Abstract
Orchestration of signaling, photoreceptor structural integrity, and maintenance needed for mammalian vision remain enigmatic. By integrating three proteomic data sets, literature mining, computational analyses, and structural information, we have generated a multiscale signal transduction network linked to the visual G protein-coupled receptor (GPCR) rhodopsin, the major protein component of rod outer segments. This network was complemented by domain decomposition of protein–protein interactions and then qualified for mutually exclusive or mutually compatible interactions and ternary complex formation using structural data. The resulting information not only offers a comprehensive view of signal transduction induced by this GPCR but also suggests novel signaling routes to cytoskeleton dynamics and vesicular trafficking, predicting an important level of regulation through small GTPases. Further, it demonstrates a specific disease susceptibility of the core visual pathway due to the uniqueness of its components present mainly in the eye. As a comprehensive multiscale network, it can serve as a basis to elucidate the physiological principles of photoreceptor function, identify potential disease-associated genes and proteins, and guide the development of therapies that target specific branches of the signaling pathway.
Keywords: protein interaction network, rhodopsin signaling, structural modeling
Introduction
The work of many different groups over the past decades has led to a detailed understanding of the molecular mechanisms underlying the initial steps of the vision process in photoreceptor cells (Palczewski, 2006; Kwok et al, 2008; reviewed in Ridge et al, 2003). Rod photoreceptor cells are neurons capable of converting light into electrical signals. They possess a specialized structure consisting of five principal regions (Figure 1A): (i) the rod outer segment (ROS) composed of ∼800 closed membrane discs where phototransduction takes place; (ii) the connecting cilium (CC) that joins the outer segment to the rest of the cell and regulates the traffic of proteins and other components in both directions; (iii) the rod inner segment (RIS) responsible for general cell metabolism, housekeeping, and protein production; (iv) the cell body with the nucleus (N); and (v) the synaptic region (SR) that makes the electrical connections to the neurons in the retina. Protein activity and turnover in the ROS are highly dynamic: about 10% of all discs are generated each day at the base of the segment, while older discs are removed at the distal end by phagocytosis of the neighboring retinal pigment epithelium cells (Boesze-Battaglia and Goldberg, 2002). To replenish, the components of the ROS and the vesicles synthesized in the RIS compartment need to be transported through the CC region, either actively or by diffusion (Reidel et al, 2008).
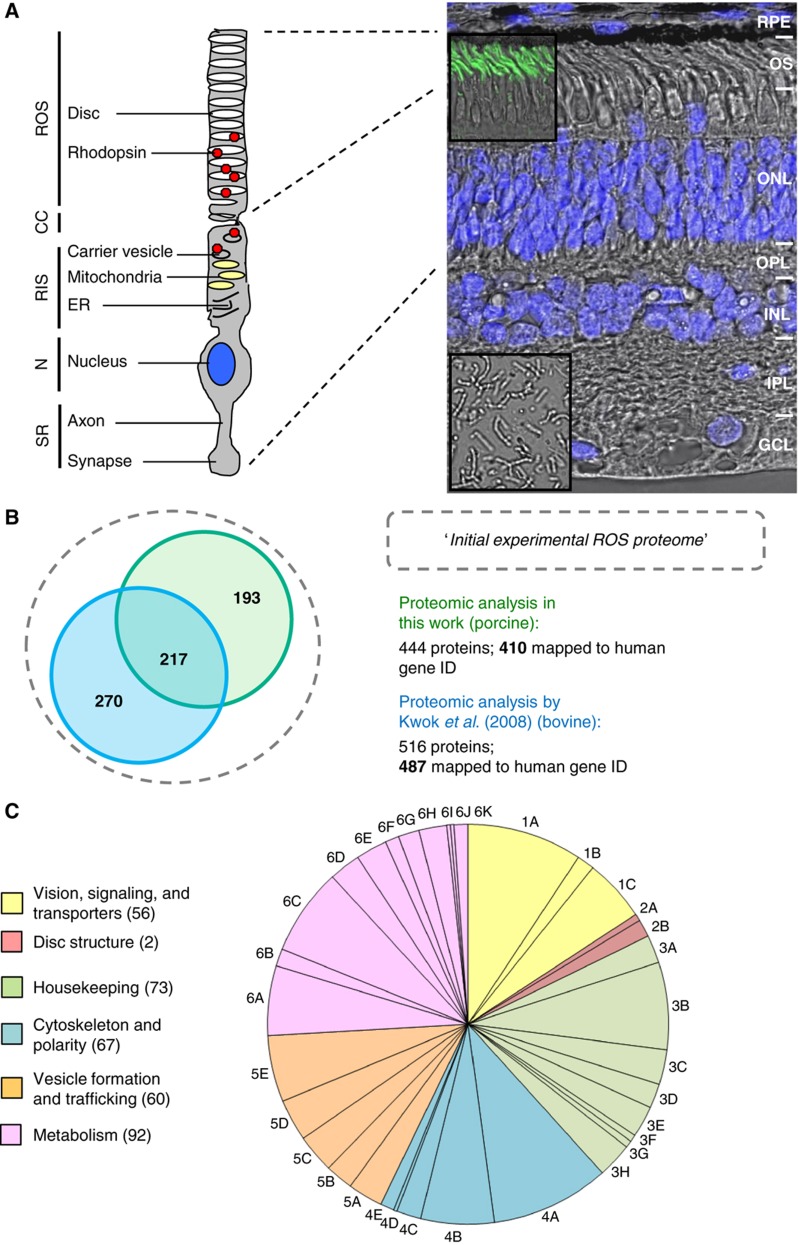
Figure 1.
Proteomic description of the retina ROS inventory and GO analysis. (A) Schematic model of a rod photoreceptor cell (left) and its corresponding location within the retina (depicted in the micrograph to the right). Segments labeled in the model are ROS with enclosed stacks of discs membranes containing the visual pigment molecules rhodopsin; CC; RIS containing mitochondria, Golgi, and ER membranes, and vesicles in which opsin molecules are assembled before transported to the outer segment; and the cell body containing the nucleus and a synaptic termini, where neurotransmission to second-order neurons occurs. The micrograph depicts the vertical porcine retina with its cytoarchitectural organization labeled as photoreceptor outer segments (OSs); the outer nuclear layer (ONL) containing cell bodies of rods and cones; the outer plexiform layer (OPL); the inner nuclear layer (INL); the inner plexiform layer (IPL), and the ganglion cell layer (GCL). The retinal pigment epithelium (RPE) is localized above the photoreceptor cell layer (for details, see http://webvision.med.utah.edu). Retinal cells nuclei were stained with DAPI (magnification × 40). Insets show micrographs of the OS immunolabeled with anti-rhodopsin with an FITC-conjugated secondary antibody (magnification × 40; top inset) and of the OS preparation (magnification × 40; bottom inset). (B) Comparison of different proteomic data sets determined in ROS, based on proteins and the protein overlap identified in the proteomic analysis from this work and that of Kwok et al (2008). The union of the two data sets was defined as the initial experimental ROS proteome. (C) Functional modules and GO analyses of the filtered core ROS proteome. By performing an automatic and a manual GO search (based on the UniProt and KEGG databases), we characterized the 355 proteins (see Supplementary Table S2) to be involved in vision, signaling, transport, and channels (56), disc structure and morphology (7), housekeeping functions (73), cytoskeleton and polarity (67), vesicle, structure, and trafficking (60), and metabolism (92). Sub-modules/sub-functions of the GO terms are indicated as described in Supplementary Table S2 (1A, phototransduction/channels (33); 1B, retinol recycling (5); 1C, calcium signaling (18); 2A, disk morphology (2); 2B, link to ECM (5); 3A, protein folding (8); 3B, chaperones/heat shock (25); 3C, ubiquitination/degradation/proteasome (10); 3D, scaffolds/adaptor proteins (7); 3E, oxidative stress/cell redox homoestasis (9); 3F, apoptosis (2); 3G, others (2); 3H, signaling (10); 4A, regulation of cytoskeleton (34); 4B, cytoskeleton proteins (21); 4C, motor proteins (7); 4D, protein transport (1); 4E, axon guidance (4); 5A, endocytosis (10); 5B, exocytosis (8); 5C, Golgi endosome (11); 5D, vesicle transport/fusion (12); 5E, Golgi/ER/trafficking (19); 6A, glycolysis (20); 6B, tricarboxylic acid (5); 6C, ATP synthesis (25); 6D, lipid/fatty acids metabolism (9); 6E, amino-acid metabolism (9); 6F, one-carbon metabolism (4); 6G, nucleotide metabolism (6); 6H, glucose/lipid/phosphate/amino acid/ion transport (8); 6I, pentose phosphate shunt (1); 6J, mevalonate (1); and 6K, others (4)).
Rhodopsin is the major visual pigment in rod photoreceptor cells. It is a prototypical seven transmembrane-spanning G protein-coupled receptor (GPCR) that contains 11-cis-retinal as its intrinsic chromophore ligand, and it is highly concentrated in the ROS discs (Liang et al, 2003; Nickell et al, 2007). Due mainly to its high endogenous expression, rhodopsin was the first structurally resolved mammalian GPCR (Palczewski et al, 2000). In disc membranes, rhodopsin is tightly packed into paracrystalline dimer arrays, enabling optimal association with the heterotrimeric G protein transducin as well as with additional regulatory components (Filipek et al, 2004; Fotiadis et al, 2004; Ciarkowski et al, 2005). Photon-activated rhodopsin promotes the activation of the associated G protein transducin, which in turn activates phosphodiesterase 6 (PDE6), leading to hydrolysis of cGMP and closure of the cGMP-gated channels. This initiates ultra-fast phototransduction (Hamer et al, 2005), translating light energy first into a biochemical signal, followed by an electrical cue that is transmitted through the neuronal network of the retina. Adaptation to different light conditions, and regeneration of rhodopsin, is regulated at multiple levels, including through differential phosphorylation, differential calcium concentrations, and regulated enzymatic cycles, e.g., when regenerating 11-cis-retinal (Lamb and Pugh, 2004). Disruption of these highly organized structures and processes by germline mutations can cause severe blinding diseases, such as retinitis pigmentosa (RP), rod-cone dystrophies, and congenital stationary night blindness (Berger et al, 2010).
Proteomic analyses of purified ROS have identified about 500 proteins (Figure 1B) that include metabolic enzymes, transport proteins, cytoskeleton elements, regulatory proteins, scaffolds, and housekeeping components, providing a detailed description of the outer segment protein repertoire (Kwok et al, 2008; Figure 1C). In addition, the relative abundance has been determined for 150 proteins (Kwok et al, 2008). Finally, studies have demonstrated that some of these proteins, such as arrestin, transducin, guanylate cyclase, and RhoA, localize differentially in the outer and inner segment in response to light/dark cycles (Hallet et al, 1996; Reidel et al, 2008; reviewed in Artemyev, 2008). To date, however, this wealth of data has not been fully analyzed nor integrated on a functional proteome-wide scale. It is anticipated that a large part of ROS proteins, and a core of the functional modules, will be common to all cells, while others will be photoreceptor specific (Hofmann et al, 2006). Yet, similar to an assembly of music instruments, proteins organized as molecular machines can function in different, context-dependent ways. Connectivity, as well as the timing and tuning of different modules, appears to be crucial for the proper orchestration of signal transduction as well as for parallel signal processing in a concerted manner: at the systems level, this results in the music of life. Although many details of the core phototransduction processes have been established, and mathematical models have been proposed (Dell'Orco et al, 2009), the overall orchestration of the outer segment functions, which include processes like disc shedding and renewal, protein transport along cilia, and light adaptation, are far from being understood. Further, how a variety of mutants of proteins primarily localized in the outer segments can cause visual impairment by perturbing the function of this organelle can only be speculated. For instance, a mutation could impair not only the proper folding of the protein but also its interactions with its partners within the physiologically functional protein networks. Therefore, identifying protein interactions and their networks is an important step toward improving our understanding of the molecular defects that underlie genetically inherited and age-related blinding diseases, and may directly lead to identifying novel disease-associated genes.
There are several aspects that cannot be clearly determined through large-scale studies, such as the network dynamics, the simultaneous regulation of several distinct higher order biological outputs by one network, and the possibility that interactions detected for a particular protein might not be compatible simultaneously (Ito et al, 2001; Gavin et al, 2002, 2006; Ho et al, 2002; Rual et al, 2005; Stelzl et al, 2005). As a consequence, information about the dynamics and the tempo-spatial resolution of networks has been limited to smaller signaling modules, such as receptor-initiated signal transduction (Olsen et al, 2006; Becker et al, 2010). Structural information can help discriminate between direct and indirect interactions in a given complex. More importantly, it can add a dynamical value to the classical interaction networks by determining if two or more predicted partners of any given protein or complex can simultaneously bind to a target, or if they instead compete for the same interaction surface (Kim et al, 2006; reviewed in Campagna et al, 2008 and Kiel et al, 2008). Integrating interaction data with protein expression information may assist in adding a dynamic dimension, and therefore a more realistic view, to the abstract ‘organism interactome’ (Hofmann et al, 2006).
Here, we have combined experimental data, literature mining, and structural information to provide a comprehensive view of the signal transduction network centered on rhodopsin (see the flowchart in Figure 2). Integrating structural information with the relative estimates of expression levels allowed us to distinguish between mutually compatible or mutually exclusive interactions, enabling us to structure a network of nodes and edges toward sub-networks and functional modules. The resulting network offers an unprecedented view of signal transduction in vision and suggests a light-dependent orchestration of the core vision pathway to functions that have so far not been related to this pathway, such as cytoskeleton dynamics, vesicle transport, and energy metabolism. The specific light-dependent connectivity of rhodopsin to these functions is likely to be conferred by small GTPases and their regulators and interacting proteins, such as the prenyl-binding protein PDEδ and other prenylated proteins. This would establish a dynamic and light-dependent mode of regulating the localization (to the membrane or cytosol), and thus the activity, of these GTPases.
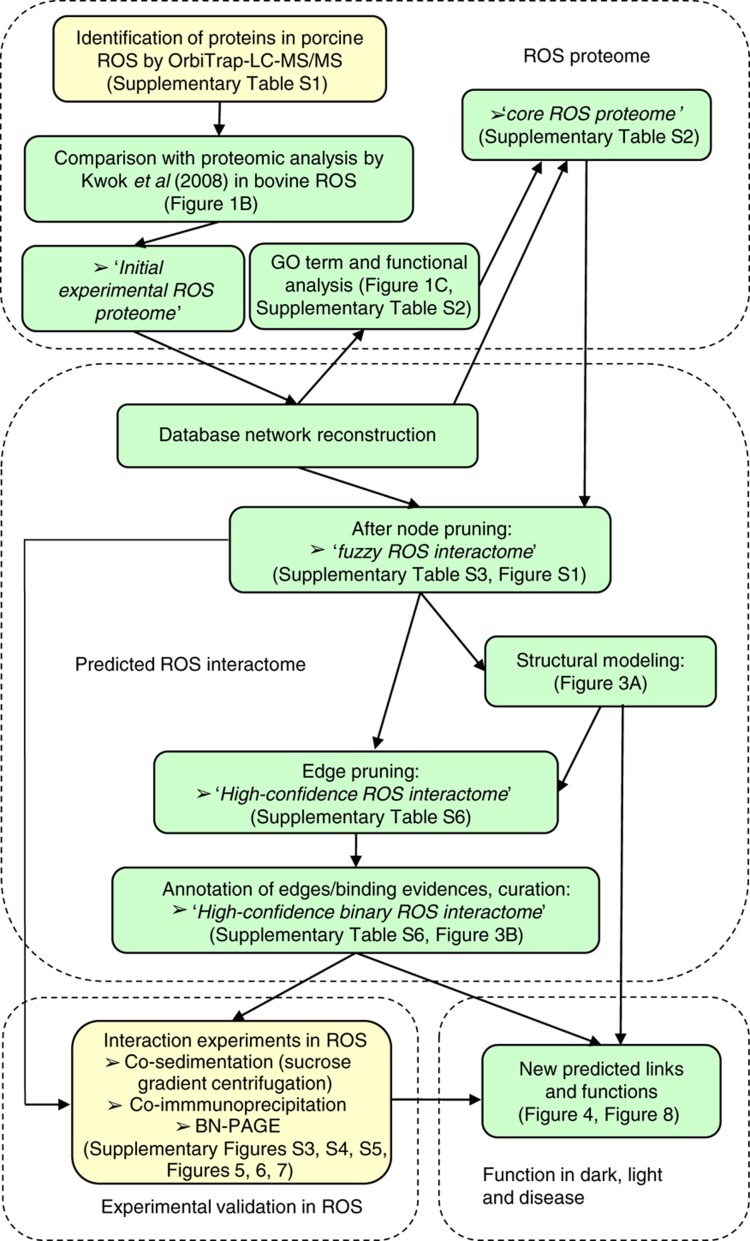
Figure 2.
Experimental and computational workflow. The flow charts of experimental (yellow boxes) and bioinformatic (green boxes) methods used in this work are shown. The initial ROS proteome was generated based on the union of proteins identified in bovine ROS in this work and those from a proteomic analysis of porcine ROS (Kwok et al, 2008). After filtering, a high-confidence ROS proteome was defined. A static ROS interactome was compiled by literature mining. In addition, new experiments were performed in ROS in this work (co-sedimentation and co-IP). Further, we performed structural analyses and homology modeling to distinguish between compatible and mutually exclusive interactions. This enabled us to break the network of nodes and edges into functional machines or sub-networks and modules. The comprehensive multiscale network highlights new predicted links and functions. Finally, disease-associated genes were identified and modeled into available structures.
Results
ROS proteome determination and contaminant removal
To determine the proteomic content of photoreceptor outer segments, dark-adapted porcine ROS and outer segments discs were isolated as previously described (Swiatek-de Lange et al, 2008; see Materials and methods). Proteins were then resolved by one-dimensional gel electrophoresis (1DE) and identified by mass spectrometry (MS). In three independent experiments, a total of 50 proteins were identified by MALDI-TOF from both ROS and ROS discs, and 434 proteins by the more sensitive Orbitrap-LC-MS/MS (Supplementary Table S1). The union of the two data sets resulted in a total of 444 proteins, of which 410 could be mapped to the human proteome. Comparing our data set to a recent proteomic study that identified 516 proteins from bovine ROS (of which 487 mapped to the human proteome; Kwok et al, 2008) resulted in an overlap of 217 proteins (Figure 1B). We then created a unified data set consisting of 680 human proteins, defined as our ‘initial experimental ROS proteome’.
To further refine the protein list presented in the initial experimental ROS proteome, we applied heuristic filtering procedures to remove proteins that might have contaminated the ROS fraction in either experiment (from the surrounding cells or from other cellular domains of the rod photoreceptor). First, we looked at the functional annotations of the initial experimental ROS proteome to identify annotations that contrasted with the expected properties of an ROS protein (such as transcription factors, nuclear proteins, and mitochondrial proteins), based on GO terms from the UniProt database (Supplementary Table S2). Second, we performed a detailed manual functional analysis based on UniProt, the KEGG database, and relevant literature. This revealed 81 putative contaminants (Supplementary Table S2); of these, 68 were found in only one of the two experimental data sets (e.g., ours and that from Kwok et al, 2008), further supporting their classification as contaminants. The 13 proteins identified in both sets are synaptic proteins and G proteins believed to be expressed only in cones (Kwok et al, 2008). We, thus, retained 605 proteins after these analyses.
We next removed all proteins for which there was no interaction data or further experimental evidence about their presence in ROS (protein group 1; Supplementary Table S2); this information was compiled from the literature and databases such as MINT (for details, see Materials and methods and http://mint.bio.uniroma2.it/mint/) (Zanzoni et al, 2002; Chatr-aryamontri et al, 2007; Ceol et al, 2010; Supplementary Table S3). A total of 347 proteins passed this filter. From the analysis of published information, eight additional proteins that lacked associated protein–protein interactions (PPIs) were nevertheless considered to be bona fide ROS proteins with important functional and/or structural roles and were thus retained in the ROS proteome; these were the retinal-specific ATP-binding cassette transporter ABCA4, the cellular retinaldehyde-binding protein RLBP1, the photoreceptor outer segment all-trans-retinol dehydrogenase RDH8, peripherin-2 (PRPH2), the ROS membrane protein ROM1, Rab11B, RP1, and fascin 2 (FSCN2). This filtering procedure left us with 355 bona fide ROS proteins or proteins that are dynamically localized to ROS (protein groups 2 and 3, respectively, in Supplementary Table S2). These proteins represent the ‘core ROS proteome’.
Functional modules of the core ROS proteome
We next classified the core ROS proteome into six functional groups based on the above information, and we annotated lipid modifications, such as prenylation and geranylation (Supplementary Table S2; Figure 1C): (1) vision, signaling, transporters, and channels: 56 proteins have functions that are either directly associated with vision or support visional functionality (i.e., visual cycle, protein homeostasis, or energy production). This module contains well-known members of the phototransduction pathway, including the core signal transduction of light (Dell'Orco et al, 2009) and the visual cycle involved in regenerating 11-cis-retinal necessary to complement photosensitive rhodopsin after photo-bleaching (Lamb and Pugh, 2004). We further included here proteins involved in Ca2+-dependent signaling and proteins associated with ion channels that regulate photoreceptor membrane conductance and polarity; (2) outer segment structure and morphogenesis: the seven proteins in this group are those implicated in outer segment structure and disc morphogenesis (Molday et al, 1987; Poetsch et al, 2001), and those that link the cytoskeleton to the extracellular matrix (ECM), such as α and β catenin; (3) housekeeping: in this group of 73 proteins, we consider protein-folding chaperones and heat-shock proteins, members of the ubiquitination/degradation-proteasome machineries, scaffold proteins such as the 14-3-3 family members, and proteins involved in oxidative stress, cell redox homeostasis, and apoptosis regulation (De La Paz and Anderson, 1992; for review, see Wenzel et al, 2005); (4) cytoskeleton and polarity (67 proteins): this group contains cytoskeleton proteins, such as actin and tubulin, as well as their respective binding proteins and molecular motors, proteins involved in regulating cytoskeleton dynamics including GTPases, and intermediate filaments. Many of these are associated with the CC and the axoneme of ROS and might therefore be present at low concentrations (reviewed in Adams et al, 2008). Proteins that are known to function in axon guidance were also added to this class; (5) vesicles formation and trafficking: we included here 60 proteins involved in Golgi function, protein and vesicle transport, and fusion, as well as the annexins that function in exocytosis and phagocytosis; (6) metabolism: we included here 92 proteins related to metabolism, in processes such as glycolysis, ATP synthesis, nucleotide, and fatty acid and carbon metabolism. Interestingly, we found that several of these are metabolic proteins involved in energy production (about 50% of the enzymes detected in this group are involved in glycolysis and about 20% in the tricarboxylic acid pathway), including ATP synthase, the activity of which has recently been demonstrated in intact discs (Panfoli et al, 2009). This suggests that the ATP used in vision signaling is indeed produced within ROS and is probably fueled by glucose transported along the cilium. Indeed, this study identified a glucose transport protein, SLC2A1, as an ROS protein, supporting this hypothesis. High energy demands of ROS, and a capacity of only limited diffusion through the interconnecting cilium, may require on-site production of ATP.
Network reconstruction and structural modeling of the ROS interactome
Information about the PPIs among the core ROS proteome was mined from protein interaction databases to assemble an ROS protein network (Supplementary Table S3; Zanzoni et al, 2002; Chatr-aryamontri et al, 2007; Kerrien et al, 2007; Ceol et al, 2010). The protein interaction degree ranges from 1 to 179, with the highest number of interaction partners for actin, tubulin, 14-3-3 family members, heat-shock protein members, and ERK (Supplementary Figure S1A).
Overall, the complete core ROS proteome PPI network consists of 5337 interactions among its members (Supplementary Table S3). The experimental evidence for most of these interactions (5047) was based on co-immunoprecipitation (co-IP) or pull-down experiments, which offers little support for their direct nature (Supplementary Figure S1B). In addition, many of the edges in the network are supported by single experimental pieces of evidence (>85% of the PPI), often derived from high-throughput approaches. Thus, we refer to this network, which represents all the interactions that we could retrieve from published data, as a ‘fuzzy ROS interactome’, since it contains many interactions supported by only one non-binary piece of evidence.
Next, we aimed at increasing the information content of the network by structural modeling. Pairs of interacting proteins often share common structural features with other interacting pairs of known structure (domains and linear motifs). We use structural information, combined with computational tools, to support low-confidence experimental interaction evidence and to determine whether two interactions involving a common partner are compatible or mutually exclusive (see Supplementary Material 1 and Supplementary Figures S2 and S3). We considered two levels of structural evidence that may support any given interaction. First, for each pair of members of the core ROS proteome, we searched the PDB database (http://www.pdb.org) for protein complexes of known structures whose elements share at least 70% homology with the query proteins. By this approach, we identified 84 complexes in the ROS core network whose structures could be confidently modeled on homologous structures (Supplementary Table S4). Most of the interactions for which there are X-ray structures, or structures from close homologs, are found between the connecting proteins in the modules 1 or 4 (vision, signaling, transporters, and channels and cytoskeleton and polarity) as well as among proteins involved in interactions connecting these two modules (Supplementary Figure S4).
Next, using a lower level of structural detail and confidence, we exploited the notion that similar domain pairs are likely to interact in a similar way (‘nature repeats itself’) (Aloy and Russell, 2002). For example, members of the Ras family and proteins containing a Ras-binding domain (RBD) are likely to use the same interaction surface when they interact (see Kiel and Serrano, 2006). To overlay a domain-level model on the ROS network, we represented each of the 355 nodes as a stack of Pfam domains (http://pfam.janelia.org/; Supplementary Table S5). We then searched the 3Did database (http://3did.irbbarcelona.org/; Stein et al, 2005, 2009) for structural evidence of pair-wise interactions between any of the domains in our database (for details, see Supplementary Material 1). Structural evidence was found for 352 pair-wise interactions, excluding pairs that had already been identified by comparison with homologous crystallized complexes (Supplementary Table S4). A confidence ‘interaction score’ of ⩾2.3 for the identification of the interacting pairs was obtained by interrogating the InterPReTS server (http://www.russelllab.org/cgi-bin/tools/interprets.pl/; Aloy and Russell, 2003). This score was validated using a yeast two-hybrid positive and negative binding data set described by Vidal and co-workers (Rual et al, 2005). We found a confidence of over 70% that two proteins containing the target domains will interact in a two-hybrid experiment when the InterPReTS score was >2.3 (see Supplementary Material 1, Supplementary Figures S2 and S3). Of the 352 interactions that had a hit in the 3Did database, 107 had InterPReTS scores higher than our chosen threshold and were therefore annotated as ‘supported by structural evidence’. A total of 191 interactions supported by structural evidence (that is, the 84 interactions with known or closely related structures, and the 107 with significant InterPReTS scores) were merged with the literature-based interaction network. Interactions that could be annotated with structural evidence were mainly found within the functional modules (Supplementary Figure S4).
To increase the confidence in the resulting network, edges that were only supported by a single piece of evidence from any type of experiment except yeast two-hybrid experiments were removed (Supplementary Table S6), with the exception of interactions for which there was also structural information available (i.e., a three-dimensional structure of the complex itself or of a highly homologous complex). This curated static network (‘high-confidence ROS interactome’) comprises 660 edges and links the majority of the nodes (with 266 proteins, as indicated in Supplementary Table S2 and Figure 3A) that were present in the original network. The missing nodes are equally distributed among the proteins with respect to their GO terms, although an enrichment for proteins assigned to the classes retinol recycling and metabolism was observed (of 80 and 50%, respectively).
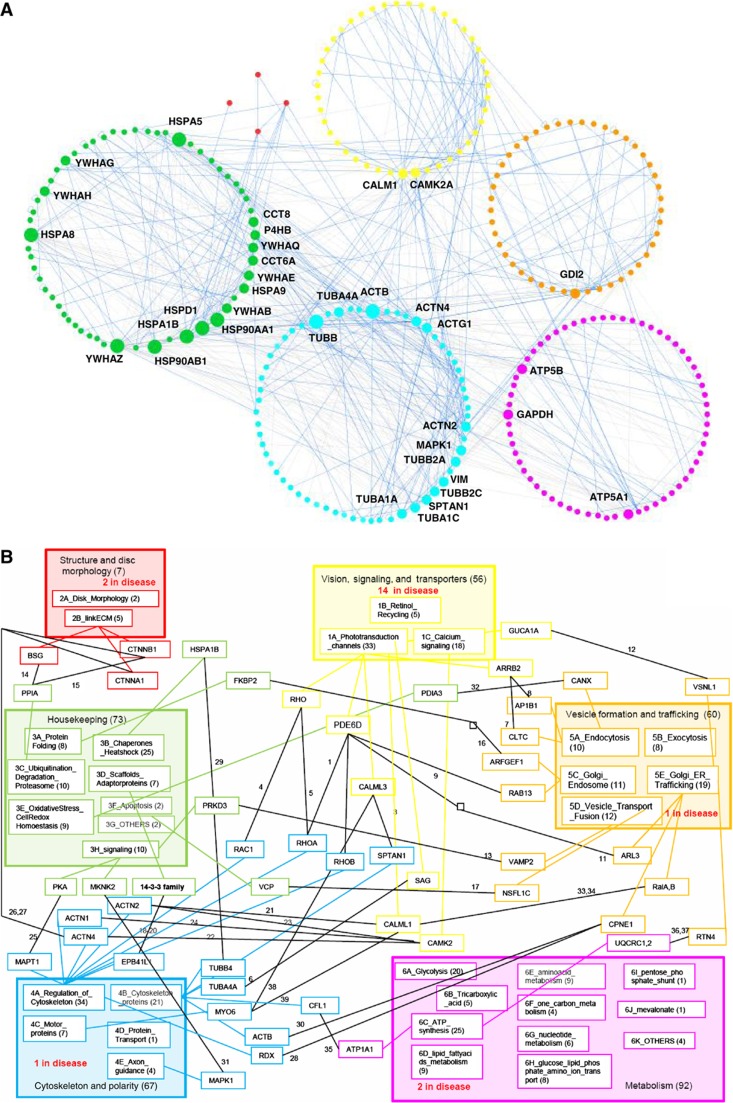
Figure 3.
The high-confidence ROS interactome and the high-confidence binary ROS interactome. (A) The high-confidence ROS interactome. The 660 higher confidence interactions of the ROS interactome are listed (Supplementary Table S6). The size of the nodes indicates the number of interaction partners for a given protein (of >10 or >20). Edges with binary evidence are indicated with blue, while edges supported by more than one piece of evidence are indicated in gray. Proteins are colored according to their function. (B) The high-confidence binary ROS interactome. Modules and sub-modules are shown, and only the interactions of proteins from two different modules are indicated (see Supplementary Material 2). The number of proteins implicated in diseases in each category is indicated.
By considering only edges supported by at least one evidence of direct binary interaction, we obtained a ‘high-confidence binary ROS interactome’ that contains 222 nodes (note that most of the nodes that were not captured by this network are annotated with metabolism ontology terms), linked by 349 edges (indicated as binary in Supplementary Table S6). Except for reactions involving guanylate kinase (GK) and nucleoside diphosphate kinase (NDPK), a nucleotide and cyclic nucleotide modifying enzyme, all interactions of the core vision pathway (Ridge et al, 2003; Wensel, 2008; Dell'Orco et al, 2009) are represented in our PPI network as true binary interactions. Thirty-five nodes have more than ten interaction partners, with a maximum degree of 55. Only 2 proteins in the vision category have >10 interaction partners (CALM1 and CAMK2A). Most of the interactions involve the heat-shock proteins, 14-3-3 family members, ERK (MAPK1), tubulins, and actin. More than ten interaction partners were found in the metabolism branch for glyceraldehyde-3-phosphate dehydrogenase (GADPH) and for the two ATPase subunits.
Additionally, 109 direct binary interactions connected the defined functional modules, and 240 binary interactions connected proteins within modules (Figure 3B; for a detailed description of the connections between the modules, see Supplementary Material 2). Out of those 240, 188 are classified within sub-boxes/sub-functions. The observation that roughly two thirds of the interactions were found within functional modules, and only a third between modules, provides confidence to our module classification and manual functional annotation. Interestingly, the most highly connected modules are module 1 (vision) and module 4 (cytoskeleton), illustrating the important crosstalk between the core vision pathway and the cytoskeleton. The less connected modules are the ones involved in the structure of the discs and in metabolism. As expected, the housekeeping module, despite having fewer connections, is linked to all other modules.
The high-confidence ROS interactome suggests new functional links
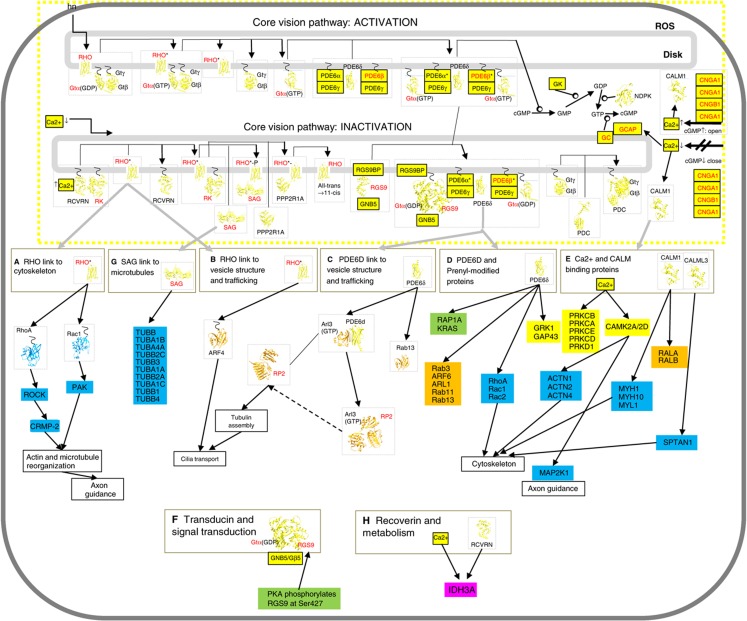
Using our curated high-confidence binary ROS interactome as a basis, we decided to analyze in more depth the core vision pathway, which is probably one of the best-studied biochemical pathways (Ridge et al, 2003; Wensel, 2008; Dell'Orco et al, 2009). We extended the published core vision pathway (Dell'Orco et al, 2009) using evidence from our high-confidence network and indicated structural coverage and outputs to different functional cellular processes emanating from the proteins in the pathway (Figure 4; for a detailed description, see Supplementary Material 2). Of these, we decided to validate the link to the GTPases RhoA and Rac1 (Figure 4, link A and link D), which suggests a link between vision activation and cytoskeleton reorganization.
Figure 4.
Structural coverage of the core vision pathway and its links to other functional modules. The published core pathway (Dell'Orco et al, 2009) was extended using evidence from our high-confidence network. Outputs to different functional cellular processes emanating from the proteins in the pathway are indicated, and the available structures are displayed by ribbon representation (see the main text and Supplementary Material 2). Proteins are colored according to their function.
Rho–Rac1 and the cytoskeleton connection
Previous work has demonstrated functional links between rhodopsin, certain GTPases (Mitchell et al, 1998) (most prominently transducin), and the cytoskeleton. S-arrestin specifically binds to activated and phosphorylated rhodopsin, inhibiting activation of transducin and terminating phototransduction (Kühn, 1978; Kühn et al, 1984; Wilden et al, 1986). Nair et al (2004) have shown interactions between S-arrestin and microtubules (Figure 4, link G).
We were able to confirm that small GTPases Rac and the GTP-bound form of RhoA bind rhodopsin, as has been previously described (Wieland et al, 1990a, 1990b; Balasubramanian and Slepak, 2003; Gray et al, 2008; Figure 4, link A). For this, we performed co-segregation/co-sedimentation experiments to reveal proteins within large complexes, as described previously to analyze the light-harvesting complex of photosystem II in plants (Swiatek-de Lange et al, 2008; Supplementary Figures S5 and S6A). These experiments indicated that Rac1, Rho, and CRMP-2 were present in a large complex that also contained cytoskeletal proteins, rhodopsin, and components of the vision pathway. Although the low resolution of the technique, and the complexity of the patterns, preclude using these experiments to add new binary interactions to the ROS network, it can be used to corroborate interactions supported by further experimental evidence or from the literature (Supplementary Tables S3 and S6). Using BN-PAGE (Schägger and von Jagow, 1991; Nijtmans et al, 2002; Camacho-Carvajal et al, 2004) or IP experiments in combination with either MS or subsequent immunoblotting, we obtained further evidence for the existence of large complexes containing rhodopsin that also included the cytoskeletal proteins actin and tubulin as well as its specific regulators such a RhoA, Rac1, and CRMP-2 (Figure 5; Supplementary Figure S6B).
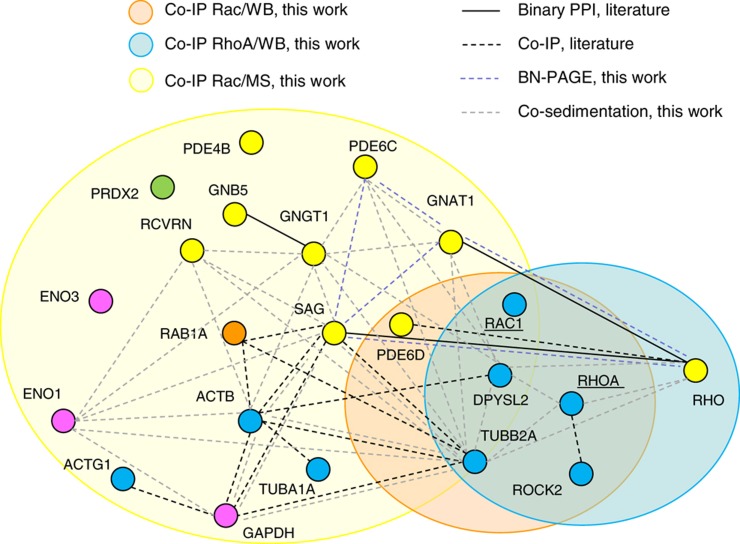
Figure 5.
Graphical representation of experiments performed in this work and its comparison with interactions described in the literature. Protein complexes that were obtained using Rac1, RhoA, or Rac1 as the bait protein are displayed within orange, blue, and yellow circles, respectively. The Rac1 and RhoA complexes were identified by western blot, and the Rac1 complex by Orbitrap. The overlap of the three circles indicates the proteins that were identified in the same complex in one of the three experiments. Connecting lines between proteins indicate either binary or co-IP interactions from the literature, or from BN-PAGE or co-sedimentation interactions as determined in this work. Proteins are colored according to their function.
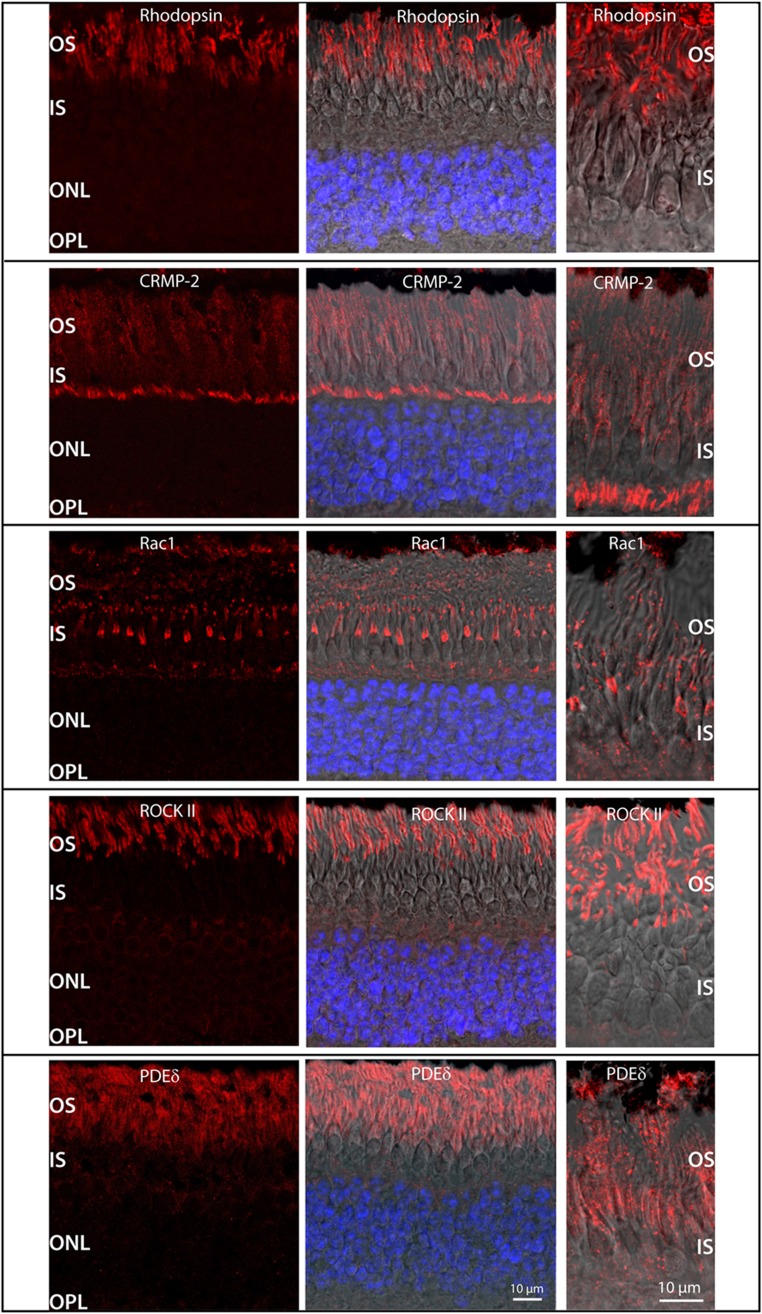
We next isolated the protein partners of the glycosylated N-terminus of rhodopsin by using concanavalin A affinity purification (Plantner and Kean, 1976; De Grip, 1982). The interactions between rhodopsin, RhoA, and CRMP-2 were confirmed by these concanavalin A pull-down experiments, and in part by additional co-IP experiments in which we detected rhodopsin associated with the core signaling complexes of the visual pathway including transducin and, again, with Rho, Rac1, and CRMP-2 (Supplementary Figure S6C and D). To confirm that CRMP-2, Rac1, and ROCK II are indeed bona fide ROS proteins rather than contaminants, we performed immunohistochemistry for these proteins. Indeed, all three proteins were constituents of ROS on cryosections of porcine retina (Figure 6). Despite considerable efforts, we were not able to confirm the presence of RhoA due to a lack of selectivity of various antibodies against RhoA in retinal sections.
Figure 6.
Immunohistochemical analyses of porcine retina. Cryostat sections of the retina were stained with primary antibodies (red) against indicated proteins, and nuclei were counterstained (blue). The images on the left were taken from the outer retina (outer segments (OS), inner segments (IS), outer nuclear layer (ONL), and outer plexiform layer (OPL)). Images in the middle are an overlay of antibody staining, nuclei staining, and DIC optics (Nomarski). Images on the right were taken with higher magnification, to focus on the OS and IS. All indicated proteins were unambigiously identified as constituents of ROS. Control sections without primary antibodies showed no staining (Supplementary Figure S7).
Functional analysis of the PDEδ–Rac1 complex
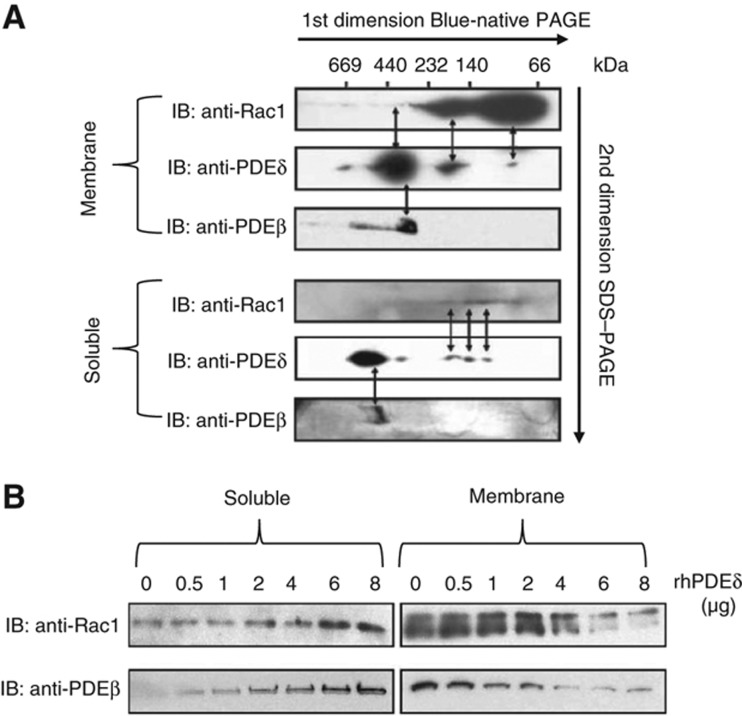
PDEδ has been reported to bind to prenyl-modified proteins, such as several small GTPases and rhodopsin kinase (Hanzal-Bayer et al, 2002; Zhang et al, 2004), and it appears as an important node within our network (Figure 4, link D). PDEδ could thus have a critical regulatory role both in facilitating the transport of prenylated target proteins along the cilia together with Arl3 (Figure 4, link C; Veltel and Wittinghofer, 2009) and in serving as an effector or guanine nucleotide dissociation inhibitor (GDI) for many GTPases, such as Arf, Rac1, RhoA, and Rab, all of which are expressed in photoreceptors (Figure 4, link D). Therefore, we tested whether PDEδ functions as a GDI for Rac1 in ROS. First, we demonstrated that PDEδ and Rac1 are colocalized in ROS using immunohistochemistry (Figure 6; Supplementary Figure S7). Second, we showed that PDEδ and Rac1 colocalize in ROS in native protein complexes, by using dark-adapted ROS separated by BN-PAGE (Figure 7A). In dark-adapted ROS, PDEδ was part of distinct complexes that ranged from high molecular weight complexes of 660 kDa to smaller complexes of around 90 kDa; and interestingly, Rac1 colocalized with PDEδ within different complexes between the soluble and membranous fractions. Third, we tested whether PDEδ could dissociate Rac1 from ROS membranes in vitro (see Materials and methods). Indeed, adding recombinant human (rh) PDEδ led to the solubilization of Rac1 from the ROS membranes (Figure 7B). Solubilization occurred in a dose-dependent manner with increasing amounts of rhPDEδ. As a positive control, we verified that PDEδ solubilized PDEβ from ROS membranes, as previously described (Florio et al, 1996). Thus, PDEδ can solubilize Rac1 from ROS membranes, a feature characteristic of GDIs.
Figure 7.
Experimental evidence that PDEδ acts as a GDI for Rac1 in ROS. (A) PDEδ and Rac1 colocalize in ROS in native protein complexes. After solubilization with β-DM, native ROS protein complexes from soluble and membranous fractions of light- and dark-adapted ROS were separated by BN-PAGE. Components of the native protein complexes were separated by SDS–PAGE for second-dimension electrophoresis. Western blots with anti-Rac1 and anti-PDEδ antibodies showed that PDEδ and Rac1 colocalized but were in different complexes in ROS depending on the dark-adapted state of the retina. Colocalization of PDEδ and Rac1 seemed to be stronger in the dark-adapted state, where both proteins colocalized to the soluble and membranous fractions. In light-adapted ROS, colocalization of PDEδ and Rac1 was detected only in the membranous fraction but not in the soluble fraction. (B) In-vitro solubilization of Rac1 GTPase from light- and dark-adapted ROS membranes. Membranes isolated from light- or dark-adapted ROS were incubated for 1 h at 37°C with different amounts of recombinant human PDEδ (rhPDEδ) or buffer alone, and the unsolubilized material was recovered by ultracentrifugation. Immunoblots with anti-Rac1 or anti-PDEδ antibodies showed that PDEδ solubilizes Rac1 from ROS membranes in a dose-dependent manner. Since it has been previously determined that PDEδ solubilizes PDEδ from ROS membranes in a dose-dependent manner (Florio et al, 1996), this was used here to demonstrate the functional activity of the rhPDEδ protein.
All of the interactions determined here—with the exception of the ones identified by co-sedimentation, as this method is considered as weak evidence for physical interactions—were added as supporting evidence to our network (Supplementary Table S6). In total, co-purification and co-elution experiments supported 60 interactions that had been included in our network based on the literature, and new evidence for 175 interactions from our co-IP results was added. Additionally, our results supported five interactions that had structural evidence (with INTERPRETS score ⩾2.3).
Restricted to a single new pathway (Rac1/RhoA–PDEδ−CRMP-2), our experimental data support the physiological relevance of our network. It should be noted, however, that these data cannot be considered to be complete or free of false positives, since the number of interactions tested and validated was small when considering the extent and complexity of the network.
Discussion
In this work, we investigated the protein interaction network in a highly specialized cellular region of the mammalian photoreceptors, the ROS. Graphs representing protein interactions are idealized descriptions of all the interactions that can possibly occur in an organism. The realization that, in any given cell type, only a fraction of these interactions can possibly occur prompted the development of approaches to combine different genome-wide information to build interaction networks that are either specific for a cell type (Bossi and Lehner, 2009) or that change dynamically, such as during the cell cycle or after specific pathways have been induced. Here, we take this one step further and propose a protein interaction network for a structurally very distinct and functionally highly specialized region of the mammalian photoreceptors: the ROS. In addition to proposing novel interactions, we present a structural model that allows us to discriminate between protein interactions that are compatible and those that are mutually exclusive.
A curated and structure-based PPI network central to rhodopsin
We first generated a ROS-specific protein interaction network by combining proteomic expression levels in ROS with interaction information, which we mined from the literature and then subsequently supplemented with our new data pertinent to the description of protein complexes in their physiological context. We next performed structural analysis of the curated network by decomposing proteins within this network into domains. This step allowed us to validate interactions at a domain level and to thereby increase the confidence in the network. By reassembling the decomposed network based on structural constraints into structure-functional modules, we were able to define logical relationships between the network nodes, and to define sub-networks that physically and functionally fit into molecular machines. Finally, we annotated these functional modules according to their respective physiological processes to derive a network of pathways and processes. Based on a compilation of experimental evidence and several layers of expert as well as automated curation, filtering, and modeling, the resulting network represents a multiscale description of wiring and physical connectivity in the ROS of photoreceptors. The extended core pathway shows how rhodopsin activation–deactivation leads to other possible functional effects in addition to its primary function of signaling for closing the cGMP-gated cation channel. Thus, in addition to its relationship with the module of (1) vision, signaling, transporters, and channels, wiring rhodopsin to (2) outer segment structure and morphogenesis, (3) housekeeping, and (4) cytoskeleton and polarity suggests a regulation of cytoskeleton assembly–disassembly and dynamics, vesicle and Golgi trafficking, and transport along the interconnecting cilium of photoreceptors by rhodopsin. Connections between active rhodopsin and Arf4 (Deretic et al, 2005; Mazelova et al, 2009), and between PDEδ and Rab13 and the GTP-bound form of Arl3 (Hanzal-Bayer et al, 2002), also link the vision cycle to vesicle trafficking and structure (Figure 4B and C). We experimentally validated two of the proposed new functional links. Our results suggest a link between rhodopsin, Rac1, RhoA, ROCK II, and CRMP-2. This points to a second, not yet experimentally tackled pathway that is influenced by light, which appears to be a delineation of an archetypical G protein-regulated pathway known to be active in growth cone dynamics and collapse (Liu and Strittmatter, 2001). RhoA binds to CRMP-2 (gene name DPYSL2, Figure 5), a scaffold protein involved in actin cytoskeleton dynamics in neurons that regulates growth cone dynamics. CRMP-2, working through the GPCR lysophospatidic acid receptor, has been described as a crucial molecule in axon guidance, where it dynamically regulates the antagonistic effects of RhoA and Rac1. Regulated by a Rho-associated kinase (ROCK), CRMP-2 promotes either outgrowth or collapse in response to active RhoA or Rac1, respectively (Hall et al, 2001). When RhoA-GTP levels are high, more CRMP-2 is phosphorylated by the Rho-effector kinase ROCK, and thus less non-phosphorylated CRMP-2 is complexed with Rac1, leading to cytoskeleton collapse (reviewed in Liu and Strittmatter, 2001). CRMP-2 can bind directly to tubulin heterodimers to promote microtubule assembly (Fukata et al, 2002). This presents the exciting possibility that GPCR rhodopsin autoregulates its own axonal/dendritic guidance and possibly regulates outer segment growth via the archetypical mechanisms of axon guidance. Based on this scenario, the outer segment would function as a continuously extending growth cone, autoregulated by light and other as-yet unidentified guidance cues that may be produced in other retinal cells, most notably in the retinal pigment epithelium.
We additionally provide experimental evidence that PDEδ could act as a GDI for the small GTPase Rac1. PDEδ could thus have a very crucial regulatory role: (1) in transporting prenylated target proteins (Zhang et al, 2004) along the cilia, together with Arl3 (Veltel and Wittinghofer, 2009) and (2) as an effector or GDI for many GTPases (Hanzal-Bayer et al, 2002), such as Arf, Rac1, RhoA, and Rab, by keeping them GDP bound and inactive. This is important since we did not find the conventional RhoGDI in ROS, suggesting that PDEδ could indeed substitute for this function in ROS (similar to that demonstrated for the small GTPase Rab13 in ROS; Marzesco et al, 1998). However, despite the structural similarity of the PDEδ and RhoGDI domains (Scheffzek et al, 2000; Hanzal-Bayer et al, 2002), we learned by superimposing the Rac1–RhoGDI with the Arl2-PDEδ structure that these two interactions depend on different moieties for binding (Supplementary Figure S8). We provide experimental evidence in this work that PDEδ could act as a GDI for Rac1. We did not find any GEFs or GAPs for small GTPases in our network but only GDIs (ARHGDI for RhoA, PDEδ for Rac1, and GDI1 and GDI2 for Rab proteins). Interestingly, this could suggest that these are not regulated by the usual switch-like mechanism of GTPase regulation, but rather by a gradient activation, in which the activity of active RhoA is determined only by the concentration of RhoGDI, keeping RhoA in the inactive form.
The role of Ca2+ in vision cycle, phototransduction, and actin cytoskeleton changes
Intracellular Ca2+ concentrations influence the activities of numerous kinases, such as different PKC isoforms, the PKA kinase, Ca2+/calmodulin-dependent kinases, and the two CaMK-II isoforms, all of which are integral to the network. Predicted kinase phosphorylation sites from CaMK-II, PKA, PKC, MAPK, and PKD are summarized in Supplementary Table S7. Several Ca2+-regulated kinases phoshorylate cytoskeletal target proteins, such as actinin and myosins, and small GTPases and their regulators. This opens the intriguing possibility that the nucleotide state and the dynamic spatial cellular distribution of several small GTPases are controlled by Ca2+. As perturbed Ca2+ homeostasis is a consequence of the activity of a perturbed visual pathway in specific forms of RP (Paquet-Durand et al, 2010), this is likely to affect a variety of critical pathways and thus generate a systemic perturbance of ROS physiology. Our network reveals several direct binary connections between Ca2+-regulated proteins and cytoskeleton proteins: CaMK2A with actinin, calmodulin with GAP43 (neuromodulin) and S1008 (tubulin polymerization initiation), and PKC with 14-3-3 family members. Calmodulin is known to have a wide range of effector binding specificity, which dynamically changes with Ca2+ binding. Calmodulins 1 and 3 were linked to about 10 proteins from the two modules, cytoskeleton and vesicle transport. Calmodulin (CALM3 or CALM1) can bind to the cytoskeleton regulator spectrin α, actinin (ACTN2 and ACTN4), and the myosin motor protein MYO6. Therefore, calmodulin proteins could provide an important link between Ca2+ signaling and regulation of the actin cytoskeleton, with spectrin having a critical role in organizing and maintaining membrane sub-domains that harbor rhodopsin (Berghs et al, 2000). Further, a Ca2+-dependent kinase, CaMK2A, was found to directly contact actinin-1, -2, and -4, and to be in a ternary complex with densin, a synaptic adhesion molecule (Walikonis et al, 2001), which is not present in our network (as it was not taken into consideration). Another link appears between calmodulin and RalA and RalB, both of which are involved in trafficking: RalA has a role in exocytosis regulating exocyst assembly, while RalB interacts with EXOC8 (a part of the exocyst complex); RALBP1 is an effector of both RalA and RalB. Ca2+ activity is also likely to regulate metabolic activities through IHD3A, recoverin, and neurocalcin: the hippocalcin-like protein 1 is a recoverin-like protein that was suggested to have an anti-apoptotic function and might protect photoreceptors from Ca2+-induced cell death (Krishnan et al, 2009).
Finally, Ca2+ could have an important role in the light-dark cycle by affecting PKA activity. Phosphorylation of RGS9-1 by PKA (Balasubramanian et al, 2001) is regulated by light and Ca2+, and results in the reduction of RGS9-1 GAP activity: with light, RGS9-1 causes rapid Tα-GTP inactivation and photoreceptor recovery, while in the dark, PKA is activated by rising concentrations of Ca2+ and cAMP, which in turn phosphorylates RGS9-1. In this way, GAP activity is reduced, the active transducin lifetime is prolonged, and the photoresponse is strengthened (Balasubramanian et al, 2001).
While it remains to be seen how all of these connections are orchestrated, and to which degree they impact vision homeostasis, there is no doubt that Ca2+ has a crucial role in ROS functionality.
Structural information, structural coverage, and ‘AND’ and ‘XOR’ gates
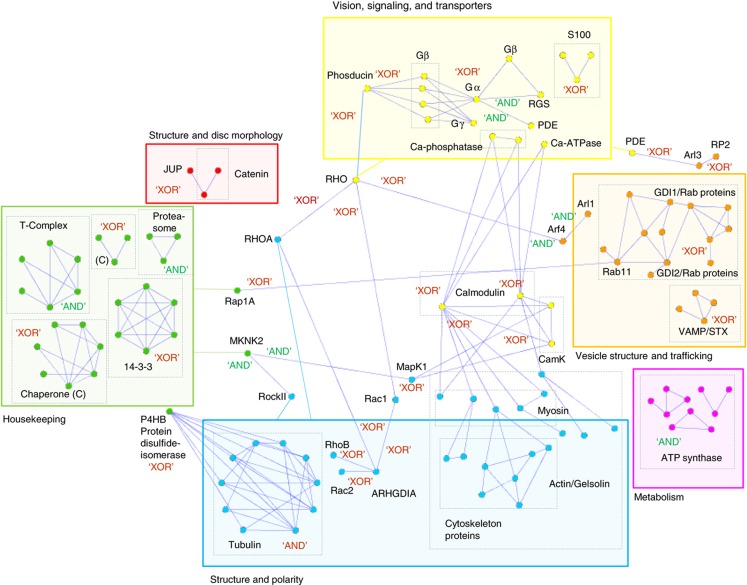
Structural information allows the confidence of any independent interaction evidence to be tested and at the same time can add topological information to the molecular level by defining sites or interaction domains. When several proteins can bind to a single protein, the various interactions can occur simultaneously or can be mutually exclusive (reviewed in Santonico et al, 2005 and Kim et al, 2006). If two or more proteins compete for the same binding site, then it seems unlikely that binding can occur simultaneously, whereas binding to topologically distinct sites may occur at the same time. At the level of graph representations within a network, structural information can thus support logical constraints. Here, it is important to mention that interactions were defined as exclusive or compatible from a structural point of view, and that this cannot be directly translated to biological terms in all cases (i.e., for competition to occur, the target protein should be at lower concentration than the competing ones). When both competitors are present at the same place and time, changes in concentration levels or additional regulatory constraints (e.g., those introduced by post-translational modifications) could regulate competition.
Structurally superimposing domains onto interactions allowed us to define ternary complex formation and, importantly, to model both the composition of macromolecular assemblies and its dynamic dissection into mutually exclusive complexes (Supplementary Figure S9). With this information, we can add dynamics to the network, using the following ‘AND’ and ‘XOR’ (‘XOR’=exclusive OR) logical gate symbols: if three or more proteins can interact at the same time, then they are compatible (indicated with ‘AND’), while if three or more proteins cannot interact simultaneously, then they are mutually exclusive (indicated with ‘XOR’) (Figure 8). Competitors are frequently found in highly dynamic processes or may dynamically connect a given protein to different signaling and functional modules. The structural and interaction analyses of the core vision pathway and its cytoskeleton branch show several examples of non-compatible (‘XOR’) interactions (Figure 8). For example, rhodopsin may interact with transducin, arrestin, or rhodopsin kinase (in the core vision pathway). It may also interact with Rac1 or RhoA (which are antagonists in cytoskeletal dynamics) or with Arf4 (involved in trafficking). Changes in rhodopsin activation, concentration, and localization, or in its activation states, may therefore switch signaling into different pathways. Further, rhodopsin localization during ciliary transport and disk formation, and dynamic changes in concentrations and activation states in response to light, can alter the array of rhodopsin binding partners, since these are determined by the phosphorylation state of rhodopsin on the one hand and the availability or concentration of binding proteins on the other hand. Interestingly, ‘AND’ gates are mainly found in the housekeeping, structure and polarity, and metabolism branches, e.g., within large functional complexes, such as the T-complex, the proteasome, tubulin, and the ATP synthase machinery. ‘XOR’ gates, which are mainly prevalent in the vesicle structure and trafficking branch, indicate switch behavior or redundant protein functions, such as for Rab GTPases (Del Conte-Zerial et al, 2008). In the vision branch, both ‘AND’ and ‘XOR’ gates synergize. This may allow dynamic tuning of light and dark states. However, all connections from the vision module to other modules are ‘XOR’ connections, suggesting that competition, together with local protein concentration changes, could be important for transmitting signals from the core vision module.
Figure 8.
Network representations distinguishing mutually exclusive from compatible interactions, based on structural information. All protein–protein interactions for which structural information was available (Supplementary Table S4), and for which structural superimpositions were performed (Supplementary Figure S9), are represented here. Mutually exclusive complexes are indicated with ‘XOR’, and compatible interactions are indicated with ‘AND’. Proteins are colored according to their function (see Figure 3B).
The vision network and disease
A large fraction of retinopathies involve the degeneration of rod photoreceptors; these include RP, syndromes incorporating retinal degeneration with different associated phenotypes (such as Usher's syndrome and Bardet–Biedl syndrome), and Leber congenital amaurosis (LCA), a congenital form of retinal degeneration. An increasing number of genes and proteins has been implicated in these pathologies (http://www.sph.uth.tmc.edu/Retnet/). These proteins include: components of the visual transduction cycle; structural components of the cytoskeleton, rod and/or cone photoreceptor outer segment disc membranes; components of synthesis and recycling of the retinoid; transcription factors (including CRX and NRL) and splicing factors; those involved in signaling and cilium maintenance, phagocytosis of the outer segment discs of the photoreceptors, and trafficking of intracellular proteins; and those with functions in pH maintenance in the retina, in metabolism, and as chaperones. The protein with by far the largest number of mutations is rhodopsin (>100 mutants), while the others range contain from 40 mutations (for the retina-specific crumbs homolog 1 [CRB1]) to 1 (for transducin α) (see Supplementary Material 3 and Supplementary Figure S10). Structural analyses of the different mutations mapped on the available structures or homology models (156 mutations) indicated that the majority of these are within the hydrophobic core of the corresponding proteins and are therefore likely to cause misfolding (see Supplementary Material 3). Mapping all proteins involved in vision-related diseases into the network made it apparent that the core visual pathway is the most susceptible to disease, and that the other functional modules are relatively robust. Out of 36 proteins considered here to be involved in retinal degeneration, the majority (20 proteins) are localized in ROS (the remaining are found in other regions of the rod cells or in other cells involved in retina homeostasis (pH control), retinal recycling, or phagocytosis of the ROS discs). We found two cases of an ROS protein also expressed in other tissues, with no other apparent phenotype; e.g., isocitrate dehydrogenase NAD-dependent subunit B is found in many cell types besides rod cells.
The prevalence of proteins from the core visual pathway in disease may have several explanations: first, mutations in other modules central for cellular function may result in a systemic all-or-nothing behavior, affecting the overall viability or proper development of an organism and thereby causing early death. This may be true for critical cytoskeletal proteins and GTPases, as e.g., those involved in vesicle trafficking and maturation, and for proteins involved in metabolic activity. Second, the lack of redundancy for the very specific functions within the visual pathway might cause it to be more susceptible. Here, evolution may have favored high-end functional properties over the robustness of the pathway. Thus, lack of redundancy may have been accepted by evolution even though it interferes with robustness as a pay-off for the high-end performance that is achieved in photoreceptors with single photon detection and with multi-color vision.
Conclusions
Taken together, this work suggests that rhodopsin is able to trigger several distinct physiological activities in addition to its primary function of closing and opening the cAMP-gated cation channel. Considering protein interactions as a result of domain interactions has allowed us to increase the resolution, define discrete functional modules, and add a spatial dimension to this network. Based on this study, we obtained a novel biological insight that offers new testable hypotheses, which have been partially validated through the experiments performed here, namely, of the connectivity of rhodopsin to small GTPases involved in cytoskeleton assembly/disassembly and dynamics, and to vesicle and Golgi trafficking. This suggests a role for rhodopsin in self-regulating and fine-tuning the structural and functional integrity of photoreceptors. Cytoskeleton changes, such as microtubule assembly reorganization, are likely to affect protein transport between the inner and outer segments during light-to-dark changes (reviewed in Reidel et al, 2008), as well as to regulate cell polarity and disc development. The involvement of rhodopsin in regulating intracellular Ca2+ levels suggests its role in an overarching Ca2+-dependent regulatory network that determines dynamic changes in kinase activity and protein complex assembly. This in turn results in higher order physiological behavior, such as cytoskeletal dynamics and vesicular trafficking tuned by light. At a systems level, these network relationships imply a concerted regulation of outer segment structure, polarity, and vesicular trafficking orchestrated via GTPase-guided signaling pathways activated by light, Ca2+-regulated processes activated by cGMP-gated channel activity (and thus also by light), and cytoskeletal and ciliary dynamics (which may also be fine-tuned by light). With respect to disease, we can conclude that, among at least four pathways driven or regulated by rhodopsin, the visual pathway is the only one highly associated with disease, whereas all others are relatively unaffected. Conceptually, our work presents a general approach applicable to the analysis of any cellular pathway. The resulting comprehensive multiscale ‘vision network’ can serve as a basis for elucidating physiological principles of photoreceptor function and may help to identify potential disease-associated proteins and to guide signaling branch-specific therapy development.
Materials and methods
Isolation of ROS and ROS discs
Porcine eyes were obtained from a local slaughterhouse. After the retinae were dissected, two approaches for ROS isolation were compared: that to Molday et al (1987) with that of Papermaster and Dreyer (1974). Briefly, for the Molday protocol, ROS was detached from the retinal tissue by gentle mechanical homogenization in cold isolation medium (20% (w/v) sucrose, 20 mM Tris, 2 mM MgCl2, 130 mM NaCl, at pH 7.2) and separated from the homogenate by loading onto a 27–50% linear sucrose density gradient. Alternatively, fresh retinae were homogenized by shaking in cold isolation medium (34% (w/v) sucrose, 65 mM NaCl, 2 mM MgCl2, and 5 mM Tris-acetate buffer, pH 7.4). ROS was then pelleted by centrifugation, and the remaining retinal tissue was rehomogenized with a teflon homogenizer. Supernatants from both homogenization steps (crude ROS) were combined and loaded onto step-density gradients of 1.15, 1.13, and 1.11 g/ml sucrose. After cold centrifugation in a Beckman SW40 rotor for 1 h at 38 000 r.p.m., purified ROS was collected from the surface of a 1.11–1.13 g/ml sucrose gradient, and the protein content was determined by Bradford assay (Bio-Rad). Osmotically intact discs were isolated from ROS according to Smith et al (1975). ROS was ruptured by osmotic shock and intact discs were separated by flotation in 10% Ficoll (Sigma). After centrifugation (120 000 g, 2 h, 4°C), intact discs were harvested from the Ficoll surface. The purity of the ROS preparations was checked either optically by microscope (Figure 1A, inset) or by immunoblot analysis for RIS markers (BIP and Tom20) (Supplementary Figure S11).
Sucrose density gradient centrifugation
ROS or intact discs were ruptured by osmotic shock, and the membranes were separated from soluble fraction by centrifugation. An amount of membrane equivalent to 1 mg protein was solubilized in 1% (w/v) β-dodecylmaltoside (DM) (Sigma) as described (Mueller and Eichacker, 1999), loaded onto linear 0.1–1.0 M sucrose gradients, and centrifuged for 17 h at 230 000 g at 4°C. Individual gradient fractions were either loaded directly for SDS–PAGE or were precipitated with methanol/chloroform as previously described (Wessel and Flügge, 1984).
SDS–PAGE and immunoblotting
SDS–PAGE and subsequent immunoblotting onto PVDF membranes (Amersham) were carried according to standard procedures. Antibody–antigen complexes were visualized using enhanced chemiluminescence detection (ECL+, Amersham) on Hyperfilm (Amersham). Immunoblots were incubated with the following antibodies: anti-RhoA 26C4 and anti-ROCK II H-85 (Santa Cruz), anti-visual arrestin, anti-transducin α, and anti-rhodopsin (Affinity BioReagents), anti-rhodopsin (Acris Antibodies), anti-CRMP-2 (C4G, a generous gift from M Morishima and Y Ihara, University of Tokyo, Japan), anti-Rac1 (BD Transduction Laboratories), anti-RhoABC (Sigma), anti-BIP (BD Bioscience), and anti-Tom20 (BD Bioscience). HRP-coupled secondary goat-anti-rabbit and goat-anti-mouse antibodies were obtained from Jackson ImmunoResearch.
Immunoprecipitation
IP was performed with anti-RhoA-agarose- or anti-Rac1-agarose-conjugated antibodies (Santa Cruz) or anti-rhodopsin (Acris Antibodies). An amount of ROS equivalent to 500 μg protein was ruptured by osmotic shock in lysis buffer (50 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl, pH 6.8) and centrifuged to separate the membrane and soluble fractions. The membrane fraction was solubilized in 1% (w/v) DM, and the soluble fraction was directly subjected to IP. For anti-rhodopsin IP, solubilized ROS (1% DM) was directly subjected to IP. Non-specific protein binding to agarose beads was prevented by pre-incubation of the fraction with 25% protein G-agarose (Santa Cruz). For IP, the ROS fractions were incubated with 5–10 μg of antibody conjugate/antibody at 4°C for 3 h or overnight with rotation. As a control for non-specific antibody binding species, specific IgGs (Sigma-Aldrich) were used.
Immunohistochemistry
Porcine eyes were obtained from a local slaughterhouse, fixed in 4% paraformaldehyde in 0.1 M phosphate buffer (PB) for 4 h, and rinsed in 0.1 M phosphate-buffered saline (PBS). Cornea, lens, and vitreous body were removed, and the retina was cut in 1.5 × 1.5 cm pieces. The fixed tissue was cryoprotected at 4°C stepwise in 10, 20, and 30% sucrose in PBS, for 1 h for the first two steps and overnight for the last step. Retina was then embedded in tissue-freezing medium (Leica Microsystems) and frozen in liquid nitrogen. In all, 12 μm sections were prepared, mounted on Superfrost glass slides, and air dried at 37°C. Retinal sections were rinsed in PBS and then non-specific binding sites were blocked with PBS containing 10% normal goat serum (NGS), 1% bovine serum albumin (BSA), and 0.3% Triton X-100 for 1 h at room temperature. Sections were incubated overnight at 4°C with the following primary antibodies diluted in blocking solution: rabbit anti-CRMP-2 (1:300; Abcam), rabbit anti-Rac1 (1:100; Sigma), rabbit anti-ROCK II (1:300; Abcam), rabbit anti-PDE6δ (1:200; ABR), or mouse anti-rhodopsin (1:200; Millipore). Sections were then washed in PBS and incubated with the appropriate fluorescent-labeled secondary antibody (goat-anti-rabbit IgG-Alexa 568 or goat anti-mouse IgG-Alexa 568; Molecular Probes) diluted 1:500 in PBS for 1 h at room temperature. Nuclei were counterstained with Sytox Green Nucleic Acid Stain (Molecular Probes). After three final washes in PBS, sections were mounted with Mowiol 4-88 (Polysciences). As negative controls, the primary antibodies were also omitted; in these cases, no staining was observed. Stained cryostat sections were analyzed and scanned with a confocal laser scanning microscope (Zeiss LSM510 META, Jena, Germany), using an argon laser at 488 nm and a He/Ne laser at 543 nm excitation with appropriate filter sets. Images were taken sequentially to assure that only one channel was detected at a time. The Sytox Green nuclear stain was allocated to the blue color channel for convenient viewing. Transmitted light images with DIC optics (Nomarski) were recorded simultaneously. Control sections without primary antibody incubation were scanned with the same laser and detection settings.
Concanavalin A pull down
For concanavalin A pull-down experiments, an amount of ROS equivalent to 300 μg protein was ruptured by osmotic shock in lysis buffer. The membrane ROS fraction was solubilized in 1% (w/v) β-DM (Sigma-Aldrich), and the soluble fraction was directly subjected to concanavalin A pull down. The ROS fractions were incubated with 50 μl of concanavalin A sepharose (Amersham Biosciences) conjugate for 3 h at 4°C. Non-specific protein binding of the rhodopsin-associated protein to concanavalin A was prevented by performing the pull down in the presence of 0.2 mM α-methylmannoside (as the presence of α-methylmannoside lowers the affinity of proteins for the beads).
Blue-native PAGE
Membranes from either ROS or intact discs corresponding to 300 μg protein were suspended in 60 μl buffer containing 750 mM ε-aminocapronic acid, 50 mM bis-Tris, pH 7.0, and 0.5 mM EDTA, and then solubilized in 1% (w/v) DM. The solubilized membrane samples were added to a buffer containing 5% (w/v) Serva Blue G in 750 mM ε-aminocapronic acid, loaded onto 4–12% PAA gradient gels, and electrophoresed (Schägger and von Jagow, 1991). To separate in the second dimension, gel lanes were incubated for 20 min in solubilization buffer containing 2% (w/v) SDS, 66 mM DTT, and 66 mM Na2CO3, and loaded onto denaturing PAA gels.
MS–MALDI-TOF
Selected spots were excised from dried silver-stained gels, destained (Gharahdaghi et al, 1999), dehydrated in 40% acetonitrile (100 μl), and subjected to tryptic proteolysis in 1 mM Tris–HCl, pH 7.5, and 0.01 μg/μl trypsin. In parallel studies, proteins excised from dried gels were subjected to SDS removal by ion-pair extraction before in-gel tryptic proteolysis as described (Zischka et al, 2004). MALDI-TOF PMFs were obtained on a Bruker Reflex III mass spectrometer (Bruker Daltonics, Bremen). Aliquots from each tryptic digest were co-crystallized with a matrix composed of 2.5-dihydroxybenzoic acid (20 mg/ml in 20% acetonitrile, 0.1% trifluoroacetic acid (TFA)) and 2-hydroxy-5-methoxybenzoic acid (20 mg/ml in 20% acetonitrile, 0.1% TFA) in a 9:1 ratio (v/v) on 400 μm AnchorChip targets (Bruker Daltonics). Alternatively, PMF and MS/MS spectra were measured on AB4700 mass spectrometer (Applied Biosystems, Darmstadt, Germany), and aliquots from each tryptic digest were co-crystallized with a matrix comprising 5% cyanohydroxycinnamic acid (in 70% acetonitrile, 0.1% TFA) on steel targets (Applied Biosystems). Database searches were performed using the Mascot software (Perkins et al, 1999).
MS–Orbitrap
LC-MS/MS analysis was performed on an Ultimate3000 nano-HPLC system (Dionex) coupled to an LTQ OrbitrapXL mass spectrometer (Thermo Fisher Scientific) by a nanospray ion source. Tryptic peptide mixtures were automatically injected and loaded with a flow rate of 30 μl/min in 95% buffer C (0.5% TFA in HPLC-grade water) and 5% buffer B (98% acetonitrile, 0.1% formic acid in HPLC-grade water) onto a nanotrap column (100 μm i.d. × 2 cm, packed with Acclaim PepMap100 C18, 5 μm, 100 Å, Dionex). After 5 min, peptides were eluted and separated on an analytical column (75 μm i.d. × 15 cm, Acclaim PepMap100 C18, 3 μm, 100 Å, Dionex) by a linear gradient from 5 to 40% of buffer B in buffer A (2% acetonitrile, 0.1% formic acid in HPLC-grade water) at a flow rate of 300 nl/min over 90 min. The remaining peptides were eluted by a short gradient of 40–100% buffer B over 5 min. Eluting peptides were analyzed by the LTQ OrbitrapXL. From the high-resolution MS pre-scan with a mass range of 300–1500, the 10 most intense peptide ions were selected for fragment analysis in the linear ion trap if they exceeded an intensity of at least 200 counts and if they were at least doubly charged. The normalized collision energy for CID was set to a value of 35, and the resulting fragments were detected with normal resolution in the linear ion trap. The lock mass option was activated, and a background signal of a mass of 445.12002 was used for the lock mass. Every ion selected for fragmentation was excluded for 30 s by dynamic exclusion. The raw data were analyzed using Sequest (Thermo Fisher Scientific) and Scaffold (Proteome Software) as described previously (Gloeckner et al, 2009) against a non-redundant pig, human, mouse, rat, and bovine protein sequence database derived in-house from Uniref100, due to an insufficient number of entries for porcine proteins in the databases. Proteins were considered to be specific when they displayed two or more peptides (with a peptide probability >95%) in at least two out of four experiments. The protein probability threshold was set to 99%. Contaminants such as keratins were removed.
Comparison of different proteomic data sets determined in ROS
All proteins identified in the three different proteomic data sets were mapped to their corresponding human ortholog gene IDs by sequence comparison (using the default BLAST value of 10) and then compared. Since the proteomic analysis of Liu et al (2007) also contains part of the cilium, our ‘near-to-complete’ proteomic data set was defined as the union of the protein set identified by Kwok et al (2008) with the one determined here (Figure 1B).
Protein interaction network analysis
All the results described in our studies were uploaded into Supplementary Tables S3 and S6 according to standard database curation rules (Zanzoni et al, 2002; Ceol et al, 2010). Results from pull-down and co-IP experiments were resolved as binary protein interactions, in which each bait protein was linked to all identified preys. Co-sedimentation and complex-purification experiments that unambiguously identified protein complexes but lacked sufficient detail to determine their exact interaction topology are represented in the database as a list of interactors (complex members). A comprehensive literature mining and database curation effort was also carried out to include as exhaustively as possible the set of rhodopsin/vision-related interactions already described in the scientific literature. The curated interaction sets were represented and analyzed by the Cytoscape visualization and analysis software (Shannon et al, 2003). PPI data from databases included interactions determined from ROS extracts (by co-sedimentation or affinity chromatography). However, in the majority of cases, interaction information was derived from in-vitro experiments, such as large-scale yeast two-hybrid screens, or tandem-affinity purifications in artificial cell systems. Additionally, low-scale PPI data from the literature include data determined with quantitative affinity methods, such as isothermal titration calorimetry, surface plasmon resonance, nuclear magnetic resonance, and peptide arrays (using purified proteins), or PPI data from non-quantitative methods, such as affinity chromatography (GST pull down), crosslinking, and enzyme assays. According to the MINT curation rules, interactions were considered to be direct if they were supported with evidence obtained with one of the following methods, as described in the PSI MI controlled vocabulary: two-hybrid, enzymatic studies, two-hybrid pooling approach, two-hybrid array, β-lactamase complementation, surface plasmon resonance, fluorescence resonance energy transfer, biochemical, biophysical, protein arrays, protease assays, bimolecular fluorescence complementation, far-western blotting, crosslinking studies, electron paramagnetic resonance, two-hybrid fragment pooling approach, protein kinase assay, GTPase assay, enzyme-linked immunosorbent assays, peptide arrays, isothermal titration calorimetry, bioluminescence resonance energy transfer, competition binding, fluorescence technologies, antibody arrays, saturation binding, fluorescence polarization spectroscopy, protease accessibility laddering, affinity technologies, protein crosslinking with a bifunctional reagent, ubiquitin reconstruction, fluorescence microscopy, β-galactosidase complementation, biochemical activity, classical fluorescence spectroscopy, fluorescence technology, phosphatase assay, and reconstituted complex.
Structural information and interaction modeling
Structural information was derived by a combined approach of comparing different domain interaction types, as listed in the 3Did database (http://3did.irbbarcelona.org/; Stein et al, 2005, 2009), between two interacting proteins for which there was experimental evidence that they could form a complex. The 3DID database was improved by analyzing all structures for crystallographic artifacts, using: (i) the interaction annotation of the author, or if this was not available and (ii) the protein quarternary structure (PQS) method (Henrick and Thornton, 1998). The confidence of two domains to mediate the interaction was then assessed using the InterPreTS (http://www.russelllab.org/cgi-bin/tools/interprets.pl/; Aloy and Russell, 2003) scoring system, which evaluated sequence similarity and amino-acid propensities in the interface. We further screened for all X-ray and NMR complex structures and homologs (with a sequence similarity threshold of 70%) among all 360 proteins of the network. For further details, see Supplementary Material 1.
PDEδ subunit activity assay
Recombinant PDEδ protein (rhPDEδ) was obtained from GenWay Biotech at a concentration of 0.7 μg/μl in storage buffer (10 mM Tris, pH 8.0, 0.1% Triton X-100, 0.002% NaN3, and 10 mM dithiothreitol). An amount of ROS (in isolation medium) corresponding to 100 μg protein was ruptured by three freeze-thaw cycles in liquid nitrogen and centrifuged at 4°C for 30 min at 100 000 g (Beckman Optima ultracentrifuge; Rotor TLA110). The resulting pellet, containing the membranous fraction, was resuspended in 100 μl incubation buffer (25 mM Hepes, 20 mM Tris–HCl, pH 7.5, 1 mM dithiothreitol, 1 mM MgCl2, 5 mM EDTA, 150 mM NaCl, and protease inhibitor cocktail (Roche)) and then incubated with different amounts (0, 0.5, 1, 2, 4, 6, or 8 μg) of rhPDEδ for 1 h at 37°C in a horizontal shaker. To rule out that the Triton X-100 in the storage media affected the recombinant PDEδ, all samples were adjusted to the same volume (volume of the sample with the highest PDEδ concentration used) with storage buffer. Samples were separated into membrane and soluble fractions by centrifugation at 4°C for 30 min at 100 000 g and analyzed by SDS–PAGE and western blot using anti-Rac1 antibodies and anti-PDEδ antibodies.
Supplementary Material
Supplementary Material 1–3, Supplementary Figures S1–11
Proteins identified in ROS using OrbiTrap-LC-MS/MS
List of all proteins annotated by gene ontology (GO)
Interactions from literature among the proteins of the initial proteome
Structural evidences from X-ray, homolog structures, 3DID and InterPReTS
Prediction of domains using Pfam
Higher confidence network. Interactions from literature (MINT) and determined in this work (labeled ‘in this work’)
Kinase Predictions using Netphorest
Acknowledgments
This work was supported by the German Federal Ministry for Education and Research through the BMBF grant QuantPro 0316865A, Dynamo 0315513A, and BMBF—IMAGING FKZ 0315508A, to MU. We like to acknowledge the EU funding for financing part of the work: INTERACTION PROTEOME, LSHG-CT-2003-505520 (to MU, GC, and LS), PROSPECTS, grant agreement number HEALTH-F4-2008-201648 (to LS) and SYSCILIA, grant agreement number HEALTH-F5-2010-241955 (to MU). MU and GC received funding from the European Community's Seventh Framework Programme FP7/2009 under grant agreement number 241481, AFFINOMICS.
Author contributions: CK, LS, GC, and MU designed the research. CK, LS, GC, AV, and MU wrote the paper. AV, MS, MB, SB, and AM performed experiments. CK, ACa, AV, NK, ACh, GC, LS, and MU analyzed the data and/or provided data analyses expertise.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams M, Smith UM, Logan CV, Johnson CA (2008) Recent advances in the molecular pathology, cell biology and genetics of ciliopathies. J Med Genet 45: 257–267 [DOI] [PubMed] [Google Scholar]
- Aloy P, Russell RB (2002) Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci USA 99: 5896–5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy P, Russell RB (2003) InterPreTS: protein interaction prediction through tertiary structure. Bioinformatics 19: 161–162 [DOI] [PubMed] [Google Scholar]
- Artemyev NO (2008) Light-dependent compartmentalization of transducin in rod photoreceptors. Mol Neurobiol 37: 44–51 [DOI] [PubMed] [Google Scholar]
- Balasubramanian N, Levay K, Keren-Raifman T, Faurobert E, Slepak VZ (2001) Phosphorylation of the regulator of G protein signaling RGS9-1 by protein kinase A is a potential mechanism of light- and Ca2+-mediated regulation of G protein function in photoreceptors. Biochemistry 40: 12619–12627 [DOI] [PubMed] [Google Scholar]
- Balasubramanian N, Slepak VZ (2003) Light-mediated activation of Rac-1 in photoreceptor outer segments. Curr Biol 13: 1306–1310 [DOI] [PubMed] [Google Scholar]
- Becker V, Schilling M, Bachmann J, Baumann U, Raue A, Maiwald T, Timmer J, Klingmüller U (2010) Covering a broad dynamic range: information processing at the erythropoietin receptor. Science 328: 1404–1408 [DOI] [PubMed] [Google Scholar]
- Berger W, Kloeckener-Gruissem B, Neidhardt J (2010) The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res 29: 335–375 [DOI] [PubMed] [Google Scholar]
- Berghs S, Aggujaro D, Dirkx R Jr, Maksimova E, Stabach P, Hermel JM, Zhang JP, Philbrick W, Slepnev V, Ort T, Solimena M (2000) BetaIV spectrin, a new spectrin localized at axon initial segments and nodes of ranvier in the central and peripheral nervous system. J Cell Biol 151: 985–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesze-Battaglia K, Goldberg AF (2002) Photoreceptor renewal: a role for peripherin/rds. Int Rev Cytol 217: 183–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi A, Lehner B (2009) Tissue specificity and the human protein interaction network. Mol Syst Biol 5: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Carvajal MM, Wollscheid B, Aebersold R, Steimle V, Schamel WW (2004) Two-dimensional blue native/SDS gel electrophoresis of multi-protein complexes from whole cellular lysates: a proteomics approach. Mol Cell Proteomics 3: 176–182 [DOI] [PubMed] [Google Scholar]
- Campagna A, Serrano L, Kiel C (2008) Shaping dots and lines: adding modularity into protein interaction networks using structural information. FEBS Lett 582: 1231–1236 [DOI] [PubMed] [Google Scholar]
- Ceol A, Chatr-aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G (2010) MINT, the molecular interaction database: 2009 update. Nucleic Acids Res 38: D532–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G (2007) MINT: the Molecular INTeraction database. Nucleic Acids Res 35: D572–D574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarkowski J, Witt M, Slusarz R (2005) A hypothesis for GPCR activation. J Mol Model 11: 407–415 [DOI] [PubMed] [Google Scholar]
- De Grip WJ (1982) Purification of bovine rhodopsin over concanavalin A-sepharose. Methods Enzymol 81: 197–207 [DOI] [PubMed] [Google Scholar]
- De La Paz MA, Anderson RE (1992) Lipid peroxidation in rod outer segments. Role of hydroxyl radical and lipid hydroperoxides. Invest Ophthalmol Vis Sci 33: 2091–2096 [PubMed] [Google Scholar]
- Del Conte-Zerial P, Brusch L, Rink JC, Collinet C, Kalaidzidis Y, Zerial M, Deutsch A (2008) Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Mol Syst Biol 4: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Orco D, Schmidt H, Mariani S, Fanelli F (2009) Network-level analysis of light adaptation in rod cells under normal and altered conditions. Mol Biosyst 5: 1232–1246 [DOI] [PubMed] [Google Scholar]
- Deretic D, Williams AH, Ransom N, Morel V, Hargrave PA, Arendt A (2005) Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4). Proc Natl Acad Sci USA 102: 3301–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek S, Krzysko KA, Fotiadis D, Liang Y, Saperstein DA, Engel A, Palczewski K (2004) A concept for G protein activation by G protein-coupled receptor dimers: the transducin/rhodopsin interface. Photochem Photobiol Sci 3: 628–638 [DOI] [PubMed] [Google Scholar]
- Florio SK, Prusti RK, Beavo JA (1996) Solubilization of membrane-bound rod phosphodiesterase by the rod phosphodiesterase recombinant delta subunit. J Biol Chem 271: 24036–24047 [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K (2004) The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett 564: 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Ménager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K (2002) CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol 4: 583–591 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM (1999) Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20: 601–605 [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Boldt K, Ueffing M (2009) Strep/FLAG tandem affinity purification (SF-TAP) to study protein interactions. Curr Protoc Protein Sci 19: Unit 19.20 [DOI] [PubMed] [Google Scholar]
- Gray SM, Kelly S, Robles LJ (2008) Rho signaling mediates cytoskeletal re-arrangements in octopus photoreceptors. Am Malacol Bull 26: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Brown M, Jacobs T, Ferrari G, Cann N, Teo M, Monfries C, Lim L (2001) Collapsin response mediator protein switches RhoA and Rac1 morphology in N1E-115 neuroblastoma cells and is regulated by Rho kinase. J Biol Chem 276: 43482–43486 [DOI] [PubMed] [Google Scholar]
- Hallett MA, Delaat JL, Arikawa K, Schlamp CL, Kong F, Williams DS (1996) Distribution of guanylate cyclase within photoreceptor outer segments. J Cell Sci 109: 1803–1812 [DOI] [PubMed] [Google Scholar]
- Hamer RD, Nicholas SC, Tranchina D, Lamb TD, Jarvinen JL (2005) Toward a unified model of vertebrate rod phototransduction. Vis Neurosci 22: 417–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC (2002) The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J 21: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrick K, Thornton JM (1998) PQS: a protein quaternary structure file server. Trends Biochem Sci 9: 358–361 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hofmann KP, Spahn CM, Heinrich R, Heinemann U (2006) Building functional modules from molecular interactions. Trends Biochem Sci 31: 497–508 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, Tyers M, Salama JJ, Moore S, Ceol A, Chatr-Aryamontri A, Oesterheld M, Stümpflen V, Salwinski L, Nerothin J, Cerami E et al. (2007) Broadening the horizon—level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C, Beltrao P, Serrano L (2008) Analyzing protein interaction networks using structural information. Annu Rev Biochem 77: 415–441 [DOI] [PubMed] [Google Scholar]
- Kiel C, Serrano L (2006) The ubiquitin domain superfold: structure-based sequence alignments and characterization of binding epitopes. J Mol Biol 355: 821–844 [DOI] [PubMed] [Google Scholar]
- Kim PM, Lu LJ, Xia Y, Gerstein MB (2006) Relating three-dimensional structures to protein networks provides evolutionary insights. Science 314: 1938–1941 [DOI] [PubMed] [Google Scholar]
- Krishnan A, Duda T, Pertzev A, Kobayashi M, Takamatsu K, Sharma RK (2009) Hippocalcin, new Ca(2+) sensor of a ROS-GC subfamily member, ONE-GC, membrane guanylate cyclase transduction system. Mol Cell Biochem 325: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn H (1978) Light-regulated binding of rhodopsin kinase and other proteins to cattle photoreceptor membranes. Biochemistry 17: 4389–4395 [DOI] [PubMed] [Google Scholar]
- Kühn H, Hall SW, Wilden U (1984) Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett 176: 473–478 [DOI] [PubMed] [Google Scholar]
- Kwok MC, Holopainen JM, Molday LL, Foster LJ, Molday RS (2008) Proteomics of photoreceptor outer segments identifies a subset of SNARE and Rab proteins implicated in membrane vesicle trafficking and fusion. Mol Cell Proteomics 7: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb TD, Pugh EN Jr (2004) Dark adaptation and the retinoid cycle of vision. Prog Retin Eye Res 23: 307–830 [DOI] [PubMed] [Google Scholar]
- Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A (2003) Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem 278: 21655–21662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BP, Strittmatter SM (2001) Semaphorin-mediated axonal guidance via Rho-related G proteins. Curr Opin Cell Biol 13: 619–626 [DOI] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN Jr, Rux JJ, Speicher DW, Pierce EA (2007) The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics 6.8: 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzesco AM, Galli T, Louvard D, Zahraoui A (1998) The rod cGMP phosphodiesterase delta subunit dissociates the small GTPase Rab13 from membranes. J Biol Chem 273: 22340–22345 [DOI] [PubMed] [Google Scholar]
- Mazelova J, Astuto-Gribble L, Inoue H, Tam BM, Schonteich E, Prekeris R, Moritz OL, Randazzo PA, Deretic D (2009) Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J 28: 183–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R, McCulloch D, Lutz E, Johnson M, MacKenzie C, Fennell M, Fink G, Zhou W, Sealfon SC (1998) Rhodopsin-family receptors associate with small G proteins to activate phospholipase D. Nature 392: 411–414 [DOI] [PubMed] [Google Scholar]
- Molday RS, Hicks D, Molday L (1987) Peripherin. A rim-specific membrane protein of rod outer segment discs. Invest Ophthalmol Vis Sci 28: 50–61 [PubMed] [Google Scholar]
- Mueller B, Eichacker L (1999) Assembly of the D1 precursor in monomeric photosystem II reaction center precomplexes precedes chlorophyll a-triggered accumulation of reaction center II in barley etioplasts. Plant Cell 11: 2365–2377 [PMC free article] [PubMed] [Google Scholar]
- Nair KS, Hanson SM, Kennedy MJ, Hurley JB, Gurevich VV, Slepak VZ (2004) Direct binding of visual arrestin to microtubules determines the differential subcellular localization of its splice variants in rod photoreceptors. J Biol Chem 279: 41240–41248 [DOI] [PubMed] [Google Scholar]
- Nickell S, Park PS, Baumeister W, Palczewski K (2007) Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J Cell Biol 177: 917–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijtmans LG, Henderson NS, Holt IJ (2002) Blue native electrophoresis to study mitochondrial and other protein complexes. Methods 26: 327–334 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Palczewski K (2006) G protein-coupled receptor rhodopsin. Annu Rev Biochem 75: 743–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289: 739–745 [DOI] [PubMed] [Google Scholar]
- Panfoli I, Calzia D, Bianchini P, Ravera S, Diaspro A, Candiano G, Bachi A, Monticone M, Aluigi MG, Barabino S, Calabria G, Rolando M, Tacchetti C, Morelli A, Pepe IM (2009) Evidence for aerobic metabolism in retinal rod outer segment disks. Int J Biochem Cell Biol 41: 2555–2565 [DOI] [PubMed] [Google Scholar]
- Papermaster DS, Dreyer WJ (1974) Rhodopsin content in the outer segment membranes of bovine and frog retinal rods. Biochemistry 13: 2438–2444 [DOI] [PubMed] [Google Scholar]
- Paquet-Durand F, Beck S, Michalakis S, Goldmann T, Huber G, Muhlfriedel R, Trifunovic D, Fischer MD, Fahl E., Duetsch G, Becirovic E, Wolfrum U, van Veen T, Biel M, Tanimoto N, Seeliger MW (2010) A key role for cyclic-nucleotide gated (CNG) channels in cGMP-related retinitis pigmentosa. Hum Mol Genet 20: 941–947 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Plantner JJ, Kean EL (1976) Carbohydrate composition of bovine rhodopsin. J Biol Chem 251: 1548–1552 [PubMed] [Google Scholar]
- Poetsch A, Molday LL, Molday RS (2001) The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J Biol Chem 276: 48009–48016 [DOI] [PubMed] [Google Scholar]
- Reidel B, Goldmann T, Giessl A, Wolfrum U (2008) The translocation of signaling molecules in dark adapting mammalian rod photoreceptor cells is dependent on the cytoskeleton. Cell Motil Cytoskeleton 65: 785–800 [DOI] [PubMed] [Google Scholar]
- Ridge KD, Abdulaev NG, Sousa M, Palczewski K (2003) Phototransduction: crystal clear. Trends Biochem Sci 28: 479–487 [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS et al. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Santonico E, Castagnoli L, Cesareni G (2005) Methods to reveal domain networks. Drug Discov Today 10: 1111–1117 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199: 223–231 [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Stephan I, Jensen ON, Illenberger D, Gierschik P (2000) The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat Struct Biol 7: 122–126 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HG Jr, Stubbs GW, Litman BJ (1975) The isolation and purification of osmotically intact discs from retinal rod outer segments. Exp Eye Res 20: 211–217 [DOI] [PubMed] [Google Scholar]
- Stein A, Panjkovich A, Aloy P (2009) 3did update: domain-domain and peptide-mediated interactions of known 3D structure. Nucleic Acids Res 37: D300–D304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Russell RB, Aloy P (2005) 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res 33: D413–D417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B et al. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- Swiatek-de Lange M, Müller B, Ueffing M (2008) Native fractionation: isolation of native membrane-bound protein complexes from porcine rod outer segments using isopycnic density gradient centrifugation. Methods Mol Biol 484: 161–175 [DOI] [PubMed] [Google Scholar]
- Veltel S, Wittinghofer A (2009) RPGR and RP2: targets for the treatment of X-linked retinitis pigmentosa? Expert Opin Ther Targets 13: 1239–1251 [DOI] [PubMed] [Google Scholar]
- Walikonis RS, Oguni A, Khorosheva EM, Jeng CJ, Asuncion FJ, Kennedy MB (2001) Densin-180 forms a ternary complex with the (alpha)-subunit of Ca2+/calmodulin-dependent protein kinase II and (alpha)-actinin. J Neurosci 21: 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensel TG (2008) Signal transducing membrane complexes of photoreceptor outer segments. Vision Res 48: 2052–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A, Grimm C, Samardzija M, Remé CE (2005) Molecular mechanisms of light-induced photoreceptor apoptosis and neuroprotection for retinal degeneration. Prog Retin Eye Res 24: 275–306 [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Wieland T, Ulibarri I, Aktories K, Gierschik P, Jakobs KH (1990a) Interaction of small G proteins with photoexcited rhodopsin. FEBS Lett 263: 195–198 [DOI] [PubMed] [Google Scholar]
- Wieland T, Ulibarri I, Gierschik P, Hall A, Aktories K, Jakobs KH (1990b) Interaction of recombinant rho A GTP-binding proteins with photoexcited rhodopsin. FEBS Lett 274: 111–114 [DOI] [PubMed] [Google Scholar]
- Wilden U, Wust E, Weyand I, Kuhn H (1986) Rapid affinity purification of retinal arrestin (48 kDa protein) via its light-dependent binding to phosphorylated rhodopsin. FEBS Lett 207: 292–295 [DOI] [PubMed] [Google Scholar]
- Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G (2002) MINT: a molecular INTeraction database. FEBS Lett 513: 135–140 [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu XH, Zhang K, Chen CK, Frederick JM, Prestwich GD, Baehr W (2004) Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J Biol Chem 279: 407–413 [DOI] [PubMed] [Google Scholar]
- Zischka H, Gloeckner CJ, Klein C, Willmann S, Swiatek-de Lange M, Ueffing M (2004) Improved mass spectrometric identification of gel-separated hydrophobic membrane proteins after sodium dodecyl sulfate removal by ion-pair extraction. Proteomics 4: 3776–3782 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1–3, Supplementary Figures S1–11
Proteins identified in ROS using OrbiTrap-LC-MS/MS
List of all proteins annotated by gene ontology (GO)
Interactions from literature among the proteins of the initial proteome
Structural evidences from X-ray, homolog structures, 3DID and InterPReTS
Prediction of domains using Pfam
Higher confidence network. Interactions from literature (MINT) and determined in this work (labeled ‘in this work’)
Kinase Predictions using Netphorest