SE and NSE.
Gene methylation was treated as a continuous variable. Univariate and multivariable logistic regression models were constructed sequentially to examine the association of gene methylation with the disease status (tumour vs normal). Odds ratios (OR) were estimated with 95% confidence intervals (CI), which quantified the strength of the association and its uncertainty. The final model was selected by using the backward selection method in multivariable logistic regression.
Correlation in methylation between genes was also examined so as to aid in the selection of genes that independently contributed to distinguishing tumour from normal samples (or the two subtypes: SEs and NSEs). Receiver-operating characteristic (ROC) analyses were conducted to evaluate the marker validity to differentiate tumour and normal. Sensitivity, specificity and area under the curve (AUC) were estimated along with 95% CI. Correlated ROC curves were compared using DeLong's method, which is a non-parametric approach (DeLong et al, 1988).
On the basis of the correlations among all the 15 genes obtained using a non-parametric approach along with the individual gene performance, the genes shown to be independent of each other by Spearman correlation test, were selected to be included in a logistic regression model. This model was used to assess the probability of tumour. The genes explored further for panel performance were those with P<0.10 in the final model. Multi-variable logistic regression was performed with genes of interest, as well as patient characteristics (age, race and site), entered the model simultaneously and were eliminated by backwards selection. An association was considered statistically significant with P-value <0.05. All P-values reported are two-sided.
The optimal cutoff was determined as the point at which it simultaneously maximised sensitivity and specificity. We used cancer cases and NT samples for generating ROC curves for individual genes, and the cut point of methylation values were established with the values that optimally differentiate the two sample groups. Representative ROC curves are available in Supplementary Figure 1.
Results
Patients with TGCT were aged between 22 and 62 years (median=33). Age range of controls was 20 to 81 (median=55). Patients with SE were aged between 22 and 62 years (median=34), and for NSE, were between 23 and 38 years (median=27.5). Control subjects were significantly older compared with patients with overall TGCT (P=0.05, Wilcoxon non-parametric test). The samples include 46 stage-I TGCT and 11 ⩾stage-II TGCT. Thirty-five TGCT analysed were from right site and 22 from left site. The majority of the samples were obtained from Caucasian. Patient characteristics included in this study are summarised in Table 1.
Overall methylation frequency in TGCTs
A total of 15 genes (AIM1, ARF, CCNA1, MGMT, hMLH1, PGP9.5, S100A2, APC, RAR-β2, ER-α, ER-β, MCAM, VGF, FKBP4 and SSBP2) were analysed for promoter methylation using QMSP in 57 tumour samples, comprising 43 SEs and 14 NSEs, and 23 NT samples. The observed frequencies of each gene using optimal cutoffs that maximised specificity and sensitivity are summarised in Table 2.
Table 2. Promoter methylation frequency in TGCT and in normal testicular samples.
| Genes | Cutoff a | Number of tumors with methylation/total number of tumors (%, 95% CI) | Number of tumors with methylation/total number of tumors (%, 95% CI) |
|---|---|---|---|
| AIM1 | 44.817 | 3/57 (5, 1.1–14.6) | 1/23 (4.3, 0.1–22.0) |
| ARF | 0.3 | 2/57 (3.5, 0.4–12.1) | 2/23 (8.7, 1.1–28.0) |
| CCNA1 | 8.173 | 5/57 (8.8, 2.9–19.3) | 0/23 (0, 0–14.8) |
| MCAM | 1073 | 23/57 (40.4, 27.6–54.2) | 9/23 (39.1, 19.7–61.5) |
| MGMT | 0.193 | 8/57 (14, 6.3–25.8) | 0/23 (0, 0–14.8) |
| MLH1 | 0.096 | 22/57 (38.6, 26.0–52.4) | 1/23 (4, 0.1–22.0) |
| PGP9 | 177.09 | 2/57 (3.5, 0.4–12.1) | 0/23 (0, 0–14.8) |
| S100 | 858.15 | 23/57 (40.4, 27.6–54.2) | 19/23 (83, 61.2–95.1) |
| SSBP2 | 623.99 | 8/57 (14, 6.3–25.8) | 16/23 (69.5, 47.1–86.8) |
| APC | 19.077 | 7/57 (12.3, 5.1–23.7) | 19/23 (83, 61.2–95.1) |
| RAR-β2 | 14.273 | 1/57 (1.8, 0–9.4) | 0/23 (0, 0–14.8) |
| VGF | 0.023 | 14/57 (24.6, 14.1–37.8) | 0/23 (0, 0–14.8) |
| ER-α | 72.072 | 6/57 (10.5, 4.0–21.5) | 10/23 (43.5, 23.2–65.5) |
| ER-β | 10.145 | 13/57 (22.8, 12.7–35.8) | 7/23 (30, 13.2–52.9) |
| FKBP4 | 35.001 | 7/57 (12, 5.1–23.7) | 0/23 (0, 0–14.8) |
Abbreviations: CI=confidence intervals; TGCT=testicular germ cell tumors.
The optimal cutoff was determined as the point at which it simultaneously maximized sensitivity and specificity.
VGF show a significant cancer-specific methylation (P=0.008, by Fisher's exact test). A total of 14 out of 57 TGCT showed methylation of the promoter region of VGF, whereas no methylation was observed in NT. hMLH1 was significantly methylated in TGCT (39% (22 out of 57)) in comparison with normal (4% (1 out of 23)); P=0.002, by Fisher's exact test).
Methylation in SE and NT
A total of 15 genes (AIM1, ARF, CCNA1, MGMT, hMLH1, PGP9.5, S100A2, APC, RAR-β2, E-Rα, ER-β, MCAM, VGF, FKBP4 and SSBP2) was analysed for promoter methylation in 23 NT and 43 SE cases by QMSP. Results for all genes in all SE cases and controls (NTs) are presented in Table 3a. Individual gene sensitivity and specificity was determined on the basis of an empiric cutoff values by maximising sensitivity and specificity, and the AUC values were calculated for each gene. At least one TSG locus was methylated in 37 out of 43 SE cases (86%), and a total of 11 out of 43 (25.6%) tumour samples showed methylation at three or more of the loci. By a non-parametric statistical approach (DeLong's method), we determined the correlation of methylation events among all the 15 genes. By Spearman correlation test, eight genes (AIM1, CCNA1, MCAM, PGP9.5, hMLH1, ARF, VGF and APC) showed to be independent of each other and were selected to be included in a logistic regression model, which predict the probability of tumour. Patient characteristics (age, race and site) were included in the multi-variable logistic regression model, together with the previously mentioned eight genes. Smoking status and alcohol consumption were not considered in this analysis, as there were too many unknowns reported. Backward selection was used and the significance level of staying in the final model was set at P<0.10. Among all eight genes, only APC and MCAM remained statistically significant and stayed in the final model (P=0.057 and P=0.035, respectively) after controlling for patient characteristics (age, race and site). We then grouped the independent genes in different combinations (AIM1, CCNA1, MCAM, PGP9.5, hMLH1, ARF, VGF and APC), considering optimal cutoffs that separated both groups, and established the maximum possible specificity at 78%, and each combination had a slight different sensitivity, with no significant improvement as shown in Table 3b.
Table 3a. Seminomas and normal testicular samples: (a) promoter methylation frequency in seminomas and normal testicular samples.
| Seminoma | Normal | ||
|---|---|---|---|
| Genes | Cutoff | Number of tumors with methylation/total number of tumors (%, 95% CI) | Number of tumors with methylation/total number of tumors (%, 95% CI) |
| AIM1 | 44.817 | 0/43 (0, 0–8.2) | 1/23 (4.3, 0.1–22.0) |
| ARF | 0.3 | 5/43 (11.6, 3.9–25.1) | 2/23 (8.7, 1.1–28.0) |
| CCNA1 | 8.173 | 2/43 (4.6, 0.6–15.8) | 0/23 (0, 0–14.8) |
| MCAM | 1073 | 14/43 (32.6, 19.1–48.5) | 9/23 (39.1, 19.7–61.5) |
| MGMT | 0.193 | 1/43 (2.3, 0.1––12.3) | 0/23 (0, 0–14.8) |
| MLH1 | 0.096 | 12/43 (27.9, 15.3–43.7) | 1/23 (4, 0.1–22.0) |
| PGP9.5 | 177.09 | 4/43 (9.3, 2.6–22.1) | 0/23 (0, 0–14.8) |
| S100 | 858.15 | 0/43 (0, 0–8.2) | 19/23 (83, 61.2–95.1) |
| SSBP2 | 623.99 | 0/43 (0, 0–8.2) | 16/23 (69.5, 47.1–86.8) |
| APC | 19.077 | 5/43 (11.6, 3.9–25.1) | 19/23 (83, 61.2–95.1) |
| RAR-β2 | 14.273 | 1/43 (2.3, 0.1–12.3) | 0/23 (0, 0–14.8) |
| VGF | 0.023 | 7/43 (16.3, 6.8–30.7) | 0/23 (0, 0–14.8) |
| ER-α | 72.072 | 8/43 (18.6, 8.4–33.4) | 10/23 (43.5, 23.2–65.5) |
| ER-β | 10.145 | 12/43 (27.9, 15.3–43.7) | 7/23 (30, 13.2–52.9) |
| FKBP4 | 35.001 | 1/43 (2.3, 0.1–12.3) | 0/23 (0, 0–14.8) |
Table 3b. Seminomas and normal testicular samples: (b) analysis of gene panels, based on logistic regression model, considering optimal cutoff for differenciating both groups.
| Panel | Cutoff based on predictive probability of tumor a | Sensitivity, % (95% CI) | Specificity, % (95% CI) | AUC (95% CI) |
|---|---|---|---|---|
| MCAM, APC | 0.643971 | 76.7 (61.4–88.2) | 78.3 (56.3–92.5) | 0.81 (0.70–0.92) |
| MCAM, APC, MLH1 | 0.620704 | 79.1 (64.0–90.0) | 78.3 (56.3–92.5) | 0.84 (0.74–0.94) |
| MCAM, APC, MLH1, AIM1 | 0.601593 | 81.4 (66.6–91.6) | 78.3 (56.3–92.5) | 0.85 (0.75–0.95) |
| MCAM, APC, MLH1, AIM1, PGP9.5 | 0.590917 | 81.4 (66.6–91.6) | 78.3 (56.3–92.5) | 0.86 (0.76–0.95) |
Abbreviations: AUC=area under the curve; CI=confidence interval.
Predictive probabilities were obtained from the logistic regression models.
Methylation in NSE and NT
The frequency of promoter methylation, including the cutoff value at each gene included in this panel is listed in Tables 4a. Briefly the methylation frequency were: AIM1 21%, ARF 0%, CCNA1 14%, MCAM 71%, MGMT 50%, MLH1 71%, PGP9.5 14%, S100A2 57%, SSBP2 57%, APC 50%, RAR-β2 21%, VGF 50%, ER-α 36%, ER-β 64% each and FKBP4 36%.
Table 4a. Non-seminomas and normal testicular samples: (a) promoter methylation frequency in non-seminomas and normal testicular samples.
| Non-seminoma | Normal | ||
|---|---|---|---|
| Genes | Cutoff | Number of tumors with methylation/total number of tumors (%, 95% CI) | Number of tumors with methylation/total number of tumors (%, 95% CI) |
| AIM1 | 44.817 | 3/14 (21.4, 4.7–50.8) | 1/23 (4.3, 0.1–22.0) |
| ARF | 0.3 | 0/14 (0, 0- 6.9) | 2/23 (8.7, 1.1–28.0) |
| CCNA1 | 8.173 | 2/14 (14.3, 1.8–42.8) | 0/23 (0, 0–14.8) |
| MCAM | 1073 | 10/14 (71.4, 41.9–91.6) | 9/23 (39.1, 19.7–61.5) |
| MGMT | 0.193 | 7/14 (50, 23.0–77.0) | 0/23 (0, 0–14.8) |
| MLH1 | 0.096 | 10/14 (71.4, 41.9–91.6) | 1/23 (4, 0.1–22.0) |
| PGP9 | 177.09 | 2/14 (14.3, 1.8–42.8) | 0/23 (0, 0–14.8) |
| S100 | 858.15 | 8/14 (57.1, 28.9–82.3) | 19/23 (83, 61.2–95.1) |
| SSBP2 | 623.99 | 8/14 (57.1, 28.9–82.3) | 16/23 (69.5, 47.1–86.8) |
| APC | 19.077 | 7/14 (50, 23.0–77.0) | 19/23 (83, 61.2–95.1) |
| RAR-β2 | 14.273 | 3/14 (21.4, 4.7–50.8) | 0/23 (0, 0–14.8) |
| VGF | 0.023 | 7/14 (50, 23.0–77.0) | 0/23 (0, 0–14.8) |
| ER-α | 72.072 | 5/14 (35.7, 12.8–64.9) | 10/23 (43.5, 23.2–65.5) |
| ER-β | 10.145 | 9/14 (64.3, 35.1–87.2) | 7/23 (30, 13.2–52.9) |
| FKBP4 | 35.001 | 5/14 (35.7, 12.8–64.9) | 0/23 (0, 0–14.8) |
Abbreviation: CI=confidence interval.
On the basis of the correlations among all the 15 genes obtained using a non-parametric approach along with the individual gene performance, six genes (APC, VGF, MGMT, hMLH1, ER-β and FKBP4) were shown to be independent of each other and each of them shows the strongest predictive potential representing the corresponding cluster, and thus, were selected to be included in a multivariable logistic regression model that modelled the probability of tumour. However, issues arose with such small sample size, that is, for VGF, MGMT and FKBP4, the maximum likelihood estimates are not possible, as there is no variation in methylation values for NT samples (e.g., all normal samples had zero methylation values for each of these three genes). Thus, only hMLH1, APC and ER-β were considered in the multivariable logistic model. It turned out that hMLH1 remained statistically significant in this final model (P=0.044), which indicated its independent effect in predicting tumour. Performance of hMLH1 combined with APC and ER-β was explored as well. The significance of hMLH1 was examined with adjustment for the current available patient characteristics (age, race and site), using the logistic regression model. The independent effect of the genes of interest was evaluated with adjustment for age only. Results indicated that association of hMLH1 hypermethylation with tumour remained positive (OR=8.29; 95% CI=0.66–103.5), albeit no statistical significance (P=0.100). Results should be interpreted with caution with an imprecise estimate for strength of association (i.e., wide CI for the OR estimate). Table 4b shows the predictive performance of the three gene panels (hMLH1, APC and ER-β), considering optimal cutoffs that separated both groups. Supplementary Figure 2 illustrates the predictive powers of hMLH1 alone, as well as combination with APC and ER-β. There was no statistically significant improvement with the combined genes compared with hMLH1 alone, based on the non-parametric approach of predictive power of DeLong et al (1988).
Table 4b. Non-seminomas and normal testicular samples: (b) analysis of gene panels, based on logistic regression model, considering optimal cutoff for differentiating both groups.
| Combined genes | Cutoff based on predictive probability of tumor a | Sensitivity (%) | Specificity (%) | AUC (95% CI) |
|---|---|---|---|---|
| MLH1, APC, and ER-β | 0.225668 | 85.7 (57.2–98.2) | 82.6 (61.2–95.1) | 0.89 (0.75 – 1.00) |
Abbreviations: AUC=area under the curve; CI=confidence interval.
Predictive probabilities were obtained from the logistic regression models.
On the basis of the cutoff value determined for SE cases, six genes (CCNA1, MGMT, PGP9.5, RAR-β2, VGF and FKBP4) showed 100% specificity (no methylation in NT) in NSE cases. From this six genes, only one gene was methylated in 5 out of 14 (35.7%) cases, two genes were methylated 4 out of 14 (28.5%) cases, one sample showed methylation in three genes, 1 out of 14 (7.1%), and only one sample showed methylation in four genes, 1 out of 14 (7.1%); no samples showed methylation in five genes, and one sample out of 14 (7.1%) had all six genes methylated. At least one of these six genes was observed methylated in 12 out of 14 (86%) of the samples with 100% specificity for NSEs.
Methylation levels and frequency across sample types
When dividing the groups into normal, SEs and NSEs, one would expect to see a differential methylation of the analysed genes, as methylation is tissue specific and cancer specific. The non-parametric test (DeLong's method) was used to test for differential methylation frequency (in a binary fashion), and five genes met statistical significance (APC, MGMT, hMLH1, ER-β and FKBP4). This test accounted for the normalised methylation level as a continuous variable. MGMT, VGF, CCNA1 and FKBP4 are interesting, as they could be specific for tumours as all of the normal samples showed no methylation, and in addition, hMLH1 that had only one positive in normal. There was methylation in normal, SE and NSE in similar frequencies for the following genes: ARF, S100A2, SSBP2, ER-α and ER-β. Interestingly, SSBP2 and ER-α showed higher levels and frequency of methylation seen in the normal samples than in SE and NSE.
As supported by the previous studies (Peltomaki, 1991; Smiraglia et al, 2002; Netto et al, 2008), we found differential methylation pattern in SE and NSE. MGMT, VGF, ER-β and FKBP4 are predominately methylated in NSEs when compared with SEs. APC and hMLH1 are shown to be methylated in both subtypes, but APC exhibits not only a higher frequency, but also higher levels, and hMLH1 a significantly higher frequency. When combining APC, hMLH1, ER-β and FKBP4 (considering empiric cutoffs), it is possible to identify 86% of the NSEs, whereas only 7% of the SEs (AUC 0.90 (0.81–1.00)).
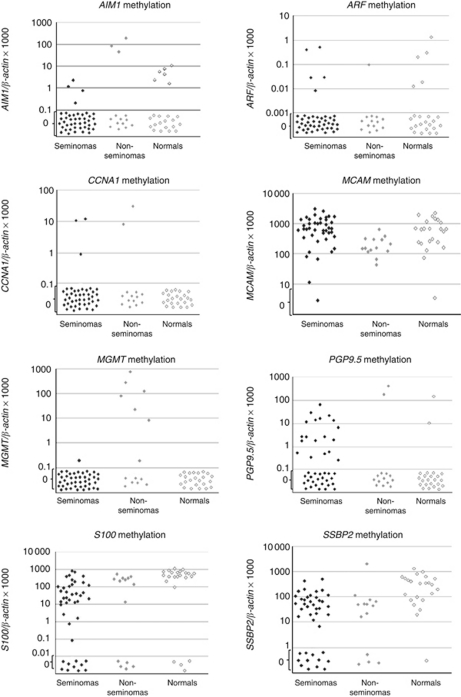
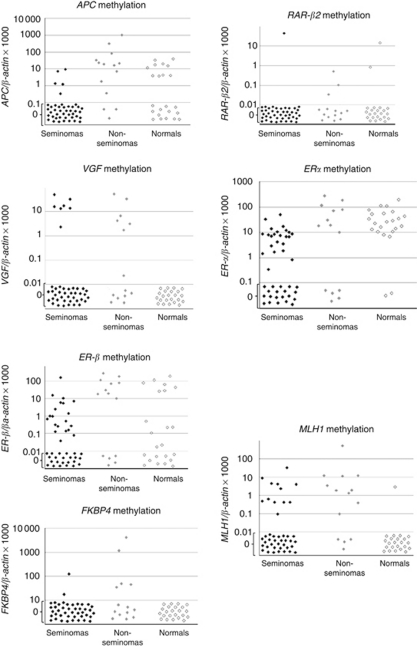
The summary data for the comparison of SEs and NSEs is in Table 5a and b Representative scatter plots of methylation of the 15 genes analysed throughout the testicular tissues are shown in Figure 1.
Table 5a. Seminomas and non-seminoma testicular samples: (a) promoter methylation frequency in seminomas and non-seminoma testicular samples.
| Non-seminoma | Seminoma | ||
|---|---|---|---|
| Genes | Cutoff | Number of tumors with methylation/total number of tumors (%, 95% CI) | Number of tumors with methylation/total number of tumors (%, 95% CI) |
| AIM1 | 44.817 | 3/14 (21.4, 4.7–50.8) | 0/43 (0, 0–8.2) |
| ARF | 0.3 | 0/14 (0, 0–6.9) | 5/43 (11.6, 3.9–25.1) |
| CCNA1 | 8.173 | 2/14 (14.3, 1.8–42.8) | 2/43 (4.6, 0.6–15.8) |
| MCAM | 1073 | 10/14 (71.4, 41.9–91.6) | 14/43 (32.6, 19.1–48.5) |
| MGMT | 0.193 | 7/14 (50, 23.0–77.0) | 1/43 (2.3, 0.1–12.3) |
| MLH1 | 0.096 | 10/14 (71.4, 41.9–91.6) | 12/43 (27.9, 15.3–43.7) |
| PGP9 | 177.09 | 2/14 (14.3, 1.8–42.8) | 4/43 (9.3, 2.6–22.1) |
| S100 | 858.15 | 8/14 (57.1, 28.9–82.3) | 0/43 (0, 0–8.2) |
| SSBP2 | 623.99 | 8/14 (57.1, 28.9–82.3) | 0/43 (0, 0–8.2) |
| APC | 19.077 | 7/14 (50, 23.0–77.0) | 5/43 (11.6, 3.9–25.1) |
| RAR-β2 | 14.273 | 3/14 (21.4, 4.7–50.8) | 1/43 (2.3, 0.1–12.3) |
| VGF | 0.023 | 7/14 (50, 23.0–77.0) | 7/43 (16.3, 6.8–30.7) |
| ER-α | 72.072 | 5/14 (35.7, 12.8–64.9) | 8/43 (18.6, 8.4–33.4) |
| ER-β | 10.145 | 9/14 (64.3, 35.1–87.2) | 12/43 (27.9, 15.3–43.7) |
| FKBP4 | 35.001 | 5/14 (35.7, 12.8–64.9) | 1/43 (2.3, 0.1–12.3) |
Abbreviation: CI=confidence interval.
Table 5b. Seminomas and non-seminoma testicular samples: (b) analysis of gene panels, based on logistic regression model, considering optimal cutoff for differenciating both groups.
| Combined genes | Cutoff based on predictive probability of tumor a | Sensitivity (%) | Specificity (%) | AUC (95% CI) |
|---|---|---|---|---|
| APC | 0.3644 | 78.6 (49.2–95.3) | 90.7 (77.9–97.4) | 0.87 (0.74–0.99) |
| APC, MLH1, ER-β and FKBP4 | 0.124726 | 85.7 (57.2–98.2) | 93.0 (80.9–98.5) | 0.90 (0.81–1.00) |
Abbreviations: AUC=area under the curve; CI=confidence interval.
Predictive probabilities were obtained from the logistic regression models.
Figure 1.
Representative scatter plots of methylation values of tested genes in seminomas, non-seminomas and normal testis samples. Genes: AIM1, ARF, CCNA1, MGMT, hMLH1, PGP9.5, S100A2, APC, RAR-β2, ER-α, ER-β, MCAM, VGF, FKBP4 and SSBP2. Calculation of the gene of interest: ratios were based values for both the gene of interest and β-actin obtained by quantitative real-time PCR analysis. The obtained ratios were multiplied by 1000 for easier tabulation. Values designated as 0.1 are 0 values, which cannot be plotted correctly on a log scale.
Association between DNA methylation changes and clinicopathological factors
There were no apparent correlations between any of the gene tested with any clinicopathological parameters, including age, site, race and disease stages, perhaps due to the limited number of sample size (data not shown). As shown before, we found differential methylation patterns in two different histological types (SEs and NSEs). Methylation values were compared as continuous variables, as well as dichotomised, and no correlation was observed in either analysis.
Discussion
In this study, we attempted to define a set of methylation markers that would allow for an accurate discrimination among the two most common types of TGCT (SE and NSE); as each type displays dissimilar clinical behaviour and successful pre-operative cytological or histological assessment is restricted. Through gene promoter methylation profiling with QMSP, six genes were found to be differentially methylated in the two tumour types. In particular, higher PGP9.5 methylation frequency was detected in SEs than NSEs, whereas high methylation frequency of MGMT, VGF, ER-β and FKBP4 were associated with NSE. Remarkably, both SE and NSE were methylated for APC and hMLH1. Netto et al (2008) showed evidence that SEs remain unmethylated, whereas the other histological types arise after de novo methylation, suggesting that they may arise in distinct periods of the development process of the germ cells, and that their genome methylation status may determine the degree of differentiation of the cells. Wermann et al (2010) also observed the same pattern of all SEs being unmethylated (as well as tumours of similar histology originating in other organs), whereas NSEs were consistently hypermethylated. Koul et al, (2002), using conventional MSP, analysed a panel of 21 genes and observed a near absence of methylation in SEs and a higher percentage in NSEs, suggesting a role for different panel of methylation-induced inactivation of TSGs in two common types of TGCT. Similar to our results, they also observed presence of methylation in the MGMT promoter in NSEs (21 vs 14% observed in this study) and absence in SEs. The other two genes that showed this same pattern were not evaluated by our study (BRCA1 and RASSF1A), and their panel did not include the other genes observed to be methylated in NSEs vs SEs (VGF, ER-β and FKBP4). Subtype-specific patterns of global methylation were also previously reported in TGCT (Smiraglia et al, 2002). Peltomaki (1991) observed hypomethylation in SEs, whereas NSEs were largely hypermethylated. The same observation was made in studies of the X chromosome (Looijenga et al, 1997). In breast cancer, Lehmann et al (2002) observed that the presence of promoter methylation in DAPK was frequent in invasive lobular cancer (53%) and not frequent in another histological subtype (9% invasive ductal carcinoma). Other reports that include different epigenetic alterations in histological subtypes of the same cancer type include: presence of methylator phenotype in low-grade gliomas when compared with de novo glioblastomas (Laffaire et al, 2011), high frequency of SFN methylation in small cell lung cancer, whereas a rare frequency in non-small cell lung cancer (Osada et al, 2002). Consistent with epigenetic heterogeneity, there is a wide search for genetic alterations that would be able to distinguish SEs and NSEs. Coffey et al (2008) reported that Kit mutations are predominant in SEs and very rarely observed in NSEs. Major accentuated genetic differences are yet to be discovered.
Previous studies did indicate a role for hypermethylation of TSG promoters in the pathogenesis of SE. In our data, when comparing SEs versus NT samples, the genes that showed higher frequency of methylation in SE were PGP9.5, hMLH1 and ER-β, when compared with the observed frequency in normal samples. In combination, the strongest pair is APC and MCAM, showing 78.3% frequency in tumours and 23.3% in normals. With the same cutoff, adding hMLH1, the frequency in tumours was 79.1%, and adding both AIM1 and PGP9.5, or only one of them, it increases to 81.4%, with the same percentage in normals (23.3%). This panel containing four genes showed reasonable percentages to distinguish normal and tumour tissue.
MCAM had not been previously evaluated in TGCT. In prostate cancer, other showed 85% methylation in tumours vs 0% in normal prostate (Hoque et al, 2008). To our knowledge, PGP9.5 and AIM1 had not been analysed in this tumour type. APC was previously observed with 10% of methylation in tumours and 0% in NT, but both positive tumours were NSE (Koul et al, 2002), and no APC methylation has been reported in Ses; it is important to emphasise that this study was performed using conventional MSP, a less sensitive technique than QMSP used in the present study (Hoque et al, 2004), which may explain the different percentages observed. In this same study, hMLH1 showed only 4% methylation in tumours and 0% in normals, occurring in both SEs and NSEs.
In respect to the comparison between NSEs and normals, there are two genes (MGMT and VGF) that stand out alone, both with a 0% frequency in normals and 50% frequency in NSEs. VGF is a gene recently reported to be methylated in cancer by our group (Hoque et al, 2008) and had never been analysed in TGCT. MGMT had been reported to be frequently methylated in TGCT (Smith-Sorensen et al, 2002), showing 69% in NSEs. In 2004, Koul et al (2004) showed methylation in MGMT in 20% of NSEs analysed by conventional MSP, a less sensitive technique as mentioned earlier. Using empiric cutoffs, at least one of the three genes (hMLH1, APC and ER-β) was methylated in 13 out of 14 (93%) of NSE cases and 10 out of 23 (43%) of the NT samples. Previously reported methylation of APC and hMLH1 were 10% to less than 5%, respectively (Koul et al, 2002). There are no reported studies exploring methylation status of ER-β in TGCT. The downregulation of ERβ was observed in SEs by Esposito et al (2010); they detected decreased expression of ERβ protein in SEs when compared with NT, but they did not evaluate promoter methylation. This gene encodes a member of the family of oestrogen receptors and superfamily of nuclear receptor transcription factors. The gene product contains an N-terminal DNA-binding domain and C-terminal ligand-binding domain, and is localised to the nucleus, cytoplasm and mitochondria. It is able to interact with specific DNA sequences to activate transcription. Some isoforms dominantly inhibit the activity of other oestrogen receptor family members.
Epigenetic inactivation of hMLH1 is found in a wide range of cancers. hMLH1 is a DNA mismatch repair (MMR) gene and is an essential component of the DNA MMR pathway, and is frequently mutated in hereditary non-polyposis colon cancer also known as Lynch syndrome. Activation of the MMR pathway may trigger DNA damage signalling, a process which induces cell cycle arrest and can lead to cell death in case of major DNA damages (for review, please see Jiricny (2006)). hMLH1 is the most prominent target of epigenetic silencing in the MMR pathway in sporadic tumours, comprising ovarian, head and neck, breast and colorectal cancer (Herman et al, 1998). However, hypermethylation of hMLH1 is often associated with other hypermethylated genes, which complicates mechanistic interpretation of associations with response to therapy in patients (Shen et al, 2007). Mechanistic investigations using in vitro system have shown that treatment of hMLH1-methylated colon cancer cell lines with the demethylating agent 5aza-2deoxycytidine (5-aza-dC) restores hMLH1 expression and subsequently renders the cells MMR proficient (Herman et al, 1998).
Promoter methylation of hMLH1 has been associated with chemoresistance to cisplatin-based therapies in ovarian cancer more than a decade ago (Strathdee et al, 1999). In our study, both SEs and NSEs displayed methylation in this locus, 27% and 71%, respectively. It is well established that NSE and SE (all TGCT) are widely responsive to cisplatin-based therapy (Bosl and Motzer, 1997), so one wouldn’t expect the high presence of hMLH1 methylation in this tumour type. Olasz et al (2005) hypothesised that this epigenetic alteration could be linked to the chemoresistance in a small group of testicular tumours, but could not observe any association. On the other hand, Honecker et al. (2009) investigated a larger cohort and observed that lack or low expression of hMLH1 (on the protein level) was significantly associated with cisplatin-resistant TGCTs, and the absence of expression was also correlated with presence of hMLH1 promoter methylation. Probably the mechanisms of resistance/sensitivity to this chemothepeutic agent are different among the ovarian and TGCT. It is widely accepted that hMLH1 dysfunction induces microsatellite instability (MSI) in various cancers, and loss of hMLH1 expression was related to its promoter methylation. However, no correlation was observed between hMLH1 methylation and MSI in TGCTs (Olasz et al, 2005). In our study, hMLH1 methylation was detected in 22 out of 57 (39%) of TGCTs. However, we did not perform expression or MSI analysis in these samples; therefore, we are not able to correlate hMLH1 methylation with expression and MSI. Esteller et al (2000) reported that promoter methylation of MGMT results in enhanced sensitivity to alkylating agents in gliomas. In the present study, MGMT was highly methylated in NSEs, which could indicate that the epigenetic silencing of this gene may be also linked to the cisplatin-based chemotherapy response, but this data needs to be further explored in a well-defined cohort consisting of cisplatin responsive group and non-responsive group.
Silencing TSG by DNA promoter methylation is an important feature of cancer. It is known that cancer types may vary in their epigenetic profiles in a tissue-specific pattern (Esteller et al, 2001). We used a candidate gene approach to test 15 promoter regions in a testicular cohort; all the markers evaluated here have shown differential methylation in malignant vs benign tissues in several cancer types (Hoque et al, 2008). Our data shows different patterns of methylation not only between tumours and normal, but also between the two histological subtypes of germ cell tumours. These distinct promoter methylation profiles of the two main subtypes of germ cell testicular cancer, SEs and NSEs, may shed a light on how they develop and differentiate from the same cell of origin. In conclusion, our study constitutes a comprehensive profile of hypermethylated genes in testicular tissues. Several of genes tested in this study were not evaluated for promoter methylation in this cancer types before, which may partially be due to rarity of the disease and limited number of samples. The observation of the higher proportion of promoter methylation of putative and established TSGs in NSEs when compared with SEs may enrich our knowledge of this tumour type. Further study using larger cohorts is needed to evaluate the potential use of these methylation markers in the detection, prognosis and therapeutic outcome of this rare tumour.
Acknowledgments
This work was supported by National Cancer Institute Grant 1R03CA128060-01A1 (MO Hoque), Career development award to Dr Hoque from Specialised Program of Research Excellence grant P50 CA098252 (PI: T-C Wu). Dr Hoque and Dr Begum are supported by a Young Clinical Scientist Award from the Flight Attendant Medical Research Institute. Dr Hoque is a paid consultant to Oncomethylome Sciences, SA. Dr Halachmi was supported by the Rosenblatt Cancer Research Fund, Technion Israeli Institute of Technology. This abstract was awarded the Scholar-in-Training Award at the 8th AACR-Japanese Cancer Association Joint Conference, Cancer Genomics, Epigenomics and the Development of Novel Therapeutics (2010).
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Al-Jehani RM, Povey S, Delhanty JD, Parrington JM (1995) Loss of heterozygosity on chromosome arms 5q, 11p, 11q, 13q, and 16p in human testicular germ cell tumors. Genes Chromosomes Cancer 13: 249–256 [DOI] [PubMed] [Google Scholar]
- Bosl GJ, Motzer RJ (1997) Testicular germ-cell cancer. N Engl J Med 337: 242–253 [DOI] [PubMed] [Google Scholar]
- Brait M, Begum S, Carvalho AL, Dasgupta S, Vettore AL, Czerniak B, Caballero OL, Westra WH, Sidransky D, Hoque MO (2008) Aberrant promoter methylation of multiple genes during pathogenesis of bladder cancer. Cancer Epidemiol Biomarkers Prev 17: 2786–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brait M, Ford JG, Papaiahgari S, Garza MA, Lee JI, Loyo M, Maldonado L, Begum S, McCaffrey L, Howerton M, Sidransky D, Emerson MR, Ahmed S, Williams CD, Hoque MO (2009) Association between lifestyle factors and CpG island methylation in a cancer-free population. Cancer Epidemiol Biomarkers Prev 18: 2984–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AL, Jeronimo C, Kim MM, Henrique R, Zhang Z, Hoque MO, Chang S, Brait M, Nayak CS, Jiang WW, Claybourne Q, Tokumaru Y, Lee J, Goldenberg D, Garrett-Mayer E, Goodman S, Moon CS, Koch W, Westra WH, Sidransky D, Califano JA (2008) Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res 14: 97–107 [DOI] [PubMed] [Google Scholar]
- Chia VM, Quraishi SM, Devesa SS, Purdue MP, Cook MB, McGlynn KA (2010) International trends in the incidence of testicular cancer, 1973-2002. Cancer Epidemiol Biomarkers Prev 19: 1151–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey J, Linger R, Pugh J, Dudakia D, Sokal M, Easton DF, Timothy Bishop D, Stratton M, Huddart R, Rapley EA (2008) Somatic KIT mutations occur predominantly in seminoma germ cell tumors and are not predictive of bilateral disease: report of 220 tumors and review of literature. Genes Chromosomes Cancer 47: 34–42 [DOI] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44: 837–845 [PubMed] [Google Scholar]
- Ellinger J, Albers P, Perabo FG, Muller SC, von Ruecker A, Bastian PJ (2009) CpG island hypermethylation of cell-free circulating serum DNA in patients with testicular cancer. J Urol 182: 324–329 [DOI] [PubMed] [Google Scholar]
- Esposito F, Boscia F, Franco R, Tornincasa M, Fusco A, Kitazawa S, Looijenga LH, Chieffi P (2010) Down-regulation of oestrogen receptor-beta associates with transcriptional co-regulator PATZ1 delocalization in human testicular seminomas. J Pathol 224: 110–120 [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG (2001) A gene hypermethylation profile of human cancer. Cancer Res 61: 3225–3229 [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG (2000) Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343: 1350–1354 [DOI] [PubMed] [Google Scholar]
- Fiegl H, Gattringer C, Widschwendter A, Schneitter A, Ramoni A, Sarlay D, Gaugg I, Goebel G, Muller HM, Mueller-Holzner E, Marth C, Widschwendter M (2004) Methylated DNA collected by tampons--a new tool to detect endometrial cancer. Cancer Epidemiol Biomarkers Prev 13: 882–888 [PubMed] [Google Scholar]
- Godmann M, Lambrot R, Kimmins S (2009) The dynamic epigenetic program in male germ cells: Its role in spermatogenesis, testis cancer, and its response to the environment. Microsc Res Tech 72: 603–619 [DOI] [PubMed] [Google Scholar]
- Herman JG (1999) Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol 9: 359–367 [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB (1998) Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 95: 6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes Jr L., Escalante C, Garrison O, Foldi BX, Ogungbade GO, Essien EJ, Ward D (2008) Testicular cancer incidence trends in the USA (1975-2004): plateau or shifting racial paradigm? Public Health 122: 862–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honecker F, Wermann H, Mayer F, Gillis AJ, Stoop H, van Gurp RJ, Oechsle K, Steyerberg E, Hartmann JT, Dinjens WN, Oosterhuis JW, Bokemeyer C, Looijenga LH (2009) Microsatellite instability, mismatch repair deficiency, and BRAF mutation in treatment-resistant germ cell tumors. J Clin Oncol 27: 2129–2136 [DOI] [PubMed] [Google Scholar]
- Honorio S, Agathanggelou A, Wernert N, Rothe M, Maher ER, Latif F (2003) Frequent epigenetic inactivation of the RASSF1A tumour suppressor gene in testicular tumours and distinct methylation profiles of seminoma and nonseminoma testicular germ cell tumours. Oncogene 22: 461–466 [DOI] [PubMed] [Google Scholar]
- Hoque MO, Begum S, Topaloglu O, Jeronimo C, Mambo E, Westra WH, Califano JA, Sidransky D (2004) Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res 64: 5511–5517 [DOI] [PubMed] [Google Scholar]
- Hoque MO, Kim MS, Ostrow KL, Liu J, Wisman GB, Park HL, Poeta ML, Jeronimo C, Henrique R, Lendvai A, Schuuring E, Begum S, Rosenbaum E, Ongenaert M, Yamashita K, Califano J, Westra W, van der Zee AG, Van Criekinge W, Sidransky D (2008) Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res 68: 2661–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich A, Shipley J, Huddart R (2006) Testicular germ-cell cancer. Lancet 367: 754–765 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics, 2010. CA Cancer J Clin 60: 277–300 [DOI] [PubMed] [Google Scholar]
- Jiricny J (2006) The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 7: 335–346 [DOI] [PubMed] [Google Scholar]
- Koul S, Houldsworth J, Mansukhani MM, Donadio A, McKiernan JM, Reuter VE, Bosl GJ, Chaganti RS, Murty VV (2002) Characteristic promoter hypermethylation signatures in male germ cell tumors. Mol Cancer 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul S, McKiernan JM, Narayan G, Houldsworth J, Bacik J, Dobrzynski DL, Assaad AM, Mansukhani M, Reuter VE, Bosl GJ, Chaganti RS, Murty VV (2004) Role of promoter hypermethylation in cisplatin treatment response of male germ cell tumors. Mol Cancer 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutanzi KR, Koturbash I, Kovalchuk O. (2010) Reversibility of pre-malignant estrogen-induced epigenetic changes. Cell Cycle 9: 3078–3084 [DOI] [PubMed] [Google Scholar]
- Laffaire J, Everhard S, Idbaih A, Criniere E, Marie Y, de Reynies A, Schiappa R, Mokhtari K, Hoang-Xuan K, Sanson M, Delattre JY, Thillet J, Ducray F. (2011) Methylation profiling identifies 2 groups of gliomas according to their tumorigenesis. Neuro Oncol 13(1): 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann U, Celikkaya G, Hasemeier B, Langer F, Kreipe H (2002) Promoter hypermethylation of the death-associated protein kinase gene in breast cancer is associated with the invasive lobular subtype. Cancer Res 62: 6634–6638 [PubMed] [Google Scholar]
- Lind GE, Skotheim RI, Lothe RA (2007) The epigenome of testicular germ cell tumors. Apmis 115: 1147–1160 [DOI] [PubMed] [Google Scholar]
- Looijenga LH (2009) Human testicular (non)seminomatous germ cell tumours: the clinical implications of recent pathobiological insights. J Pathol 218: 146–162 [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, van Gurp RJ, Verkerk AJ, Oosterhuis JW (1997) X inactivation in human testicular tumors. XIST expression and androgen receptor methylation status. Am J Pathol 151: 581–590 [PMC free article] [PubMed] [Google Scholar]
- Lothe RA, Peltomaki P, Tommerup N, Fossa SD, Stenwig AE, Borresen AL, Nesland JM (1995) Molecular genetic changes in human male germ cell tumors. Lab Invest 73: 606–614 [PubMed] [Google Scholar]
- Lutzker SG, Barnard NJ (1998) Testicular germ cell tumors: molecular understanding and clinical implications. Mol Med Today 4: 404–411 [DOI] [PubMed] [Google Scholar]
- Manton KJ, Douglas ML, Netzel-Arnett S, Fitzpatrick DR, Nicol DL, Boyd AW, Clements JA, Antalis TM (2005) Hypermethylation of the 5′ CpG island of the gene encoding the serine protease Testisin promotes its loss in testicular tumorigenesis. Br J Cancer 92: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew S, Murty VV, Bosl GJ, Chaganti RS (1994) Loss of heterozygosity identifies multiple sites of allelic deletions on chromosome 1 in human male germ cell tumors. Cancer Res 54: 6265–6269 [PubMed] [Google Scholar]
- Mostert MC, Verkerk AJ, van de Pol M, Heighway J, Marynen P, Rosenberg C, van Kessel AG, van Echten J, de Jong B, Oosterhuis JW, Looijenga LH (1998) Identification of the critical region of 12p over-representation in testicular germ cell tumors of adolescents and adults. Oncogene 16: 2617–2627 [DOI] [PubMed] [Google Scholar]
- Muller C, Readhead C, Diederichs S, Idos G, Yang R, Tidow N, Serve H, Berdel WE, Koeffler HP (2000) Methylation of the cyclin A1 promoter correlates with gene silencing in somatic cell lines, while tissue-specific expression of cyclin A1 is methylation independent. Mol Cell Biol 20: 3316–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netto GJ, Nakai Y, Nakayama M, Jadallah S, Toubaji A, Nonomura N, Albadine R, Hicks JL, Epstein JI, Yegnasubramanian S, Nelson WG, De Marzo AM (2008) Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol 21: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olasz J, Mandoky L, Geczi L, Bodrogi I, Csuka O, Bak M (2005) Influence of hMLH1 methylation, mismatch repair deficiency and microsatellite instability on chemoresistance of testicular germ-cell tumors. Anticancer Res 25: 4319–4324 [PubMed] [Google Scholar]
- Oosterhuis JW, Looijenga LH (2005) Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer 5: 210–222 [DOI] [PubMed] [Google Scholar]
- Osada H, Tatematsu Y, Yatabe Y, Nakagawa T, Konishi H, Harano T, Tezel E, Takada M, Takahashi T (2002) Frequent and histological type-specific inactivation of 14-3-3sigma in human lung cancers. Oncogene 21: 2418–2424 [DOI] [PubMed] [Google Scholar]
- Peltomaki P (1991) DNA methylation changes in human testicular cancer. Biochim Biophys Acta 1096: 187–196 [DOI] [PubMed] [Google Scholar]
- Rajpert-De Meyts E (2006) Developmental model for the pathogenesis of testicular carcinoma in situ: genetic and environmental aspects. Hum Reprod Update 12: 303–323 [DOI] [PubMed] [Google Scholar]
- Shen L, Catalano PJ, Benson III AB, O’Dwyer P, Hamilton SR, Issa JP (2007) Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res 13: 6093–6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiraglia DJ, Szymanska J, Kraggerud SM, Lothe RA, Peltomaki P, Plass C (2002) Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene 21: 3909–3916 [DOI] [PubMed] [Google Scholar]
- Smith-Sorensen B, Lind GE, Skotheim RI, Fossa SD, Fodstad O, Stenwig AE, Jakobsen KS, Lothe RA (2002) Frequent promoter hypermethylation of the O6-Methylguanine-DNA Methyltransferase (MGMT) gene in testicular cancer. Oncogene 21: 8878–8884 [DOI] [PubMed] [Google Scholar]
- Sokoloff MH, Joyce GF, Wise M (2007) Testis cancer. J Urol 177: 2030–2041 [DOI] [PubMed] [Google Scholar]
- Strathdee G, MacKean MJ, Illand M, Brown R (1999) A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene 18: 2335–2341 [DOI] [PubMed] [Google Scholar]
- Wermann H, Stoop H, Gillis AJ, Honecker F, van Gurp RJ, Ammerpohl O, Richter J, Oosterhuis JW, Bokemeyer C, Looijenga LH (2010) Global DNA methylation in fetal human germ cells and germ cell tumours: association with differentiation and cisplatin resistance. J Pathol 221: 433–442 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.