Abstract
Proteome-scale protein interaction maps are available for many organisms, ranging from bacteria, yeast, worms and flies to humans. These maps provide substantial new insights into systems biology, disease research and drug discovery. However, only a small fraction of the total number of human protein–protein interactions has been identified. In this study, we map the interactions of an unbiased selection of 5026 human liver expression proteins by yeast two-hybrid technology and establish a human liver protein interaction network (HLPN) composed of 3484 interactions among 2582 proteins. The data set has a validation rate of over 72% as determined by three independent biochemical or cellular assays. The network includes metabolic enzymes and liver-specific, liver-phenotype and liver-disease proteins that are individually critical for the maintenance of liver functions. The liver enriched proteins had significantly different topological properties and increased our understanding of the functional relationships among proteins in a liver-specific manner. Our data represent the first comprehensive description of a HLPN, which could be a valuable tool for understanding the functioning of the protein interaction network of the human liver.
Keywords: human liver, network, protein–protein interaction, yeast two hybrid
Introduction
Large-scale human protein–protein interaction maps provide new insights into protein functions, pathways, molecular machines and functional protein modules. However, only a fraction of the total number of human protein–protein interactions has been identified (Rual et al, 2005; Stelzl et al, 2005; Stumpf et al, 2008; Venkatesan et al, 2009). Enhancing the assembly rate of the human interactome remains among the most important goals of current research. Moreover, studies have indicated that tissue-specific networks are vital to understanding tissue specificity, given that each cell has an identical proteome (Bossi and Lehner, 2009; Kirouac et al, 2010).
In this study, we map the interactions of an unbiased selection of 5026 human liver expression proteins by a yeast two-hybrid (Y2H) technology and establish a human liver protein interaction network (HLPN) composed of 3484 interactions among 2582 proteins. Computational biological analyses and independent biochemical assays validated the overall quality of the Y2H interactions. The network is highly enriched for metabolic enzymes and liver-specific (LS), liver-phenotype (LP) and liver-disease (LD) proteins that are individually critical for the maintenance of liver functions. The liver enriched proteins had significantly different topological properties and, therefore, increased our understanding of functional relationships of proteins in a liver-specific manner. This network can also help to predict genes that are related to liver phenotype and liver diseases in mice and humans. In addition, we determined that GIT2 (G-protein-coupled receptor (GPCR)-kinase interacting protein 2) recruits the TNFAIP3 (tumor necrosis factor, α-induced protein 3) ubiquitin-editing complex to IKBKG (inhibitor of κ light polypeptide gene enhancer in B cells, kinase γ) and is involved in the regulation of the NF-κB pathway.
Results and discussion
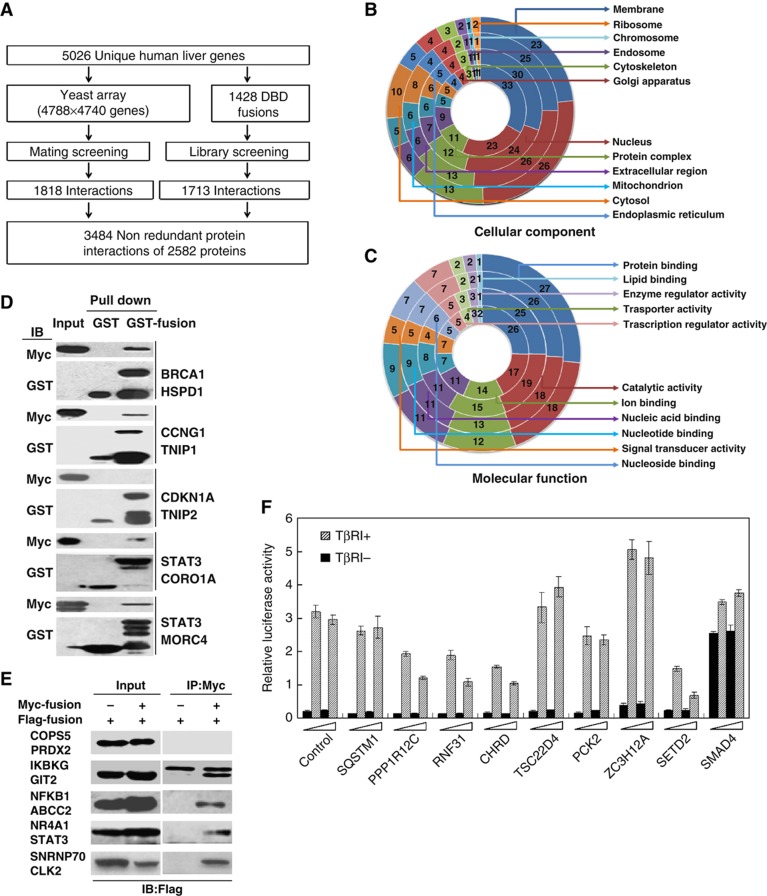
To better understand the regulatory and functional relationships between the proteins expressed in the liver, we developed a strategy for constructing a liver protein interaction network (Figure 1A). The human liver expresses >18 000 genes, and, therefore, a complete mapping of its interactome remains beyond current capabilities. Therefore, we selected 5026 proteins based on the characteristics of the human liver proteome (CNHLPP Consortium, 2010) for interaction screening (Supplementary Table S1), which includes the functional and regulatory proteins that play important roles in liver development, regeneration, metabolism, biosynthesis and diseases. The data set includes 684 metabolism enzymes (ME), consisting of liver-specific bile acid-, bilirubin- and drug-ME, 201 LS proteins, 337 LP proteins (mouse-homologous proteins, knockouts of which cause liver phenotypes) and 488 LD-related proteins. Unbiased, these proteins represent the human liver proteome through Gene Ontology (GO) (Ashburner et al, 2000) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (Kanehisa et al, 2004) (Figure 1B and C; Supplementary Figure S1). These molecules are involved in 84 of the 85 KEGG metabolic pathways, including those of carbohydrates, lipids, nucleotides, amino acids, vitamins, hormones, bile acid and drugs (Supplementary Figure S1A). The selected proteins covered all 114 human regulatory pathways in the KEGG, including the ErbB, MAPK and TGF-β signaling pathways, which have been shown to play key roles in the regulation of liver function (Supplementary Figure S1B).
Figure 1.
Construction of the proteome-scale HLPN. (A) Strategy of high-throughput Y2H screening. (B, C) Distribution of the GO categories of cellular component (CC) and molecular function (MF). From inside to outside, the rings represent all of the human proteins (16 022 with CC and 15 259 with MF annotations), human liver proteins (12 066 with CC and 10 863 with MF annotations), Y2H matrix proteins (4553 with CC and 4374 with MF annotations) and HLPN proteins (2342 with CC and 2252 with MF annotations). Each section reflects the percentage of proteins assigned to the given GO category. (D) GST pull-down assay. Bacteria-expressed GST or GST-tagged proteins were immobilized on glutathione-Sepharose 4B beads, and the beads were subsequently incubated with the Myc- or Flag-tagged proteins expressed in the HEK293T cell lysates. The proteins were detected using the indicated antibodies. (E) Co-IP assay. Flag- or Myc-tagged plasmids were transfected into HEK293T cells. Immunoprecipitations were performed using anti-Myc or anti-Flag antibodies and protein A/G-agarose. The lysates and immunoprecipitates were detected using the indicated antibodies. Myc-, GST- and Flag-fusions mean the fusion proteins with Myc, GST or Flag tags. (F) Luciferase reporter gene assays of SMAD3. Data are presented as mean values±s.d. (n=3). The results are representative of three independent experiments.
To screen the protein interactions using a high-stringency Y2H system, a matrix with 4788 × 4740 unique genes (based on 5026 selected proteins) was successfully constructed by a bacterial homologous recombination method (Zhu et al, 2010) and was screened as described (Rual et al, 2005). After detecting >2.26 × 107 combinations (covering 1.13 × 107 unique pairs), 1818 interactions among 1777 proteins were obtained (Supplementary Table S2). We aimed to construct a protein interaction network reflecting the characteristics of the liver. Because the Y2H array that we constructed was unable to catch all of the proteins expressed in the liver, the information that we obtained may be limited. Additionally, previous research has shown that different Y2H screening strategies can obtain more interaction information. Therefore, we randomly selected 1428 baits from 5026 proteins for Y2H library screening. The functional classification of 1428 baits is consistent with the 5026 proteins, which are involved primarily in such functions as liver metabolism, apoptosis, cell proliferation, transcription, signal transduction, transport and biosynthesis. We screened an adult liver cDNA library using 1428 constructed baits, and we obtained 1713 non-redundant protein interactions involving 1239 proteins (Supplementary Table S2). Only 47 interactions overlapped between the two screens, which suggest the necessity of performing array and library screening in parallel. In total, 3484 protein interactions involving 2582 proteins were obtained, and only 258 interactions were reported previously in the Human Protein Reference Database (HPRD) (http://www.hprd.org).
To evaluate the reliability of the interactions, we assigned a confidence score to each interaction by a bioinformatics tool, the PRINCESS (Li et al, 2008), which uses Bayesian network approaches to combine multiple heterogeneous biological findings to assign reliability score to protein–protein interactions. In the 2940 interactions identifiable by the PRINCESS, we found 1105 high-confidence interactions (i.e. score >2), thereby indicating that 37.6% of the interactions were supported by bioinformatics evidence. The rate was higher than that of the other two large-scale data sets (Rual et al, 2005; Stelzl et al, 2005) (30.6 and 22.0%, respectively). Additionally, we adopted a reported method to evaluate the confidence of the HLPN data set (Yu et al, 2008). It is estimated that the false positive rate of the HLPN is 58.9%. This finding might be because the analysis method greatly exaggerates the false positive rate. There are many true interactions in the golden negative interaction data set (nucleus–membrane protein pairs). Even in the HPRD, there are 192 interactions between 1044 nucleus proteins and 783 membrane proteins. Other than the bioinformatics evaluation of the confidence of the HLPN, the more convincing strategies are to validate the interactions by conducting independent experiments. Thus, we validated randomly selected interactions by performing independent biochemical or cellular assays (Supplementary Table S2). A total of 47 interactions were tested by a GST pull-down assay with a verification rate of 72.3% (Figure 1D). Another 94 interaction pairs were verified by a co-immunoprecipitation assay with a verification rate of 76.6% (Figure 1E). We also examined 117 interactions by a co-localization assay and found that 84.6% were co-localized (Supplementary Figure S2). To confirm the functional interactions, we validated selected SMAD3-interacting candidates by a luciferase reporter gene assay. Mutant TβRI that phosphorylated SMAD3 constitutively was added to activate the reporter gene. We found that six of the factors affected the reporter gene in a dose-dependent manner (Figure 1F). Taken together, our results show that a large percentage of the Y2H interaction screenings can be verified by other independent biochemical approaches and thus provide clear evidence to support the low rate of false positives in the obtained interactions. At least 72% of the interactions were confirmed by independent biochemical or cellular assays (Supplementary Table S2). Therefore, the false positive rate might be <28%, which is similar to previous reports of human interactome data sets (Rual et al, 2005; Stelzl et al, 2005). Another common feature of large-scale two-hybrid screening is the high frequency of false negatives or missed interactions. It has been estimated that <20% of the interactions could be identified by Y2H technology (Venkatesan et al, 2009). In our study, two different screening strategies were used to ensure that more protein interactions were obtained. However, a false negative ratio of ∼60% of the HLPN is estimated based on the previous report of the size of the human interactome (Stumpf et al, 2008). This result might be because of the technical limitations of Y2H technology.
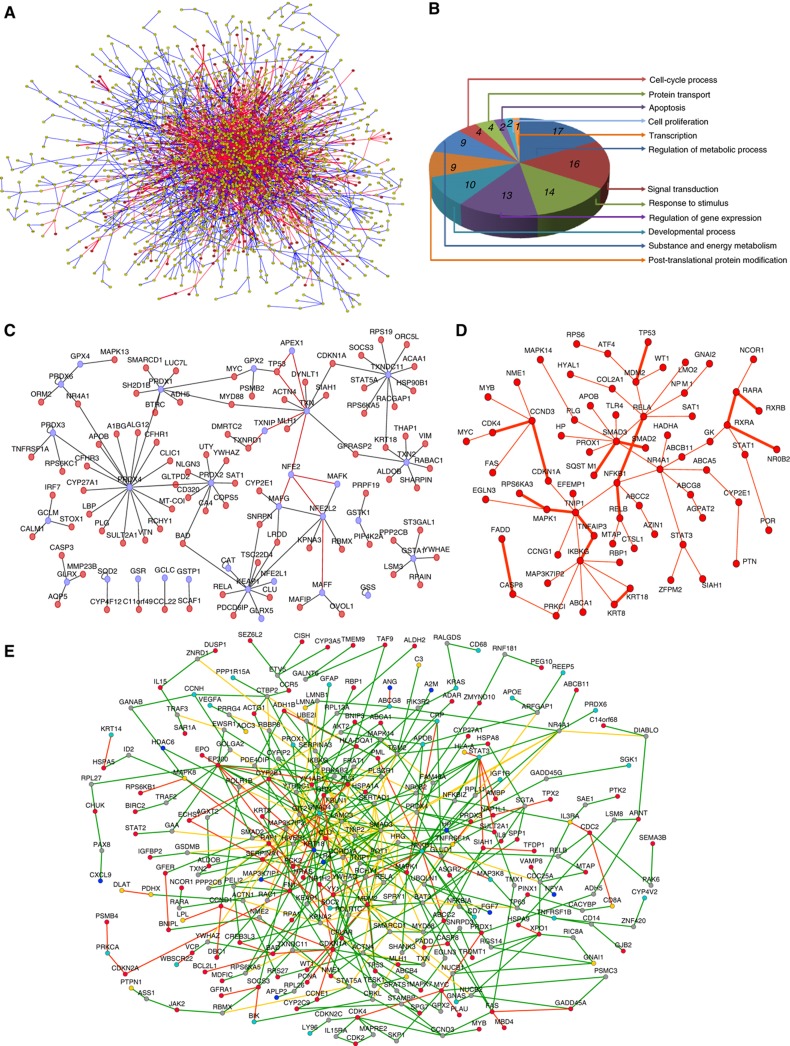
Using computational analysis, we found that the HLPN is composed of a large, connected subnet containing 2215 proteins and 134 smaller networks composed of fewer than 10 proteins (Figure 2A). The global properties of the HLPN were similar to the features found in previous reports (Supplementary Figure S3) (Rual et al, 2005; Stelzl et al, 2005). However, several features suggest that the network could be specifically used to understand the human liver proteome. First, the HLPN includes 324 ME, 154 LS, 218 LP and 175 LD, which have been shown to be specifically expressed in the liver or required individually for controlling liver functions. The HLPN indicated functional properties similar to those of the initial set of bait genes. Moreover, the distribution of the GO categories of HLPN proteins was consistent with that of the liver proteome (Figure 1B and C).
Figure 2.
Network views of the HLPN. (A) Visualization of the HLPN. Yellow circles, selected Y2H proteins; red lines, interactions identified by library screening; blue lines, interactions identified by array screening; green lines, overlapping interactions of library and array screening; bold edges, interactions reported in the HPRD. (B) Distribution of the GO categories of biological processes for metabolic enzymes and their partners in the HLPN. The sections reflect the percentage of proteins assigned to the given GO category. (C) The ROS subnetwork. Blue circles, ROS proteins; red lines, data from the HPRD. (D) The liver-phenotype proteins network. Bold edges, interactions reported in the HPRD. (E) Liver-disease-associated proteins network. Red circles, HCC; cyan circles, cholangiocellular carcinoma; blue circles, liver fibrosis; yellow circles, other liver-disease proteins; gray circles, proteins without human liver disease annotation in the Library of Molecular Associations database (http://www.medicalgenomics.org/databases/loma); red lines: interactions among liver-disease proteins; yellow lines, interactions among non-liver-disease proteins; green lines, other interactions.
Second, the HLPN revealed the different topological properties of liver enriched proteins. To analyze the local network characteristics, we extracted subnetworks for ME, LS, LP and LD. Next, we compared these subnetworks with the randomly generated networks. We examined two important network measurements: degree and betweenness centrality. The global properties of the four protein sets were summarized (Table I). We found that the degree centrality and the betweenness centrality values of LP and LD were significantly higher than for other HLPN proteins. This result is consistent with a previous report of phenotype proteins in yeast (Said et al, 2004) and other human-disease proteins (Goh et al, 2007). In the ME and LS subnetworks, we found that ME or LS did not have vital network positions in the HLPN. However, the degree and the betweenness centrality of their partners were significantly greater than the expected values. These results indicated that ME and LS tended to interact with the proteins that occupied important network positions. Significantly, the GO analysis revealed that the neighbors of LS are primarily involved in development, regulation of gene expression and apoptosis. Accumulated evidence shows that these proteins play a key role in liver development and formation. In fact, among the 200 proteins that interact with LS, 44 are LP and 27 are LD (Supplementary Table S3). This observation led us to propose the hypothesis that the expression of a few LS proteins might be an economical and effective means of regulating liver cell development and functional formation through protein interactions. The HLPN contains only part of the entire human liver network, which might lead to sampling bias. To address this concern, we compiled a virtual liver protein–protein interaction network with the data from HPRD databases using a reported method (Bossi and Lehner, 2009). All of the proteins that expressed in the human liver were included in constructing the network. In the compiled liver protein interaction network, all topological properties of ME, LS, LP and LD remained true (Supplementary Table S4). Moreover, we randomly added or removed 5–20% of the edges and found that all of the conclusions still held, which suggests that the topological features of ME, LS, LP and LD of the HLPN are not artifacts of the biased data sets.
Table 1. Topological analysis of metabolic enzymes and liver-specific, liver-phenotype and liver disease-associated proteins in the HLPNa.
| Classification | ME | MEP | LS | LSP | LP | LPP | LD | LDP |
|---|---|---|---|---|---|---|---|---|
| LD, liver disease-associated proteins; LDP, interaction partners of LD; LP, liver-phenotype proteins; LPP, interaction partners of LP; LS, liver-specific proteins; LSP, interaction partners of LS; ME, metabolic enzymes; MEP, interaction partners of ME. | ||||||||
| aDegree centrality and betweenness centrality values of the indicated group of proteins are shown. The non-parametric Mann–Whitney U-test was used to compare the indicated group of proteins with the other proteins of HLPN. A P-value of <0.01 was regarded as statistically significant. | ||||||||
| bNumbers in parentheses are the values for the other proteins in the HLPN. | ||||||||
| Degree centrality | 2.3 (3.0)b | 5.7 (2.3) | 2.9 (2.9) | 7.0 (2.4) | 4.7 (2.7) | 4.4 (2.4) | 4.0 (2.8) | 4.5 (2.3) |
| P-value | 3.3 × 10−2 | 4.6 × 10−56 | 2.8 × 10−1 | 1.6 × 10−46 | 5.9 × 10−7 | 2.8 × 10−33 | 6.3 × 10−9 | 4.7 × 10−36 |
| Betweenness centrality ( × 10−3) | 1.6 (2.5) | 6.5 (1.4) | 2.6 (2.3) | 7.9 (1.6) | 5.5 (2.1) | 4.2 (1.7) | 3.6 (2.2) | 4.7 (1.5) |
| P-value | 9.0 × 10−2 | 5.5 × 10−52 | 1.7 × 10−1 | 7.9 × 10−42 | 8.8 × 10−6 | 1.6 × 10−28 | 6.6 × 10−9 | 1.8 × 10−31 |
Third, novel interactions of ME connect their biological functions to form multiple cellular processes. The HLPN contains 324 ME that are involved in 637 interactions. Among them, 74 interactions are between two ME, of which 32 interactions are in the same KEGG metabolic pathway, and the others are enzymes from different metabolic pathways (Supplementary Table S5). We detected six interactions (UGT1A1/UGDH, NAGK/GNPNAT1, NME1/POLR1C, GK/GPD1, CYP4F12/SOD2 and RDH13/CYP3A5) that are direct neighbors in the human metabolic network, which might allow the channeling of metabolic intermediates from one active site to the next. Interestingly, we found that only 17% of the 446 ME partners participated in the regulation of the metabolic process by GO classification. Most of the ME partners are involved in various cellular processes (Figure 2B), which suggests that ME might directly play roles in multiple cellular functions other than metabolism in the liver. For example, 4-hydroxyphenylpyruvate dioxygenase and sorbitol dehydrogenase were found to connect with the NF-κB pathway and regulate its transcription activity (Supplementary Figure S4). Additionally, glycerol kinase (GK) was found to associate with nuclear receptors NR4A1 and retinoid X receptor A (Perlmann and Jansson, 1995). The overexpression of GK inhibited the binding of NR4A1 to its specific DNA-binding sequence and the transcription activity (Supplementary Figure S5).
Reactive oxygen species (ROS) are created in normal hepatocytes and are critical for its normal physiological processes. To maintain an appropriate level of ROS, cells have developed an enzymatic antioxidative system. We found that these enzymes bind strongly with each other (Figure 2C). PRDX4 binds directly to PRDX1, 2 and 3, which suggests that various peroxiredoxins might form a homo- or hetero-polymer. Moreover, these enzymes were also found to interact with many proteins that are involved in multiple cellular processes, such as the interaction of PRDX4 with APOB, LBP, CYP27A1 and SULT2A1. All of these enzymes play a role in cholesterol metabolism. Interestingly, GPX2 was found to interact with TP53 and MYC, both of which are key transcription factors for the regulation of ROS homeostasis (Sharpless and DePinho, 2002; Prochownik, 2008). It has been reported that PRDX1 interacts with c-Myc and reduces the ability of c-Myc to activate transcription and, presumably, limits its induction of ROS (Mu et al, 2002). Thus, this regulatory model might play a significant role in the regulation of ROS homeostasis.
Fourth, we can predict LP and LD based on their connectivity features (Figure 2D and E). Both LP and LD proteins tend to interact with LP and LD proteins, respectively, in the HLPN, which is a condition that could be used to predict the potential LP and LD proteins. A total of 218 LP are involved in 895 interaction pairs in the HLPN. Of these pairs, 94 LP (43.1%) interact with each other via 93 interactions, which is significantly greater than the expected value in a random network (empirical P-value <0.001, permutation test). Among the 93 interaction pairs, 77 pairs composed a connective cluster with an average short path length of 4.8 (Figure 2D). It is worth noting that, in the LP subnetwork, several proteins, such as IKBKG, SMAD3, RELA and CCND3, are critical for the network topology. Moreover, 166 proteins were found that interacted with two or more known LP proteins, among which 31 proteins were LP. This finding represents a seven-fold enrichment compared with all of the proteins encoded by the human genome (Supplementary Table S6). For example, EP300 is an acetyltransferase that interacts with seven liver-phenotype proteins. EP300 is highly expressed in the human liver. Knockout of EP300 results in defects of the heart, lung and small intestine and death at midgestation. Recently, it was reported that acetylation of metabolic enzymes of the liver is ubiquitous and is important for their function (Zhao et al, 2010). Thus, it is reasonable that EP300 might contribute to liver-specific functions and change the phenotype of the liver. Moreover, 50/175 (28.6%) LD proteins interact with each other to produce 31 non-self interactions (27 were not reported). This interaction was significantly greater than in the random network (empirical P-value <0.001, permutation test). In the HLPN, cross-validation using the known LP and LD as benchmarks revealed that the enrichment is 10.6- and 31.2-fold, respectively, which are greater values than those of randomly selected proteins. These features indicate that the HLPN information alone may offer a simple, efficient means by which to annotate protein function and prioritize candidate genes for complex human diseases. For example, we found that 27 proteins were connected to more than two hepatocellular carcinoma (HCC)-related proteins, and six of these proteins were reported to be related to HCC (Supplementary Table S7). The chance of these 27 proteins being HCC proteins was 3.4-fold higher than for randomly selected proteins from the HLPN (P-value=0.006, hypergeometry distribution test). For example, the immune response and inflammation signaling pathway is closed relative to liver cancer (He and Karin, 2011). A few members of the NF-κB signaling pathway, such as IKBKG, MYD88 and NFKB1, were identified as potential HCC candidates that might play certain roles during the pathogenesis of HCC.
Finally, many interactions were observed that involve critical signal transduction factors. In the HLPN, 279 proteins were distributed among 11 signal transduction pathways in the KEGG; these proteins were mainly involved in MAPK, ERbB, VEGF, Wnt, TGF-β and other signaling pathways that participate in the regulation of liver functions. The 279 proteins participated in 1211 interactions with 1057 proteins, among which 778 proteins were not annotated as signal transduction cofactors in the KEGG. A total of 76 pairs of interactions are annotated in a same KEGG signaling pathway, and 37 pairs are supported by the literature. This finding suggests that another 39 pairs have a high reliability and indicates new mechanisms by which proteins participate in the regulation of corresponding signaling pathways (Supplementary Table S8). In the HLPN, 141 unannotated proteins are linked to two or more proteins in the same signaling pathway, suggesting that they are candidate regulators of these signaling pathways. For example, PHC2, SHANK3, KHDRBS1, FAM59A and ARHGEF5 can be linked to two or more proteins in the MAPK pathway, of which KHDRBS1, SHANK3 and FAM59A had been reported to participate in the MAPK pathway (Martin-Romero and Sanchez-Margalet, 2001; Schuetz et al, 2004; Tashiro et al, 2009). Moreover, 135 proteins were identified as cross-talk proteins in the HLPN, which links two or more different signal transduction pathways (Supplementary Table S9). Furthermore, 53 interactions were identified between proteins involved in two different signaling pathways and were considered potential cross-talk of different pathways (Supplementary Table S10). In addition, our studies confirmed that the transcriptional activity of STAT3 is negatively regulated through binding with NFKBIZ (Wu et al, 2009), and NUMBL inhibits TNF-α and IL-1β-induced activation of NF-κB through interaction with MAP3K7IP2 (Ma et al, 2008). The latter results link the NF-κB pathway to the JAK-STAT pathway and the Notch pathway. We also found that RELA negatively regulates the Nrf2-Keap1 oxidative stress signaling pathway through interaction with KEAP1 (Yu et al, 2011).
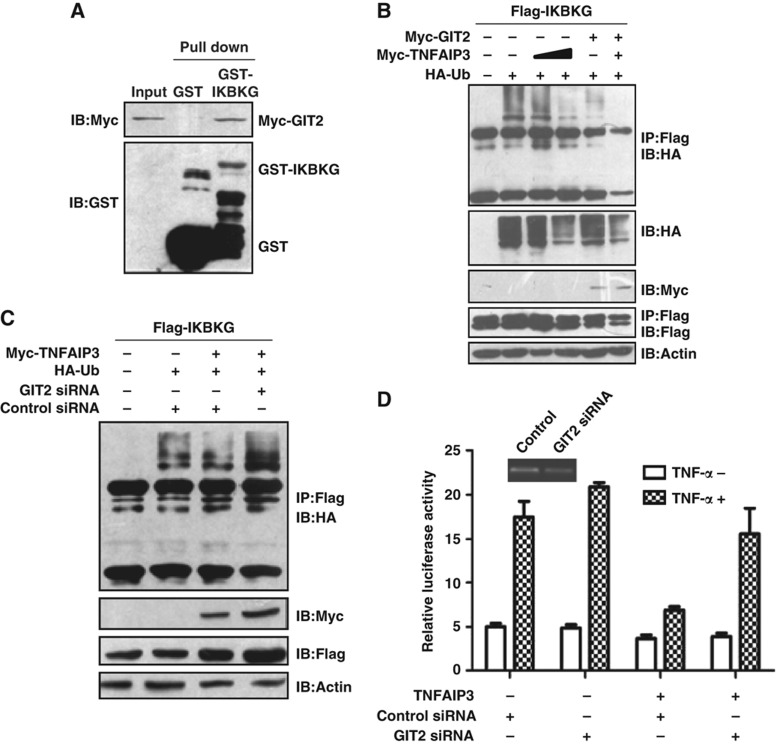
SPOP, TNIP1, TRAF1, IKBKG, TNFAIP3 and NFKBIB were identified as putative binding partners of the GPCR-kinase interacting protein 2 (GIT2). Of these partners, TNFAIP3 and TNIP1 are subunits of the TNFAIP3 ubiquitin-editing complex, which mediates the deubiquitination of IKBKG and negatively regulates the NF-κB pathway (Wertz et al, 2004; Oshima et al, 2009). GIT2 is a ubiquitous multidomain protein that has an important role in the scaffolding of signaling cascades (Hoefen and Berk, 2006). Therefore, we proposed that GIT2 may be involved in the NF-κB pathway through regulation of the interaction between IKBKG and TNFAIP3. To test this hypothesis, we confirmed the interaction between GIT2 and IKBKG (Figures 1E and 3A). The overexpression of GIT2 enhanced the deubiquitination activity of TNFAIP3 toward IKBKG, and siRNA targeted at GIT2 abrogated the TNFAIP3-dependent deubiquitination of IKBKG and impaired the ability of TNFAIP3 to inhibit NF-κB activation (Figure 3B–D). This finding suggests that endogenous GIT2 plays a role in negatively regulating inducible NF-κB activity.
Figure 3.
GIT2 is a negative regulator of the NF-κB signaling pathway. (A) Confirmation of the interaction between GIT2 and IKBKG by the GST pull-down assay. (B) GIT2 enhances the TNFAIP3-dependent deubiquitination of IKBKG. HEK293 cells were transfected with the indicated plasmids. Cell extracts were immunoprecipitated with anti-Flag antibody and detected by western blotting. (C) Knockdown of GIT2 by siRNA impairs the TNFAIP3-dependent deubiquitination of IKBKG. HEK293 cells were cotransfected with the indicated plasmids or the GIT2 siRNA. Cell extracts were immunoprecipitated with an anti-Flag antibody and detected by western blotting. (D) Knockdown of GIT2 impairs the TNFAIP3-mediated inhibition of NF-κB activity. HEK293 cells were cotransfected with the indicated plasmids. One day after transfection, the cells were stimulated with TNF-α for 6 h. Relative reporter activity was measured. Data are presented as mean values±s.d. (n=3). The results are representative of three independent experiments.
Materials and methods
Y2H screening
The interactome data set of the human liver was generated using the large-scale Y2H method. The full-length ORFs or fragments of genes were amplified by PCR from cDNA and subcloned into pDBleu and pPC86 vectors (Invitrogen) in frame with GAL4 DNA-binding domains or activation domains. Detailed information regarding the constructs is provided in Supplementary Table S1. The GAL4-based Proquest Y2H system was used (Invitrogen). To create an array for automated Y2H screening, the bait plasmids (with Gal4 DNA-binding domains) and the prey plasmids (with Gal4 activation domains) were transformed into the yeast strains MaV203 (MATα) and MaV103 (MATa), respectively. The colonies that did not pass the self-activation test were removed. The remaining yeast colonies were assembled in 96-well plates. Each set of 12 preys was assembled into a pool and screened against the baits by yeast mating methods using a liquid handling robot (Biomek FX). Diploid yeast colonies that activated the HIS3, URA3 or LacZ reporter gene were selected for further expanded Y2H screening. All of the positive clones in the first round of screening were mated with each of the 12 preys in the second round of screening. Those colonies that grew on SC–Trp–Leu–His–Ura plates and activated the LacZ reporter gene were recorded as positives. The interactions that passed two independent screens were considered true positives. For Y2H library screening, the bait plasmids were transformed into yeast MaV203. To test whether the bait could self-activate reporter genes without the presence of interaction partners, the bait constructs were transformed into the yeast strain MaV203 and were grown on an SC–Leu–Trp–His medium containing 0, 25, 50, 75 or 100 mM 3-AT for 1 week. The lowest concentration of 3-AT was used for library screening. Appropriate amounts of 3-AT were added to inhibit self-activation. For Y2H screens, the yeast strain MaV203 was sequentially transformed with the bait constructs and human liver cDNA library fusions containing the GAL4 activation domain according to the user manual. At least 1 × 106 transformants were screened. The yeast transformants were selected in high-stringency medium. The plates were incubated at 30 °C for 5–10 days. The positive colonies grown on SC–Trp–Leu–His–Ura were restreaked onto new SC–Trp–Leu–His and SC–Trp–Leu–Ura plates to grow for another 3 days. Colony-lift filter assays were used to test the expression of the reporter gene LacZ following the manufacturer's protocol. Positive colonies that activated at least two of the three reporter genes (His, Ura or LacZ) were picked up and transferred to another new plate. The interactions were confirmed by a retransformation assay in yeast. Next, the prey plasmids were extracted and sequenced to identify the encoded genes.
Publicly available data sets
A PPI human reference set containing 39 142 PPIs between 9673 proteins was obtained from the HPRD (status 13 April 2010) (Keshava Prasad et al, 2009). Mouse phenotypic data were obtained from the Mouse Genome Database 59 (9 April 2009) (http://www.informatics.jax.org). Liver-specific expressed genes were downloaded from the Tiger (Liu et al, 2008), EHCO (release 14 February 2008) (Hsu et al, 2007) and HUGE index (Misra et al, 2002). Liver-disease-related genes were obtained from the LOMA database (Buchkremer et al, 2010).
The protein interaction data set from this publication has been submitted to the IMEx (http://imex.sf.net) consortium and assigned the identifier IM-15364.
Bioinformatics analyses of the PPI map
Using gene_info (NCBI, 13 November 2009, release), proteins were mapped to an Entrez Gene namespace through their accession numbers, which were obtained in a BLASTP search against the nr (NCBI) databases. The network graphs were produced using Cytoscape software (Shannon et al, 2003), and their topological parameters, such as degree centrality, betweenness centrality, clustering coefficients, shortest path lengths and closeness, were determined using the Cytoscape plug-in Network Analyzer (Assenov et al, 2008). GO assignments were made using NCBI gene2go (17 November 2009) and the GO consortium's OBO (9 November 2009). Pathway assignment was performed using the KEGG data set (Release 53.0). The enrichment of specific GO terms was tested using a hypergeometry test, followed by the Bonferroni multiple testing correction to control for the false discovery rate (Li et al, 2005). GO co-annotations of interacting proteins were evaluated with a previously described method (Stelzl et al, 2005). To analyze the interconnection tendency between proteins of the same or different categories, we used empirical P-values to evaluate the statistical significance of an enrichment number against 1000 random networks, which were generated by randomizing the corresponding relationships between proteins and their category assignment in the real network. The empirical P-value was calculated as the fraction of random networks in which the number of certain kinds of interaction was not less than (upper tail) nor larger than (lower tail) the fraction in the real network. Significantly enriched or depleted cases in the real protein–protein interaction network existed when the upper-tailed/lower-tailed P-value was <0.05.
To estimate the false positives of our data set, we adopted a method developed by Vidal and colleagues (Yu et al, 2008). A golden standard negative data set (GSN) was constructed by selecting the proteins with different cellular locations (Rhodes et al, 2005). There are 455 nucleus proteins and 242 membrane proteins in the HLPN. After the manual analyses, we found that 68 interactions between nucleus and membrane proteins had a relatively high probability of being negative (we do not mean that any of them were false positive). Considering the whole negative interaction space for the 2582 HLPN proteins, there might be 68 × (2582 × 2581/2−8348)/(455 × 242)=2053 negative interactions. The false positive rate is 2053/3484=58.9%.
We evaluated the false negative ratio of HLPN based on the previous report of human interactome size (Proc Natl Acad Sci USA, 2008, 105(19):6959–6964); we estimated the false negative ratio of the HLPN to be 1−3484 × (1–20%)/(650 000 × (2582 × (2582−1)/2))/(25 000 × (25 000−1)/2)=60%. The ratio of false negatives might be attributable to the technical limitations of any given large-scale method.
Co-IP, pull-down and co-localization assays
For co-IP assays, the corresponding genes were cloned into pFlag-CMV2 and pCMV-Myc vectors. HEK293T cells were transfected with the indicated plasmids to express the proteins. After 24–48 h of growth following transfection, the cells were harvested. Cell lysates were prepared in a lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Tween 20, 0.2% NP-40 and 10% glycerol) and supplemented with a protease inhibitor cocktail (Roche) and phosphatase inhibitors (10 mM NaF and 1 mM Na3VO4). Immunoprecipitations were performed using anti-Myc or anti-Flag antibodies and protein A/G-agarose (Santa Cruz) at 4 °C. The lysates and immunoprecipitates were detected using the indicated primary antibodies and then the appropriate secondary antibody, followed by detection with the SuperSignal chemiluminescence kit (Pierce). For pull-down assays, the corresponding genes were cloned into pGEX-4T-2 and pCMV-Myc or pFlag-CMV2 vectors. Bacterially expressed GST or GST-fusion proteins were immobilized on glutathione-Sepharose 4B beads (GE, UK) and washed. Next, the beads were incubated with Myc- or Flag-fusion proteins expressed in HEK293T cell lysates for 3 h at 4 °C. Beads were washed with a GST-binding buffer (100 mM NaCl, 50 mM NaF, 2 mM EDTA, 1% NP-40 and protease inhibitor cocktail), and proteins were eluted, followed by western blotting. For co-localization assays, HEK293T cells were transfected with RFP and GFP expression plasmids or with Myc and Flag expression plasmids. One day after transfection, the cells were fixed with 5% paraformaldehyde for 30 min. The cells were visualized with a confocal microscope.
Luciferase reporter gene assays
HEK293 cells were transfected with the reporter NF-κB-Luc (for NF-κB pathway) or CAGA6-Luc (for the TGF-β reporter gene), with or without the indicated stimulation. After transfection for 24–36 h, the cells were lysed with a passive lysis buffer (Promega). The luciferase activity was measured with the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol. The plasmid pRL-TK (Promega) was used as an internal transfection control. Reporter assays were performed three times in parallel, and each experiment was repeated at least three times.
In vivo ubiquitination assays
For the in vivo ubiquitination assays, HEK293 cells were cotransfected with plasmids expressing Myc-GIT2, Myc-TNFAIP3, Flag-IKBKG, HA-Ub or GIT2 siRNA (5′-CGUUGAUUAUGCAAGGCAA-3′) in various combinations. At 24–36 h post-transfection, the cell extracts were prepared and analyzed for polyubiquitination of IKBKG, either by western blotting of total extracts or by immunoprecipitating Flag-IKBKG with anti-Flag beads followed by western blotting with an anti-HA-Ub antibody.
RNAi assays
The siRNA oligos for GIT2 (sense 5′-CGUUGAUUAUGCAAGGCAATT-3′; antisense: 5′-UUGCCUUGCAUAAUCAACGGG-3′) were synthesized by GenePharma Biotechnology (Shanghai, China). The siRNA oligos against GIT2 and the indicated plasmids were transfected into the HEK293 cells using Lipofectamine 2000 reagent. After 24–36 h, cells were harvested and subjected to western blotting or reporter gene assays.
Statistical analysis
All data from reporter gene experiments were presented as mean values±s.d.
Supplementary Material
Supplementary Figures S1–5
Acknowledgments
We thank Yujun Di, Liping Zhang, Jing Zhao, Wei Qian, Yating Ma, Donghua Wu, Jingxuan Shan, Yan Shi, Gang Wu, Junyong Wang, Yujie Shi, Huiling Wu, Wei Zhang, Juan Zhou, Kan Ye, Xin Yan and Xiaoming Dong for technical assistance. This work was supported by the Special Funds for Major State Basic Research of China (2006CB910802, 2011CB910600, 2011CB910202), the National High-Tech Research and Development Program (2006AA02A310, 2004BA711A19), the National International Cooperation Project (2011DFB30370), the National Key Technologies R&D Program for New Drugs (2009ZX09301-002) and the Chinese National Natural Science Foundation (30621063, 30900755, 30971402).
Author contributions: XY and FH conceived the projects and planned the experiments and analysis. High-throughput ORF cloning and yeast two-hybrid screens were performed by JW, KH, LM, LT, YY, CL, WW, WG, HC, CJ, WZ, YY, QL, YZ, CZ, ZW, WX, YZ, TL, DY, YZ, LC, DZ, XZ, LK, XG, XY, QM, JY and LZ. Other experiments were performed by XH, JW, CL and TZ. Computational analyses were performed by JW, LT, DL, ZL, YZ and XY. The manuscript was written by JW, LT, DL, FH and XY.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M (2008) Computing topological parameters of biological networks. Bioinformatics 24: 282–284 [DOI] [PubMed] [Google Scholar]
- Bossi A, Lehner B (2009) Tissue specificity and the human protein interaction network. Mol Syst Biol 5: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchkremer S, Hendel J, Krupp M, Weinmann A, Schlamp K, Maass T, Staib F, Galle PR, Teufel A (2010) Library of molecular associations: curating the complex molecular basis of liver diseases. BMC Genomics 11: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CNHLPP Consortium (2010) First insight into the human liver proteome from PROTEOME(SKY)-LIVER(Hu) 1.0, a publicly available database. J Proteome Res 9: 79–94 [DOI] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL (2007) The human disease network. Proc Natl Acad Sci USA 104: 8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Karin M (2011) NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell Res 21: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefen RJ, Berk BC (2006) The multifunctional GIT family of proteins. J Cell Sci 119: 1469–1475 [DOI] [PubMed] [Google Scholar]
- Hsu CN, Lai JM, Liu CH, Tseng HH, Lin CY, Lin KT, Yeh HH, Sung TY, Hsu WL, Su LJ, Lee SA, Chen CH, Lee GC, Lee DT, Shiue YL, Yeh CW, Chang CH, Kao CY, Huang CY (2007) Detection of the inferred interaction network in hepatocellular carcinoma from EHCO (Encyclopedia of Hepatocellular Carcinoma genes Online). BMC Bioinformatics 8: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M et al. (2009) Human protein reference database—2009 update. Nucleic Acids Res 37: D767–D772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac DC, Ito C, Csaszar E, Roch A, Yu M, Sykes EA, Bader GD, Zandstra PW (2010) Dynamic interaction networks in a hierarchically organized tissue. Mol Syst Biol 6: 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Li J, Ouyang S, Wu S, Wang J, Xu X, Zhu Y, He F (2005) An integrated strategy for functional analysis in large-scale proteomic research by gene ontology. Prog Biochem Biophys 32: 1026–1029 [Google Scholar]
- Li D, Liu W, Liu Z, Wang J, Liu Q, Zhu Y, He F (2008) PRINCESS, a protein interaction confidence evaluation system with multiple data sources. Mol Cell Proteomics 7: 1043–1052 [DOI] [PubMed] [Google Scholar]
- Liu X, Yu X, Zack DJ, Zhu H, Qian J (2008) TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics 9: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Zhou L, Shi H, Huo K (2008) NUMBL interacts with TAB2 and inhibits TNFalpha and IL-1beta-induced NF-kappaB activation. Cell Signal 20: 1044–1051 [DOI] [PubMed] [Google Scholar]
- Martin-Romero C, Sanchez-Margalet V (2001) Human leptin activates PI3K and MAPK pathways in human peripheral blood mononuclear cells: possible role of Sam68. Cell Immunol 212: 83–91 [DOI] [PubMed] [Google Scholar]
- Misra J, Schmitt W, Hwang D, Hsiao LL, Gullans S, Stephanopoulos G (2002) Interactive exploration of microarray gene expression patterns in a reduced dimensional space. Genome Res 12: 1112–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu ZM, Yin XY, Prochownik EV (2002) Pag, a putative tumor suppressor, interacts with the Myc Box II domain of c-Myc and selectively alters its biological function and target gene expression. J Biol Chem 277: 43175–43184 [DOI] [PubMed] [Google Scholar]
- Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, Shifrin N, Lee B, Benedict Yen TS, Woo T, Malynn BA, Ma A (2009) ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature 457: 906–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann T, Jansson L (1995) A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev 9: 769–782 [DOI] [PubMed] [Google Scholar]
- Prochownik EV (2008) c-Myc: linking transformation and genomic instability. Curr Mol Med 8: 446–458 [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan AM (2005) Probabilistic model of the human protein-protein interaction network. Nat Biotechnol 23: 951–959 [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S et al. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Said M, Begley T, Oppenheim A, Lauffenburger D, Samson L (2004) Global network analysis of phenotypic effects: protein networks and toxicity modulation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 101: 18006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz G, Rosario M, Grimm J, Boeckers TM, Gundelfinger ED, Birchmeier W (2004) The neuronal scaffold protein Shank3 mediates signaling and biological function of the receptor tyrosine kinase Ret in epithelial cells. J Cell Biol 167: 945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA (2002) p53: good cop/bad cop. Cell 110: 9–12 [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B et al. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, Wiuf C (2008) Estimating the size of the human interactome. Proc Natl Acad Sci USA 105: 6959–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro K, Tsunematsu T, Okubo H, Ohta T, Sano E, Yamauchi E, Taniguchi H, Konishi H (2009) GAREM, a novel adaptor protein for growth factor receptor-bound protein 2, contributes to cellular transformation through the activation of extracellular signal-regulated kinase signaling. J Biol Chem 284: 20206–20214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, Yildirim MA, Simonis N, Heinzmann K, Gebreab F, Sahalie JM, Cevik S, Simon C, de Smet AS, Dann E, Smolyar A et al. (2009) An empirical framework for binary interactome mapping. Nat Methods 6: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430: 694–699 [DOI] [PubMed] [Google Scholar]
- Wu Z, Zhang X, Yang J, Wu G, Zhang Y, Yuan Y, Jin C, Chang Z, Wang J, Yang X, He F (2009) Nuclear protein IkappaB-zeta inhibits the activity of STAT3. Biochem Biophys Res Commun 387: 348–352 [DOI] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C et al. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Li H, Liu Q, Liu F, Tang L, Li C, Yuan Y, Zhan Y, Xu W, Li W, Chen H, Ge C, Wang J, Yang X (2011) Nuclear factor p65 interacts with Keap1 to repress the Nrf2-ARE pathway. Cell Signal 23: 883–892 [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q et al. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Zhong X, Tan R, Chen L, Huang G, Li J, Sun X, Xu L, Chen J, Ou Y, Zhang T, Yuan D, Zhang Z, Shu W, Ma L (2010) High-throughput cloning of human liver complete open reading frames using homologous recombination in Escherichia coli. Anal Biochem 397: 162–167 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–5