Abstract
Numerous lineage-specific expansions of the transcription factor B (TFB) family in archaea suggests an important role for expanded TFBs in encoding environment-specific gene regulatory programs. Given the characteristics of hypersaline lakes, the unusually large numbers of TFBs in halophilic archaea further suggests that they might be especially important in rapid adaptation to the challenges of a dynamically changing environment. Motivated by these observations, we have investigated the implications of TFB expansions by correlating sequence variations, regulation, and physical interactions of all seven TFBs in Halobacterium salinarum NRC-1 to their fitness landscapes, functional hierarchies, and genetic interactions across 2488 experiments covering combinatorial variations in salt, pH, temperature, and Cu stress. This systems analysis has revealed an elegant scheme in which completely novel fitness landscapes are generated by gene conversion events that introduce subtle changes to the regulation or physical interactions of duplicated TFBs. Based on these insights, we have introduced a synthetically redesigned TFB and altered the regulation of existing TFBs to illustrate how archaea can rapidly generate novel phenotypes by simply reprogramming their TFB regulatory network.
Keywords: evolution by gene family expansion, fitness, niche adaptation, reprogramming of gene regulatory network, transcription factor B
Introduction
The evolutionary success of an organism depends on its ability to continually adapt to changes in the patterns of constant, periodic, and transient challenges within its environment. This process of ‘niche adaptation’ requires reprogramming of the organism's environmental response networks by reorganizing interactions among diverse parts including environmental sensors, signal transducers, and transcriptional and post-transcriptional regulators. Gene duplications have been discovered to be one of the principal strategies in this process, especially for reprogramming of gene regulatory networks (GRNs). In all, 90% of all regulatory interactions in Escherichia coli and yeast are believed to have arisen through duplication of either transcription factors (TFs) or target genes (Teichmann and Babu, 2004). The fate of the duplicated copies of a TF is dependent upon its functional role, structural complexity, and subsequent mutational events that can lead to gene loss, subfunctionalization (sharing ancestral function), or neofunctionalization (acquiring new functions). It is clear from lineage-specific expansions within diverse TF families that this process has occurred in all domains of life (Nowick and Stubbs, 2010).
Archaea, in particular, have experienced an intriguing expansion of two families of general transcription factors (GTFs). Similar to sigma factors in bacteria (reviewed in Gruber and Gross, 2003), GTFs in eukaryotes and archaea (reviewed in Thomas and Chiang, 2006) are required for the assembly of the preinitiation complex at all transcriptional promoters. Whereas eukaryotes require dozens of factors for recruitment of RNA polymerase, archaea require just two GTFs that are orthologous to eukaryotic TFIIB (transcription factor B (TFB) in archaea) and TATA-binding protein (TBP) (Bell et al, 1998). Historically, the functions of GTFs in eukaryotes and archaea have been discussed almost exclusively in the context of basal transcription and their possible role in regulation of physiology has been under-appreciated. Contrary to this view, ethanol production in yeast was enhanced through the mutagenesis of TFIIB, suggesting that altering the function of a GTF can have significant phenotypic consequences (Alper et al, 2006). Furthermore, several studies have unearthed a possible regulatory role for GTFs in cell-specific differentiation and development in eukaryotes (reviewed in D’Alessio et al, 2009; Goodrich and Tjian, 2010; Juven-Gershon and Kadonaga, 2010) and potentially in mediating environmental responses (e.g. heat shock and oxidative stress) of archaea (Thompson et al, 1999; Coker and DasSarma, 2007; Facciotti et al, 2007, 2010; Paytubi and White, 2009; Kaur et al, 2010). Along these lines, the exceptional success of many archaea in environmental extremes raises the hypothesis that expansion of GTFs in these organisms might partly or fully explain their extraordinary niche adaptation capability.
Characterizing the process by which expansion of these GTFs reorganizes GRNs is complicated in metazoans as the duplicated copies tend to function in different cell types (D’Alessio et al, 2009). In contrast, the fact that the entire set of duplicated GTFs functions in the same cell much like multiple sigma factors do in bacteria makes archaea especially attractive model systems for characterizing evolution of GRNs by GTF expansion. We have previously demonstrated that variations in the expanded set of GTFs in Halobacterium salinarum NRC-1 manifests at the level of physical interactions within and across the two families, their DNA-binding specificity, their differential regulation in varying environments, and, ultimately, on the large-scale segregation of transcription of all genes into overlapping yet distinct sets of functionally related groups (Facciotti et al, 2007). However, these data by themselves did not reveal whether expanding and altering combinatorial activities of TFBs and TBPs is a recipe for niche adaptation. Here, we present a systematic survey of the fitness consequences of perturbing the TFB network of H. salinarum NRC-1 across 17 environments. (‘Fitness’ is defined as the success of an organism in a given environment and determined as growth rate in pure cultures or abundance in competition cultures (Table I; Vasi et al, 1994; Shi and Xia, 2003; Pekkonen et al, 2011).) We relate these fitness changes to phylogenetic histories, expression profiles, protein–DNA, protein–protein, and genetic interactions to conclusively demonstrate a role for TFB expansion in strategies for niche adaptation. We reprogram the network with a synthetically redesigned TFB variant to generate novel adaptive capabilities and demonstrate the importance of both protein-coding and cis-regulatory mutations in this process. Finally, we also demonstrate how novel phenotypes can rapidly arise upon merely altering the regulation of existing TFBs.
Table 1. Definition of key terms and abbreviations.
| Term or abbreviation | Definition |
|---|---|
| Fitness | We define fitness as the success of an organism in a given environment. Success is determined as growth rate in pure cultures or abundance in competition cultures |
| Regulatory program | A ‘regulatory program’ or ‘program’ is defined as a set of instructions for the differential regulation of a group of genes. A TFB program then refers to a set of instructions specified by that TFB. A program is encoded in the regulation of a TFB and its interactions (protein–protein–DNA) with other genes (including other transcription factors and regulators) |
| Niche adaptation program | A program that is essential for adaptation to a particular environment or niche |
| Reprogramming | Reprogramming refers to changes in either the regulation of a TFB or its interactions that result in changes to differential regulation of genes |
| Relative importance of a TFB | Percent contribution of a TFB toward fitness in a particular environment |
| GRN | Gene regulatory network |
| GTF | General transcription factor |
| TF | Transcription factor |
| TFB | Transcription factor B |
| TBP | TATA-binding protein |
In this study, we have performed exhaustive phylogenetic comparisons of 258 TFB proteins from 82 archaeal genomes to reveal a complex evolutionary history during which the TFB family has expanded several times especially in halophilic archaea. We have investigated how this expansion correlates with environment-specific fitness traits by analyzing growth of TFB deletion strains in 17 environments with single and combinatorial perturbations in temperature (25–42°C), Cu (0.4–1.0 mM), pH (5–9.5), and salinity (2.5–5.0 M) in 1996 growth experiments. Through analysis of fitness landscapes from these experiments, we demonstrate the generalized and specialized roles of TFBs in adaptation to different environmental challenges. By performing competition experiments among the TFB deletion strains and mapping genetic interactions in varying environments, we show that different TFBs are essential under dynamically changing growth conditions and that there also exists a division of labor among TFBs to explain why multiple copies have been maintained during evolution. In order to reconstruct the functional evolutionary history of TFBs, we correlate the relationships of their fitness landscapes to their genome-wide binding locations and their gene expression patterns in 361 microarray experiments that probe cellular responses to a wide array of environmental perturbations. This integrated system analysis revealed that evolution of both protein-coding and promoter sequences of TFBs has been important in encoding environment-specific regulatory programs. We experimentally demonstrate the importance of these two classes of mutations by analyzing the fitness and transcriptional consequences of rewiring a novel synthetic TFB and altering the regulation of native TFBs. Remarkably, these experiments show that promoter mutations alone are sufficient to generate completely new environment-dependent regulatory programs for rapid adaptation to new environmental niches.
Results and discussion
An explosion of GTFs among archaea
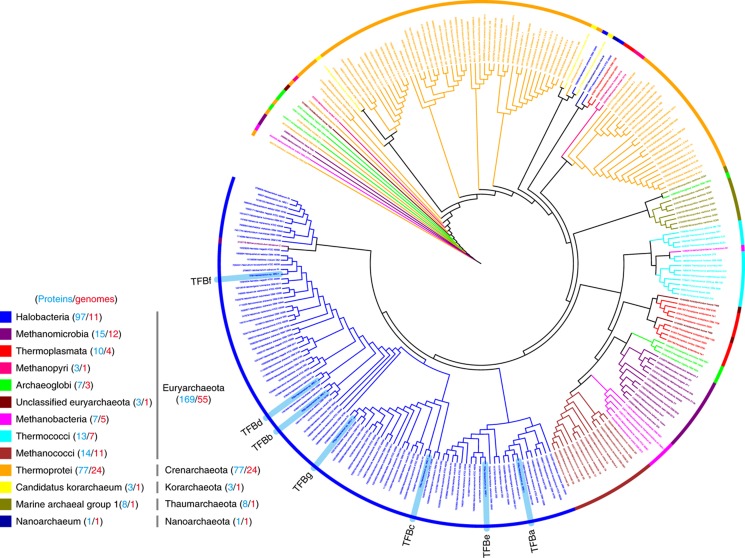
As public databases continue to be populated with fully sequenced genomes, it is indisputable that expansion of GTFs is widespread across the archaeal domain and likely to have important evolutionary implications. In all, 56 of the 82 fully sequenced archaeal genomes encode at least two or more copies of TBP or TFB (Supplementary Table S1). This is analogous to the observation that over two-thirds of all fully sequenced bacterial genomes encode more than one sigma factor (Supplementary Table S2). Comparative analysis of archaeal TFBs alone reveals a complex evolutionary history during which expansions have occurred through duplication events that are both deeply rooted and also much more recent (Figure 1). The two TFB copies in most Thermoprotei emerged post-divergence of Crenarchaeota and Euryarchaeota. TFBs in the Euryarchaeal branch further expanded within Halophilic archaea, Thermococci and more recently in Methanomicrobia, Archaaeoglobi, and Thermoplasmata. These lineage-specific expansions suggest that TFBs encode functionally specialized gene regulatory programs for the unique environments to which these organisms have adapted. (A ‘regulatory program’ or ‘program’ is defined as a set of instructions encoded in the interactions and regulation of a TFBs for the differential regulation of a group of genes (see Table I).) This hypothesis is particularly appealing when we consider that the greatest expansion is observed within the group of halophilic archaea whose habitats are associated with routine and dynamic changes in a number of environmental factors including light, temperature, oxygen, salinity, and ionic composition (Rodriguez-Valera, 1993; Litchfield, 1998).
Figure 1.
Lineage-specific expansion of the TFB family in Archaea. Phylogenetic analysis of TFB proteins in Archaea highlights the extent of lineage-specific expansion particularly in halophilic archaea. Amino-acid sequences for TFBs from 82 complete archaeal genome sequences (MicrobesOnline (Dehal et al, 2010)) were aligned with MUSCLE (Edgar, 2004) and a phylogenetic tree was constructed as described in Materials and methods. Branches belonging to the same phylum and class are colorized based on taxonomy using Archaeopteryx (Han and Zmasek, 2009) and iTOL (Letunic and Bork, 2011). Tree is outlined with the same colors to highlight expansions in the similar class ranges. Color code for each class, corresponding phylum, number of genomes (red color), and number of proteins (blue color) are given in the legend. Halophilic archaeal TFBs are highlighted in blue background. Sequences used in this analysis are listed in Supplementary Table S1.
Generalized and specialized roles for TFBs in adaptation to hypersaline environments
Our hypothesis that TFB expansion might be related to niche adaptation is supported by cursory evidence for functional association of some TFBs with specific environmental challenges such as high temperature, UV irradiation, and oxidative stress (Thompson et al, 1999; Coker and DasSarma, 2007; Gotz et al, 2007; Micorescu et al, 2008; Paytubi and White, 2009; Kaur et al, 2010). However, analysis of protein–DNA and protein–protein interactions along with system-wide changes in gene expression resulting from deletion of TFBs revealed extensive crosstalk among these GTFs (Facciotti et al, 2007). Therefore, although it is tempting to associate each TFB to an environment-specific regulatory program, our data demonstrated that functions of different TFBs are overlapping and that each TFB oversees several such programs. To investigate the phenotypic consequences of these overlapping functions, we calculated in different environments the maximum growth rate of each TFB knockout, as this property has been demonstrated to be a robust proxy for fitness (Vasi et al, 1994; Shi and Xia, 2003; Pekkonen et al, 2011). All growth rate measurements were normalized to maximum growth rate of the parent strain (Δura3) in the same environment to obtain a relative estimate of the fitness contribution of each TFB in a given environmental condition.
Using this procedure we analyzed growth curves from 1996 experiments that were performed in high throughput to quantify environment-specific fitness traits associated with various TFB deletions across 17 environmental conditions differing in salinity (2.5–5.0 M), temperature (25–42°C), pH (5–9.5), and Cu (0.4–1.0 mM) (Supplementary Table S3). Our selection of environments was deliberate, and specifically intended to investigate whether there is a distinction between TFBs that mediate adaptation to wide variations in salinity—a hallmark characteristic of all halophilic archaea, and those associated with handling other types of stresses. Analysis of fitness landscapes for each of the five TFBs that could be deleted under standard laboratory conditions supported our hypothesis that the TFBs have complex overlapping functions albeit with some recognizable trends in certain environmental contexts (Figure 2A; Supplementary Figure S1). Notably, each TFB conferred fitness in two or more environmental conditions tested, and the relative fitness contributions (see Table I) of the five TFBs varied significantly by environment (Figure 2B). The increased variability in growth characteristics in certain environments further suggested that deletion of TFBs had decreased the robustness of some cellular responses.
Figure 2.
Fitness contributions of TFBs across diverse environments reveal their complex and overlapping functions. Growth assays were performed in high throughput by tracking cell density at OD600 using the Bioscreen C instrument as described in Materials and methods. We determined the maximum growth rates (fitness) from smooth spline fitted growth curves after depositing cell density measurements into a database with relevant meta-information and associated plate layout information. Maximum growth rate of each TFB knockout was normalized to appropriate controls and log2 ratios were reported as normalized maximum growth rates or fitness (Supplementary Table S3). (A) Distinct trends in fitness contribution of TFBs across specific environmental gradients. The condition-specific fitness trends (normalized maximum growth rate) of each TFB knockout strain can be viewed as evidence for complex patterns of subfunctionalizations. (B) Relative order of fitness contributions of TFBs changes with environmental context. Fitness of each TFB knockout was subtracted from fitness of the parent to obtain degree of fitness contributed by that TFB in each environment (plotted on the y axis as ‘TFB Fitness’). Statistical significance of fitness differences among pairs of TFBs was calculated using t-test (Supplementary Figure S1). Starting with the lowest fitness contributing TFB on the left boxplots of the TFBs are rank ordered with increasing fitness contributions going rightward. The different orderings of the TFBs in these rank-ordered plots demonstrate how TFBs take turns in assuming a primary role across the 17 environmental conditions. (C) Distribution of different clades of TFBs across all of the 11 fully sequenced halophilic archaeal genomes. Clade membership of TFBs was assigned based on similarity to H. salinarum NRC-1 family members. Numbers in parenthesis indicate total number of TFB proteins in each species. While TFBf- and TFBc/TFBg-like proteins are present in all archaea, TFBb/TFBd- and TFBa/TFBe-like proteins are limited to certain species (Supplementary Table S1).
From an evolutionary perspective, the relationships among these fitness landscapes reveal a fascinating history of expansions in the TFB family in the context of regulating ‘core’ and ‘accessory’ functions for adaptation of H. salinarum NRC-1 to challenges of a hypersaline environment. In our prior work, inability to construct chromosomal deletions had already demonstrated the essentiality of two of seven TFBs (TFBf and TFBg) in H. salinarum NRC-1. Consistent with its known importance under oxidative stress (Kaur et al, 2010), in this study we have discovered that chromosomal deletion of tfbC significantly decreased fitness across 11 of 17 environmental conditions (Figure 2B). Interestingly, orthologs of all three functionally important TFBs (c, g, and f) are also present in all fully sequenced halophilic archaeal genomes (Figures 1 and 2C). Together these data suggest that two classes of TFBs (c/g- and f-type) appear to have played an important role in the evolution of halophilic archaea by overseeing regulation of core physiological capabilities in these organisms. On the other hand, TFBs of the other clades (b/d and a/e) were dispensable in most environments (Figure 2B) and, their distribution across the halophilic archaea is also spotty (Figure 2C). The most likely explanation is that these TFBs emerged much more recently through gene duplications or horizontal gene transfers and are being utilized for adaptation to specialized environmental conditions (Figures 1 and 2).
Higher-order organizational structure of the TFB network
It is clear from the fitness analysis that each TFB oversees several niche adaptation programs, and that several TFBs can be associated to the same program. (A ‘niche adaptation program’ is a gene regulatory program that is essential for adaptation to a particular environment or niche (see Table I).) When considered in the context of the high degree of cross-connectivity in protein–protein and protein–DNA interactions of the TFBs with each other and their targets, these data suggest that the expanded set of TFBs must work together in a combinatorial scheme (Facciotti et al, 2007). The significant variations in environment-dependent genomic binding locations of each TFB (Koide et al, 2009) further explains how the combinatorial scheme and, therefore, the order of relative importance of TFBs changes with environmental context (Figure 2B; Supplementary Figure S1). However, since the fitness landscapes for each TFB were determined one-at-a-time, these data are unable to shed light on epistasis, multiplicative and non-additive interactions that indicate hierarchy, collaboration, or competition among TFBs. In the following two sections we present results from experiments that were specifically designed to investigate such complex relationships among TFBs and assess whether they are affected by environmental context.
TFBs divide and conquer
In our analysis of fitness landscapes, we made an intriguing observation that deletion of most TFBs, with the exception of TFBc, improved fitness in several environments. Gene loss is known to be beneficial in fixed environments, especially when the loss of function is buffered by some functional redundancy in other genes (Frank et al, 2002). In the case of expanded TFBs, we posit that relieving the regulation of a group of genes by deleting a TFB that is not essential in a relatively stable environment might help to decrease the associated energy burden (Valentine, 2007). This is also independently supported by the observation that the number of genes including regulators such as sigma factors tends to be lower in organisms living in stable environments (Konstantinidis and Tiedje, 2004). Along these lines, in the work presented here we note that TFBe has gained a specialized role in adaptation to a low temperature environment that is also associated with either high pH or low salinity. However, deleting tfbE from the genome significantly improves fitness under 1.0 mM Cu stress (Figure 2B). This bolsters the hypothesis that many of the duplicated TFBs (especially of the b/d and a/e clades) have specialized roles in adaptation to specific environmental conditions but are dispensable in other environments.
Given that conditions in a natural environment, such as a hypersaline lake, are constantly changing, we predict that the relative importance of TFBs must also constantly change making the function of each essential at varying times. We tested this hypothesis by competing the TFB knockout strains in standard batch culture conditions wherein H. salinarum NRC-1 experiences large-scale physiological readjustment during growth in rich medium (Facciotti et al, 2007). Importantly, changes in conditions (e.g., oxygen (Schmid et al, 2007) and oxidative stress (Kaur et al, 2010)) during growth cause differential regulation of all TFBs (Facciotti et al, 2010) and alter their genome-wide distribution of DNA binding (Koide et al, 2009). Accordingly, we predict that in order to alter a cell's physiology to match changes in culture conditions, the relative importance of TFBs must vary through different phases of growth in batch culture. If our prediction is correct, then the competition experiment should reveal additional functional hierarchies among TFBs beyond what is observable in pure cultures.
The five TFB knockout strains were mixed in equal proportion (2 ml of each strain normalized to OD600: 0.05 with a systematic photometric error ±1% at Absorbance=1), and cultured together under standard laboratory conditions (DasSarma et al, 1995) to an OD600 of 0.4 at which point an aliquot was transferred to fresh medium (final OD600 0.05). Relative proportions of the five strains were tracked through four serial passes (22 generations) with qPCR using strain-specific primers (Supplementary Table S4; Materials and methods). Consistent with its behavior in pure culture, the TFBc knockout was almost entirely depleted in the first iteration of the competition experiment reaffirming the essentiality of this TFB (Figure 3A). In contrast, the importance of TFBa during growth was revealed only in the competition experiment where the abundance of the TFBa knockout significantly decreased. Similarly, the relative fitness of the other TFB deletion strains did not follow the same trends observed in the pure cultures (e.g. see relationship between ΔtfbD and ΔtfbE (Figure 3A and B)). Although deletion of four of the five TFBs improved fitness in pure culture at 37°C, the competition experiment revealed that there was indeed hierarchy to the fitness contributions of TFBs beyond what could have been predicted from fitness studies conducted in pure cultures (Figure 3A). We speculate that limiting nutrients and dynamically changing growth conditions exaggerate subtle fitness differences among TFBs when they are made to compete. Interestingly, there were significant differences in functional hierarchies of TFBs at 37 and 25°C, possibly reflecting variations in the types of environmental challenges incurred at the two growth temperatures (e.g. see relative fitness of ΔtfbB and ΔtfbD in competition experiments performed at 37 versus 25°C (Figure 3A and B)). We conclude from these data that expansion of TFBs in H. salinarum has resulted in ‘division of labor’ such that no TFB is individually capable of handling the entire workload under dynamically changing environmental conditions. Conversely, the non-redundant functions of the various TFBs in dynamically changing environments makes them all essential albeit at different times and explains why multiple copies have been maintained in H. salinarum NRC-1 and other halophilic archaea.
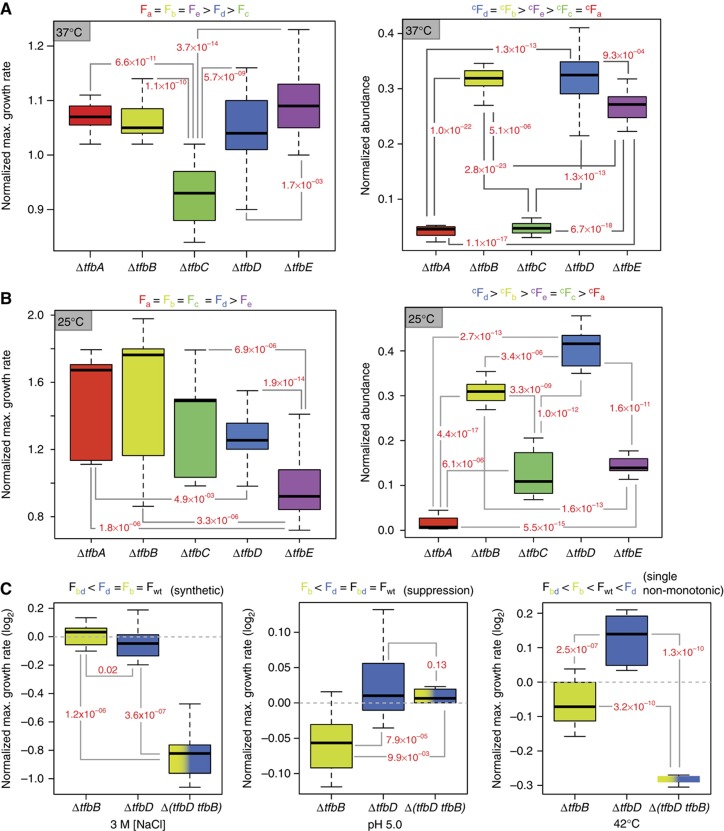
Figure 3.
Functional hierarchies and genetic interactions of TFBs change with environmental context. Relative fitness levels of TFB knockouts in pure cultures at 37°C (A, left) and 25°C (B, left) were determined as described in Figure 2. Competition experiments were performed by mixing equal numbers of cells of each TFB knockout grown to mid-log phase of growth. The mixed cultures were incubated at 37°C (A, right) or 25°C (B, right) to OD600∼0.4 when they were serially diluted into fresh medium to a final OD600 of 0.05. The competition was performed over ∼22 generations and relative success of each TFB was determined by tracking the relative abundance of the knockout strains with qRT–PCR. Significance of fitness differences between pairs of TFBs was determined using two-sample t-test and P-values for significant changes are reported in red font adjacent to lines connecting respective TFB pairs. Ranking of relative fitness of each TFB knockout is indicated on top of each plot. (F: fitness in pure cultures; cF: fitness in competition.) Difference in rank order of F and cF of knockouts in the same environment suggest division of labor among the TFBs that is not at all apparent when they are cultured individually. Consistent with the results in Figure 2B, difference in cF across environments (25 and 37°C) further demonstrates that the TFBs switch their relative roles (primary, secondary, tertiary, etc.) depending on context. (C) Functional (genetic) interactions among TFBs vary by environmental context. Genetic interactions between tfbB and tfbD were determined by assessing fitness differences (t-test, P<0.01) of single (ΔtfbB or ΔtfbD) and double (ΔtfbBΔtfbD) knockout strains. Mode of genetic interactions was assigned based on fitness inequalities indicated on top of each graph (Fb: fitness of ΔtfbB; Fd: fitness of ΔtfbD; Fbd: fitness of ΔtfbB ΔtfbD; Fwt: fitness of WT) per the scheme devised by Carter et al (2009).
The architecture of functional interactions among TFBs changes with environmental context
The fitness analyses showed that functional importance and relationships among TFBs changes with environmental context. For instance, TFBs b and d have similar fitness contributions in some environments (e.g. see fitness at 37, 25°C, 4.5 M NaCl, and 1.0 mm Cu in Figure 2B) but they have opposing effects on fitness in other conditions (e.g. 4 M NaCl, 42°C, 0.4 mM Cu, pH 5.0, and pH 6.5). There was clear hierarchy to the functions of the two TFBs at 25°C but not at 37°C (Figure 3B). Similarly, TFBs of different clades such as TFBd and TFBe had similar fitness contributions in certain environments (again, see fitness at 42°C, 0.8 mM Cu and 2.5 M NaCl/25°C in Figure 2B) but different functional hierarchies in the competition experiment (Figure 3A). These data support the hypothesis that the seven TFBs operate in a combinatorial scheme wherein their regulatory interactions dynamically reorganize depending on environmental context. As a further test of this hypothesis, we mapped the genetic interactions between two pairs of TFBs (TFBb and TFBd; and TFBd and TFBe) in six environmental conditions by comparing fitness landscapes of their single and double knockout strains. These data confirmed that despite belonging to the same phylogenetic clade, the nature of the genetic interactions between TFBb and TFBd differed significantly depending on environmental context. For example, the importance of TFBb and TFBd at 3 M salinity was revealed only when both were deleted from the genome (a synthetic interaction); deletion of TFBd suppressed the ΔtfbB phenotype at pH 5.0 (a suppressor interaction); and deletion of TFBd had opposing consequences on fitness at 42°C in the wild-type (WT) relative to the ΔtfbB genetic background (a single non-monotonic interaction) (classification of genetic interactions was done according to the scheme proposed by Carter et al (2009) (Figure 3C; Supplementary Figure S2A). This example illustrates that depending on environmental context, the same two TFBs interact in three completely different ways. Likewise, we also observed at least two different types of environment-dependent interactions (suppression and non-interactive) between TFBd and TFBe (Supplementary Figure S2B).
Recently, it was shown in yeast that a different set of genetic interactions could be identified with and without DNA damage (Bandyopadhyay et al, 2010). Here, we have shown that the nature of genetic interactions between the same pair of TFBs can vary significantly in different environmental contexts. Not only does this confirm our hypothesis that the arrangement of collaborations among TFBs changes with environmental context, but it also explains why just seven TFBs are able to encode a much larger set of programs for adaptation. The combinatorial activity of the TFBs might be encoded in their (1) physical interactions with each other at the protein level, (2) interactions with each other's promoters, (3) competition for binding sites throughout the genome, (4) differential control of transcriptional regulators, and/or (5) shared interactions with a similarly expanded set of TBPs and other regulators encoded in the genome. We and others have previously presented experimental evidence for these mechanisms (Facciotti et al, 2007; Paytubi and White, 2009). Here, we have connected the mechanisms to phenotypic consequences under dynamically changing environmental conditions.
The reconstructed evolutionary history of the TFB family reveals an important role for promoter evolution in generating novel niche adaptation programs
To elucidate the mechanisms by which novel phenotypes are generated by the expanded TFBs, we reconstructed their functional evolutionary history by correlating the relationships of their fitness landscapes to their genome-wide binding locations and their gene expression patterns in 361 experiments representing perturbations in diverse environmental factors (Figure 4A). The different data types used in this reconstruction are listed in Supplementary Table S5. Relationship at the level of sequence, expression, and fitness was determined by hierarchical clustering using euclidean distance/average linkage method. Relationships at the level of DNA-binding specificity (under the same growth condition) were determined by hierarchically clustering the matrix of hypergeometric P-values for significance of shared binding across all pairs of TFBs (Figure 4A; Supplementary Table S6) (see Materials and methods).
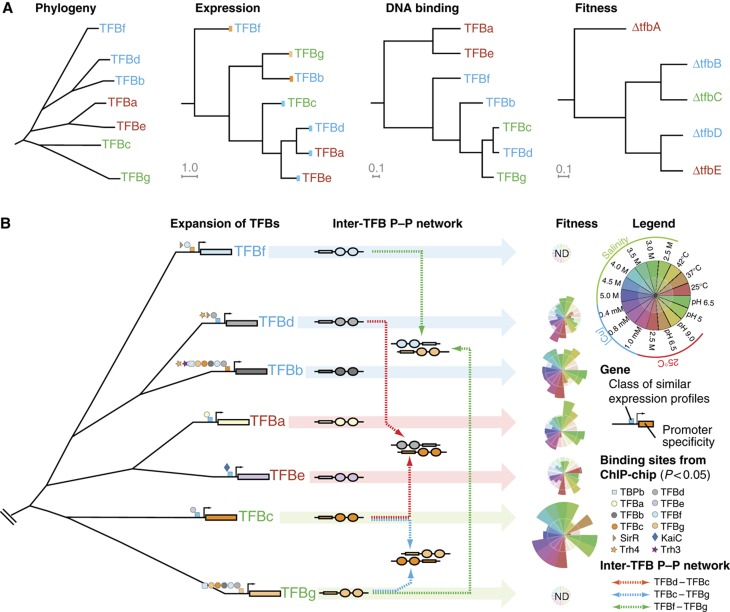
Figure 4.
Reconstruction of evolutionary events responsible for the extant architecture of the seven TFB GRN in H. salinarum NRC-1. (A) Relationships among TFBs at the level of their phylogeny, regulation, distribution of their DNA-binding locations, and fitness contributions. Font coloring of TFBs indicates their clade membership. The first tree shows phylogenetic relationships of TFBs based on the amino-acid sequence similarities. The second tree illustrates relationships in regulation (‘cis-mutations’) of TFBs that were determined by hierarchical clustering of their transcript level changes across 361 environmental conditions. It is clear from this tree that TFBs from the same clade (see b/d/f and g/c clades) are expressed under very different regulatory schemes. The blue and orange color bars on the leaves of this tree indicate related expression profiles; this color code is also utilized in (B) to help the reader relate these data across the two panels. Relationships at the level of DNA binding (‘trans-mutations’) were determined by clustering the hypergeometric P-values for shared-binding sites among pairs of TFBs (Supplementary Table S6). This plot reveals that similarity of DNA-binding specificity is mostly consistent with TFB relationships at the primary sequence level with some important exceptions (see text for details). Finally, similarities in fitness contributions of TFBs across 17 different environments are explained by a combination of cis- and trans-mutations (see text for details). (B) Changes to both cis and trans segments of TFBs need to be considered to explain current day architecture of the seven TFB GRN. This reconstruction was done in the framework of gene duplication events that were inferred from phylogenetic analysis. Promoter evolution was reconstructed by integrating experimentally mapped TF-binding sites (Facciotti et al, 2007) of eight GTFs and four regulators in the TFB promoters, and transcript level changes (A; see inset key). This reconstruction explains subtle differences in the regulation of phylogenetically related TFBs in context of gain and loss of TF-binding sites (for instance, relative to TFBb, the TFBd promoter has gained a TF-binding site for SirR but lost TF-binding sites for six GTFs and Trh3). This reconstruction also reveals convergent evolution of promoters for TFBs from different clades (for instance, TFBc and TFBe); notably, the set of TFs whose TF-binding sites were mapped do not explain the similar expression profiles of TFBc and TFBe. An intra-TFB protein–protein network occurs away from DNA and is speculated to modulate recruitment of these factors to cognate promoters. Coupled changes in DNA-binding specificities of TFBs, their regulation and their protein interactions mediates transcriptional segregation of different aspects of physiology and corresponding environment-specific subfunctionalization of individual TFBs (height of a colored sector in each star plot is proportional to the normalized fitness contribution of that TFB in a particular environment; see inset).
As expected, similar chromosomal binding patterns of TFBs could be explained by sequence-based phylogenetic relationships. However, sequence-similarity alone did not explain why TFBd-binding distribution is more like that of TFBc and TFBg (similarity in binding pattern of TFBd and TFBc: 90 shared-binding sites with hypergeometric P-value: 3.0 × 10−35; TFBd and TFBg: 73 shared-binding sites with hypergeometric P-value: 3.0 × 10−18) (Supplementary Table S6). Furthermore, despite sharing chromosomal binding locations with TFBs c and g, the fitness landscape of TFBd resembles that of TFBe. Similar functional divergence was also observed for TFBa and TFbe, which belong to the same phylogenetic clade. Clearly, sequence-similarity and binding distributions do not fully explain relationships among the fitness landscapes of the TFBs (Figure 4B). Interestingly, the convergent and divergent evolution of promoters discovered from analysis of expression patterns of TFBs helps to explain some of these confounding observations. The similar fitness landscapes of TFBs d and e could be better explained by their coexpression across diverse environmental conditions (Pearson correlation: 0.853; P-value: 2.2 × 10−16) (Supplementary Table S7) due to convergent evolution of their promoters. In a similar vein, the divergent promoter evolution of TFBb and TFBd (Pearson correlation: −0.148; P-value: 4.8 × 10−03) explains why they have different fitness landscapes despite being related at a primary sequence level, and also in their DNA-binding specificity (similarity in binding pattern of TFBb and TFBd: 144 binding sites, hypergeometric P-value: 2.8 × 10−53) (Supplementary Table S6). There is at least one example where none of the data (interactions, regulation, and phylogeny) explains fitness relationships between TFBs adequately. Specifically, TFBa and TFBb belong to different phylogenetic clades yet they are tightly correlated in their fitness properties especially in response to changing temperatures. The most likely explanation is that these TFBs regulate unrelated pathways that are affected in similar ways under these conditions. Regulation of different pathways by the two TFBs is supported by the substantially different fitness of the two knockout strains in competition experiments (Figure 4). However, given that TFBb potentially regulates far more genes than TFBa, the lower fitness of the tfbA knockout demonstrates that the importance of a TFB might not be determined just by the total number of genes they regulate but also by the specific functions they regulate.
With the exception of this one example, rest of the integrated analysis of physical interactions, regulation, and fitness landscapes of TFBs revealed that evolution of both their protein-coding sequence and their promoter has been instrumental in the encoding of environment-specific regulatory programs (Figure 4B). In other words, a duplicated TFB can confer novel fitness capability not just through alterations to its DNA- and protein-binding properties (trans-mutations), but also via mutations that change when it is expressed (cis-mutations). As changes to cis-elements can happen faster than evolution of protein interaction interfaces (Stone and Wray, 2001; Lercher and Pal, 2008), for which the constraints are far greater, we predict that promoter evolution of a duplicated TFB is an important mechanism for rapid adaptation when an organism migrates to a new environment.
Gene conversions among expanded TFBs accelerates GRN evolution for niche adaptation
Previous work in yeast has demonstrated that mutating TFIIB can have significant phenotypic consequences (Alper et al, 2006). Unlike yeast that has a single copy of TFIIB, the situation here is different due to expansion of the TFB family, which not only increases the combinatorial space of regulatory programs but also accelerates the process by which novel TFB variants can arise. Specifically, the convergent and divergent evolution of regulation and binding properties of TFBs suggests that, aside from horizontal gene transfer (HGT) and random mutations, a third plausible (and perhaps most interesting) mechanism for acquiring a novel TFB variant is through gene conversion (Santoyo and Romero, 2005). A fundamentally interesting question regarding this process is whether it simply transfers and recombines fitness properties across TFBs or, as suggested by our data, it actually generates a novel fitness landscape beyond what is encoded by the parent TFBs. The latter would allow an organism to rapidly explore a larger space of possible solutions to adapt to a new environment by randomly recombining information across members of the TFB family. We investigated the feasibility of such a mechanism by attempting artificial network rewiring through the functional integration of novel TFBs that recombined coding sequence and promoter variations of two phylogenetic lineages. We also explored the influence of the host genetic background and environmental context on the fate of the novel TFB. We selected TFBd as the backbone in which to construct the novel TFB (designated as tfbX for gene and TFBx for protein), and the TFBa/e clade as the source of mutations because these TFBs were determined to be non-essential and utilized for specialized niche adaptation programs.
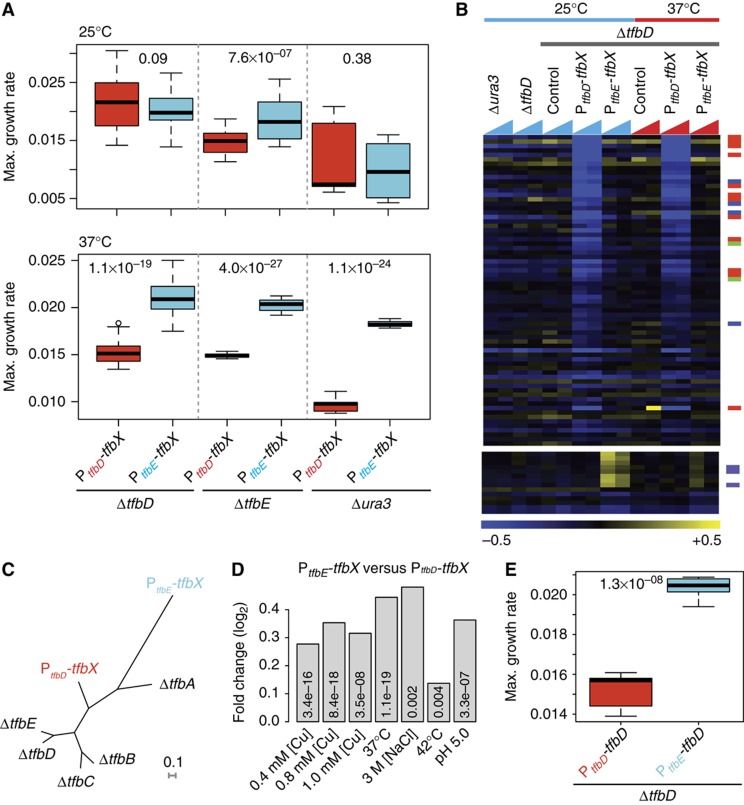
We created a scenario wherein subsequent to its split from TFBb, TFBd acquires 23 mutations characteristic of the TFBa/e lineage with no selective pressure and independent of all other TFBs, in accordance with Ohno's model (Ohno, 1970) (Supplementary Figure S3). Alternatively, this procedure can also be seen as modeling the acquisition of a novel TFB through HGT. This synthetic TFB construct contains 23 amino acids that are characteristic of the TFBa/e lineage substituted into the TFBd-coding sequence. Next, we placed the synthetic TFB under the control of either the TFBd promoter (PtfbD-TFBx) or the TFBe promoter (PtfbE-TFBx) in a plasmid vector. As mentioned earlier, expression profiles of tfbE and tfbD have few differences (Figure 4A). Therefore, this experimental design allows us to investigate whether subtle changes to regulation of a TFB have any consequence on overall fitness of the host. Finally, we introduced the two variants independently into three different genetic backgrounds: the WT, the ΔtfbE background, and ΔtfbD backgrounds, to investigate whether variations in the architecture of the GRN of the host could also influence the fate of a newly acquired TFB. This is important as microbial populations in the natural environment are known to be a complex mix of diverse genomic variants (Boucher et al, 2001). High-throughput growth assays in a range of environmental conditions (Supplementary Table S3) showed that the synthetic TFB had significantly enhanced fitness in many environmental conditions but only when it was expressed under transcriptional control of PtfbE (Figure 5A).
Figure 5.
The importance of cis- and trans-mutations in altering fitness programs specified by TFBs. (A) Fitness benefits gained from rewiring the synthetic TFB are a function of its regulation, genetic background, and environment. A synthetic TFB (TFBx) was synthesized by transferring TFBa/e clade-specific residues to the TFBd backbone to simulate acquisition of a novel TFB through gene conversion across members of this expanded gene family. Two plasmids harboring a copy of TFBx transcriptionally fused to either the tfbD or tfbE promoter (PtfbD or PtfbE) were transformed into the Δura3 (WT), ΔtfbD, and ΔtfbE genetic backgrounds (altogether six strains). The fitness consequences of introducing TFBx into the resident GRN were evaluated by analyzing growth characteristics of these six strains at 37 and 25°C. This revealed that all controlled parameters—regulation of TFBx, genetic background of the host, and environment—significantly influenced how TFBx altered the host phenotype. Remarkably, the fitness contributions of TFBx were significantly greater at 37°C when it was expressed under the control of PtfbE. (B) Novel regulatory programs resulting from incorporation of the synthetic TFB into GRN are conditional on its regulation and environmental context. Global transcriptional changes of the six strains described above and the control (each of the hosts harboring just the plasmid vector) were determined during growth at 25 and 37°C by hybridizing fluorescently labeled total RNA to Agilent custom design 8X60K tiling arrays as described in Materials and methods. Δura3 (WT), ΔtfbD (tfbD knockout); PtfbD-tfbX/ΔtfbD: plasmid carrying synthetic TFB controlled by tfbD promoter; PtfbE-tfbX: plasmid carrying synthetic TFB controlled by tfbE promoter; control: plasmid without the synthetic TFB construct. Significant changes in transcript levels were identified using significance analysis for microarrays (SAM) within the MEV package (Saeed et al, 2006). The rewiring via transcriptional fusion to PtfbD resulted in differential expression of 67 genes at 25°C and 82 genes at 37°C. These data demonstrate that incorporation of TFBx into the GRN generated both environment-dependent (see genes differentially regulated by PtfbD-TFBx) and -independent (genes enriched for thioredoxin-related functions (purple bars)) novel regulatory programs. Notably, the differentially regulated genes also included two TBPs (TBPc and TBPd—indicated with green bars adjacent to the heatmap), numerous transcriptional regulators (blue bars), and putative non-coding RNAs (orange bars) (Koide et al, 2009), implicating additional secondary mechanisms by which rewiring of the synthetic TFB had completely altered the transcriptional network. (C) Fitness landscape of the synthetic TFB is unlike those specified by any of the resident naturally evolved TFBs. Analysis of growth characteristics across 10 environmental conditions revealed that the synthetic TFB encoded completely novel fitness landscapes that bore no similarity to fitness landscapes of any of the parents (TFBd or TFBa/e) (Supplementary Table S8). This illustrates the striking ability of the TFB network to generate completely novel niche adaptation capability. (D) Transcriptional fusion to PtfbE consistently improves fitness conferred by the synthetic TFB across all environments. Although the transcriptional analysis revealed that transcription fusion to PtfbD altered the regulatory programs in a unique manner, transcriptional fusion to PtfbE was consistently associated with enhanced fitness. (E) Replacing the native promoter of tfbD with PtfbE improves fitness. Relative fitness contributions of TFBd (log2 ratios) across seven environmental conditions is higher when it is under the transcriptional control of PtfbE relative to when it is transcribed from its native promoter. This result confirms that changes to regulation of a TFB alone can significantly improve fitness.
To understand how TFBx had altered fitness under some configurations and not others, we measured global transcriptional profiles and mapped transcription start sites and termination sites of all genes. We made these measurements during early and mid-log growth phase at 25 and 37°C, as TFBx had significantly different consequences on fitness in these environments (Figure 5A) (see Materials and methods). Our microarray experimental design included WT (Δura3), tfbD knockout (ΔtfbD), plasmid vector in ΔtfbD background (control), and synthetic TFBx variants (PtfbD-tfbX or PtfbE-tfbX) in ΔtfbD background. Figure 5B shows significant changes in transcript levels upon introduction of synthetic TFB variants into ΔtfbD background. We made three insightful observations: first, the patterns of differential regulation revealed that different regulatory programs were generated when TFBx was expressed from PtfbD or PtfbE, upon altering genetic background, and upon changing environmental context (Figure 5B); second, differential regulation of two TBPs, a significant number of TFs (6) and ncRNAs (11) (hypergeometric enrichment P-value: 5.2 × 10−6) (Koide et al, 2009) (Figure 5B) explained why a single TFB variant had system-wide consequences and generated fitness landscapes that were unlike any of the native TFBs (Figure 5C); and finally, despite altering 23 amino acids, not a single transcription start site or transcription termination site was affected—even for genes whose regulation was altered—revealing that the preinitiation complex can tolerate enormous sequence variation in a TFB (Supplementary Figure S4). In sum, gene conversion events spanning the coding sequence and the promoter, environmental context, and genetic background of the host are all extremely influential in the functional integration of a TFB into the GRN. These results suggest that over 50% of archaea that possess multiple GTFs might use this simple gene conversion strategy for rapidly generating completely novel fitness capabilities.
Altering just the regulation of a TFB generates completely novel regulatory programs
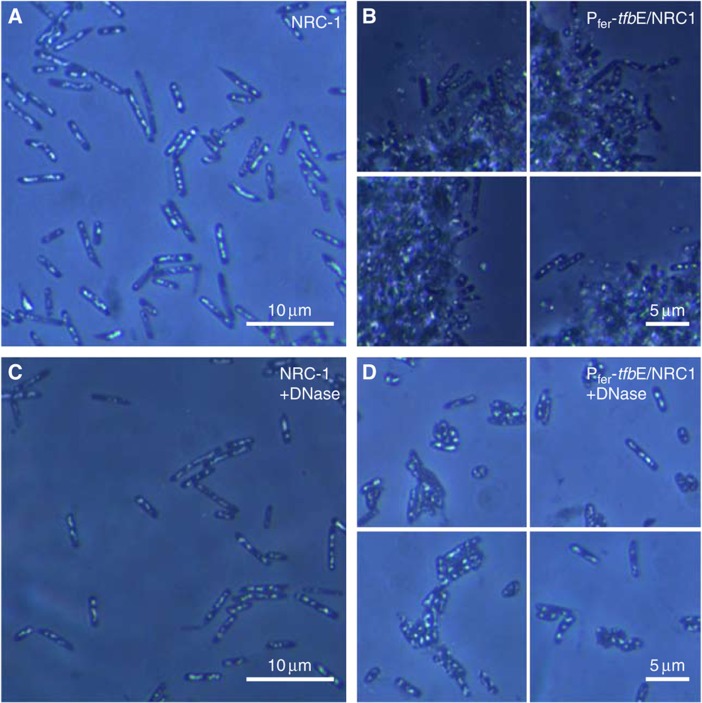
While evolution of protein interaction interfaces are known to take a very long time, promoter changes are known to occur at a significantly faster pace (Stone and Wray, 2001; Lercher and Pal, 2008) and driven by positive selection (Kostka et al, 2010; He et al, 2011). Consistent with this rationale, our data reveal that altering the regulation of an existing set of expanded TFBs might be an efficient mechanism to reprogram the GRN to rapidly generate novel niche adaptation capability. (‘Reprogramming’ refers to changes in either the regulation of a TFB or its interactions that result in changes to differential regulation of genes (see Table I).) We tested this hypothesis by (1) placing tfbD under transcriptional control of PtfbE and (2) overexpressing each of the seven TFBs. Remarkably, placing the native tfbD under transcriptional control of PtfbE significantly improved growth rate (P-value: 1.3 × 10−8) under standard laboratory conditions (Figure 5E). In our second experimental test, we increased the absolute abundance of the TFBs by replacing each of their promoters one-at-a-time with the substantially stronger ferredoxin (fer2) promoter (whereas the native TFB promoters rank among the weakest in the genome, the fer2 promoter is in the top five (unpublished data and Gregor and Pfeifer, 2005). Although artificial-upregulation of six of the seven TFBs did not alter phenotype, transcriptional fusion of tfbE to the fer2 promoter resulted in a phenotype that was previously reported only in the presence of Ca2+ ions (Kawakami et al, 2005). We observed flocculation of cells in a manner that was reminiscent of biofilm formation in other organisms (Kjelleberg and Givskov, 2007). Subsequent analysis revealed that these floccules were comprised of a large number of cells entangled in a mesh of DNA (Figure 6; Supplementary Figure S5). It is possible that by overexpressing TFBe, we unmasked one of its regulatory programs by overriding the need for a specific environmental context (i.e. Ca2+ ions). Nonetheless, these results emphasize the significance of cis-regulatory mutations of duplicated TFs in evolution of GRNs. Above all, they validate our hypothesis that archaea can rapidly generate novel niche adaptation programs by simply altering regulation of duplicated TFBs. This is significant because expansions in the TFB family is widespread in archaea, a class of organisms that not only represent 20% of biomass on earth but are also known to have colonized some of the most extreme environments (DeLong and Pace, 2001). This strategy for niche adaptation is further expanded through interactions of the multiple TFBs with members of other expanded TF families such as TBPs (Facciotti et al, 2007) and sequence-specific regulators (e.g. Lrp family (Peeters and Charlier, 2010)). This is analogous to combinatorial solutions for other complex biological problems such as recognition of pathogens by Toll-like receptors (Roach et al, 2005), generation of antibody diversity by V(D)J recombination (Early et al, 1980), and recognition and processing of odors (Malnic et al, 1999).
Figure 6.
Overexpression of tfbE results in biofilm formation. Phase contrast microscopy (oil immersion, × 100) of WT H. salinarum NRC-1 illustrates its typical cellular morphology in liquid cultures (A). In contrast, overexpression of tfbE resulted in formation of white flocculent structures in liquid cultures that were discovered to be because of cell clumping (B). Addition of DNase I to culture media had no effect on the WT but resulted in disassembly of these clumps, suggesting that DNA is a major component of the matrix that holds cells together within the clumps (C: NRC-1+DNase ( × 100); D: Pfer-tfbE/NRC-1+DNase ( × 100).
Conclusion
Gene family expansions underlie many dramatic events during the course of evolution (David and Alm, 2011). This process has been fairly well documented for a large number of regulators (Demuth et al, 2006; Degnan et al, 2009; Emerson and Thomas, 2009; Janga and Perez-Rueda, 2009; Nowick and Stubbs, 2010), enzymes (Alm et al, 2006; Demuth et al, 2006; De Grassi et al, 2008; da Fonseca et al, 2010), and even for sigma factors in bacteria (Gruber and Gross, 2003; Chiang and Schellhorn, 2010). Owing to its shared ancestry with eukaryotic TFIIB, expansion of the TFB family in archaea is somewhat unusual in that these GTFs are typically associated with a highly restricted role in basal transcription. Our discovery that the TFB family as well could play a role in generating new regulatory programs begs the question of why this seems to have exclusively happened in archaea—not as isolated events but on numerous occasions, in diverse lineages, and at different times in evolution. A counter argument could be that there are yet to be discovered expansions of this protein family in eukaryotes, whose genomes have thus far not been sequenced. That said, other GTFs in eukaryotes (e.g. TATA-box-binding protein and TBP-associated factors) have expanded and been associated with developmental programs, cellular differentiation, and mitotic bookmarking (reviewed in Freiman, 2009; Goodrich and Tjian, 2010). The important functional consequences of tissue-specific expression of GTFs is consistent with our model and suggests that even eukaryotes have exploited the multiplicity of GTFs by reprogramming their promoters to generate novel capabilities.
Materials and methods
Strains, media composition, and culture conditions
All TFB single and double knockout strains were derived from H. salinarum NRC-1 Δura3 parental strain via two-step in-frame gene replacement strategy as described previously (Kaur et al, 2006). All strains were cultured in complex medium (CM: 250 g/l NaCl, 20 g/l MgSO4, 2 g/l KCl, 3 g/l sodium citrate, 10 g/l Oxoid brand bacteriological peptone) at 25, 37, or 42°C with continuous shaking (∼220 r.p.m.). Gene knockout strains were cultured with 50 mg/l uracil to compensate for their uracil deficiency due to the Δura3 counter-selectable marker. Strains carrying recombinant plasmids were cultured with 0.02 mg/ml Mevinolin. Additional perturbations were administered by changing CM composition to vary salinity (2.5–5.0 M), pH (pH 5.0, pH 7.0, and pH 9.0), or Cu concentration by adding CuSO4•5H2O to a final concentrations of 0.4, 0.8, or 1.0 mM.
Fitness calculations
Growth assays were performed using two Bioscreen C instruments (Growth Curves USA, Piscataway, NJ), with a throughput of up to 400 cultures (200 μl each) in each run. The experimental design included multiple biological and technical replicates spread across different runs to account for biological and technical variation. In all cases, the starter cultures were grown to OD600: ∼0.8 and used as preinoculum to adjust the final cell density in the desired culture medium to OD600 of 0.05 and grown with shaking at 25, 37, or 42°C (∼200 r.p.m.). OD was measured in every 30 min for the duration of 6 days. Each Bioscreen run included appropriate control strains to be able to compare growth across multiple experiments.
We have developed a custom R package, ‘Growth Curve Analysis Function’ to automate the analysis of growth curves. Briefly, cell density measurements were deposited into a database with relevant meta-information and associated plate layout information to enable rapid calculation of maximum growth rate (μ) from smooth spline fitted growth curves (Kahm et al, 2010). Maximum growth rate was normalized to appropriate controls and log2 ratios were reported as normalized maximum growth rates (Supplementary Table S9). We found that maximum growth rate was reproducible across replicates and was not affected from fluctuations at high optical densities during stationary phase (Supplementary Figure S6). Boxplots and barplots used in representing the data were plotted in R.
Phylogenetic tree constructions
Phylogenetic analysis of TFBs within all fully archaeal sequenced genomes was done by using sequence data and tools available at MicrobesOnline (Dehal et al, 2010). Specifically, 258 TFB amino-acid sequences from 82 complete archaeal genomes were aligned to each other using MUSCLE multiple sequence alignment algorithm (Edgar, 2004). The resulting alignment was then processed with Geneious Software Package to construct phylogenetic tree by using Jukes-Cantor Genetic Distance Model (Jukes and Cantor, 1969) with Neighbour Joining tree building method. Archaeopteryx (Han and Zmasek, 2009) and iTOL (Letunic and Bork, 2011) was used for visualization and coloring tree braches based on the taxonomy. Detailed information for all of the archaeal TFB sequences used in this analysis are listed in Supplementary Table S1.
Calculation of relationships between fitness landscapes, transcript level changes, and DNA-binding specificities of TFBs
Transcript level changes for all TFBs across 361 microarray experiments representing diverse environmental conditions were collated using Gaggle (Shannon et al, 2006) and exported to MeV (Saeed et al, 2006). Within MeV, the expression data were hierarchically clustered using Euclidean distance/average linkage. Relationships among fitness landscapes of TFB knockout strains in 17 conditions were calculated in a similar manner.
TFB-binding sites were determined with ChIP-chip, that is, by immunoprecipitating c-myc-tagged TFBs and localizing enriched DNA fragment by microarray analysis (Facciotti et al, 2007). We analyzed this data using the MeDiChI algorithm (Reiss et al, 2008) to locate all statistically significant DNA-binding locations (P-value <0.05) for all TFBs. Next, we identified statistically significant shared-binding sites for all TFB pairs within 100 bp proximity to each other. The distribution of these protein–DNA binding maps was analyzed to calculate statistical significance (using the hypergeometric distribution) of shared-binding locations for each TFB pair (Supplementary Table S6). The matrix of P-values for shared-binding across all TFB pairs was then hierarchically clustered as described above. All trees were visualized with Archaeopteryx. All data sources used in this analysis are listed in Supplementary Table S5.
Construction of synthetic TFB
Multiple sequence alignment of TFBa, e, b, d, and f was performed using ClustalW to identify clade-specific amino-acid residues. Twenty-three conserved amino-acid residues that differ between the TFBa/e and TFBb/d/f clades were transferred to the TFBd backbone via gene synthesis and cloned into pUC57 vector (GenScript, Piscataway, NJ) to yield pUC57_tfbX.
The TFBd promoter was PCR amplified from H. salinarum NRC-1 genomic DNA with forward primer 5′-GTA ATT GGT ACC GAT GGT CGT CTC GGT GAT G-3′ and reverse primer 5′-ATT AGC ATA TGT GTG GGG CTG GCT GCG-3′. The PCR products were digested with KpnI and NdeI whose sites were engineered into the two primers (recognition sites for the two enzymes are underlined in the two primers). The TFBe promoter was also amplified and processed in a similar manner; the sequence for the two primers were as follows: forward primer 5′-GAT AAC GGT ACC CGC ATC ACC AAC TGG CGA C-3′ and reverse primer 5′-TAG CGC CAT ATG CGG TCT CAC CTG ATT GAG-3′. The processed PCR products were cloned into NdeI+KpnI digested pMTF-c-myc(Stu) vector to yield vectors pMTF_PtfbD_1.2 and pMTF_PtfbE_7.3, respectively. Subsequently, the synthetic TFB was amplified from pUC57_tfbX with forward primer 5′-GTG CGG CAT ATG ATG ACC AAC CAG CGG ACC AC-3′ with NdeI site and reverse primer 5′-AAT TAT GGA TCC TCA GGC CTC GAC GCC GGG CTC-3′ with BamHI site (underlined). The PCR product was digested with BamHI+NdeI and cloned into BamHI+NdeI digested pMTF_PtfbD_1.2 and pMTF_PtfbE_7.3 to yield PtfbD-tfbX and PtfbE-tfbX, respectively.
Two promoter constructs for an episomal copy tfbD were constructed by amplifying the tfbD gene from H. salinarum NRC-1 genomic DNA using PCR and primers tfbD-wt-Nde2 containing NdeI restriction site (5′-GCGCATATGATGACAAACCAGCGCACAAC-3′) and tfbD-wt-Xba-R containing XbaI restriction site (5′-CAGTCTAGATTACGCTTCCACGCCGGGTTC-3′). The XbaI–NdeI digested PCR product was used to replace the tfbX gene fragment within the two aforementioned vectors pMTF_PtfbD_1.2 and pMTF_PtfbE_7.3 to yield PtfbD-tfbX and PtfbE-tfbXto create PtfbD-tfbD and PtfbE-tfbD, respectively.
Competition experiments and quantitative RT–PCR
Equivalent proportions of pure cultures for all TFB knockout strains grown to late-log phase in CM at 37°C were mixed to a final cell density OD600: ∼0.025 in a total volume of 40 ml in 125 ml flasks. The mixed cultures were incubated at 37°C with shaking and serially diluted into fresh CM medium at a cell density of OD600: ∼0.4. The serial dilutions were repeated four times and abundance of each strain was tracked through the serial passes by quantitative RT (qRT)–PCR using strain-specific primers (Supplementary Table S6). In brief, genomic DNA was isolated from 200 μl of culture using DNeasy Genomic DNA isolation kit (Qiagen, Valencia, CA). DNA quality and quantity was determined using the Nanodrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Strain-specific primers that uniquely amplify the deleted loci for each of the TFB knockout strains were designed using Primer3Plus software (Untergasser et al, 2007). qRT–PCR analyses were performed in 96-well-Fast plates with Power SYBR Master mix (Applied Biosystems) in 7900HT Fast Real-Time PCR instrument (Applied Biosystems). Standard curves for each PCR amplified product were determined by using as template known concentrations of genomic DNA for each knockout strain. The experiment was done using biological replicates and each qRT–PCR reaction was performed in quadruplicate and data analysis was performed via SDS 1.2 software (Applied Biosystems) (Supplementary Table S10).
Tiling array construction and transcriptome structure analysis
The relative changes in transcript levels and transcriptome structure at 37 and 25°C were determined for WT, tfbD, and tfbE in-frame deletion knockouts and all recombinant strains transformed with a plasmid carrying the synthetic TFB constructs. The strains were batch cultured in flasks at either 25 or 37°C with constant shaking, culture aliquots (∼4 ml) were collected over early (OD600: ∼0.2), mid (OD600: ∼0.4), and late (OD600: ∼0.8) phases of growth, centrifuged (16 000 g, 90 s), and flash frozen. Total RNA was prepared from the cell pellets using the mirVANA RNA kit (Ambion, Austin, TX) according to the manufacturer's instructions. Whole-genome tiling arrays for H. salinarum NRC-1 were designed with e-Array (Agilent Technologies), using strand-specific 60 mer probes with 24 nt spacing between adjacent probes for the main chromosome (NC_002607) and the plasmids pNRC200 (NC_002608) and pNRC100 (NC_001869). Altogether the array contained a total of 60 K probes, including the manufacturers’ controls. The microarrays were printed by Agilent Technologies. Labeling with Cyanine 3 (Cy3) and Cyanine5 (Cy5) dyes (Molecular Probes and Kreatech BV), hybridization, and washing were performed as described earlier (Baliga et al, 2004). Arrays were scanned in ScanArray (Perkin-Elmer) and spot finding was done using Feature Extraction (Agilent Technologies). Normalization and statistical analysis were performed as described before (Koide et al, 2009). Transcript boundaries were mapped using multivariate segmentation as reported previously (Koide et al, 2009). Interactive data visualization was done in the Gaggle Genome Browser (Bare et al, 2010).
The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (GEO accession no. GSE31308).
Statistical analysis
Hierarchical clustering of TFBs based on fitness and expression was performed by using Euclidean Distance metric with Average Linkage criteria in MeV package. Significance of fitness differences between WT and each TFB and between TFBs were determined by using two-sample t-test. Genetic interactions reflected as fitness inequalities between single and double TFB knockouts were assigned by using classification rules proposed by Carter et al (2009). Fitness inequalities were tested by using t-test. Significant expression and fitness correlations of TFB pairs across environments were calculated as Pearson correlation coefficient and associated P-values in R (Supplementary Tables S7 and S8). Statistically significant TFB DNA-binding sites were identified by using MeDiChI (Reiss et al, 2008). The matrix of P-values was constructed by assigning a hypergeometric P-value for significant shared-binding sites between each pair of TFBs based on binding site distribution calculated by MeDiChI. Hierarchical clustering of the final matrix was done as described above. All statistical analyses were performed in R Statistical Computing Software (http://www.r-project.org).
Accession codes
The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (GEO accession no. GSE31308).
Supplementary Material
Supplementary information file containing supplementary Figures, Tables and Legends.
Supplementary Table in excel worksheet format showing copy number of TFBs across fully sequenced archaeal genomes.
Supplementary Table in excel worksheet format showing copy number of sigma factors across fully sequenced bacterial genomes.
Acknowledgments
We thank W Lee Pang, Sung Ho Yoon, Aaron N Brooks and Karlyn Beer for help and discussions. This work was supported by grants from NIH (P50GM076547 and 1R01GM077398-01A2), and NSF (DBI-0640950). This work conducted by ENIGMA—Ecosystems and Networks Integrated with Genes and Molecular Assemblies was supported by the Office of Science, Office of Biological and Environmental Research, of the US Department of Energy under Contract No. DE-AC02-05CH11231.
Author contributions: ST and NSB designed the experiments, analyzed the data, and wrote the manuscript. ST performed the experiments. MP did tiling array experiments. WLS and ST performed tfbE overexpression experiments. DJR developed the algorithm for determining the transcriptome structure. DJR and GG developed the algorithm for growth curve analysis. ST and CLP helped with statistical analysis. CJB helped with other data analysis.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alm E, Huang K, Arkin A (2006) The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol 2: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314: 1565–1568 [DOI] [PubMed] [Google Scholar]
- Baliga NS, Bjork SJ, Bonneau R, Pan M, Iloanusi C, Kottemann MC, Hood L, DiRuggiero J (2004) Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res 14: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, Fiedler D, Dutkowski J, Guenole A, van Attikum H, Shokat KM, Kolodner RD, Huh WK, Aebersold R, Keogh MC, Krogan NJ et al. (2010) Rewiring of genetic networks in response to DNA damage. Science 330: 1385–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bare JC, Koide T, Reiss DJ, Tenenbaum D, Baliga NS (2010) Integration and visualization of systems biology data in context of the genome. BMC Bioinformatics 11: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SD, Jaxel C, Nadal M, Kosa PF, Jackson SP (1998) Temperature, template topology, and factor requirements of archaeal transcription. Proc Natl Acad Sci USA 95: 15218–15222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher Y, Nesbo CL, Doolittle WF (2001) Microbial genomes: dealing with diversity. Curr Opin Microbiol 4: 285–289 [DOI] [PubMed] [Google Scholar]
- Carter GW, Galas DJ, Galitski T (2009) Maximal extraction of biological information from genetic interaction data. PLoS Comput Biol 5: e1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SM, Schellhorn HE (2010) Evolution of the RpoS regulon: origin of RpoS and the conservation of RpoS-dependent regulation in bacteria. J Mol Evol 70: 557–571 [DOI] [PubMed] [Google Scholar]
- Coker JA, DasSarma S (2007) Genetic and transcriptomic analysis of transcription factor genes in the model halophilic Archaeon: coordinate action of TbpD and TfbA. BMC Genet 8: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio JA, Wright KJ, Tjian R (2009) Shifting players and paradigms in cell-specific transcription. Mol Cell 36: 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca RR, Johnson WE, O’Brien SJ, Vasconcelos V, Antunes A (2010) Molecular evolution and the role of oxidative stress in the expansion and functional diversification of cytosolic glutathione transferases. BMC Evol Biol 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasSarma S, Fleischmann EM, Rodriguez-Valera F (1995) Media for halophiles. In Archaea: A Laboratory Manual, Robb FT, Place AR, Sowers KR, Schreier HJ, DasSarma S, Fleischmann ME (eds), Vol. 1, pp 225–230. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- David LA, Alm EJ (2011) Rapid evolutionary innovation during an Archaean genetic expansion. Nature 469: 93–96 [DOI] [PubMed] [Google Scholar]
- De Grassi A, Lanave C, Saccone C (2008) Genome duplication and gene-family evolution: the case of three OXPHOS gene families. Gene 421: 1–6 [DOI] [PubMed] [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS (2009) Early evolution of metazoan transcription factors. Curr Opin Genet Dev 19: 591–599 [DOI] [PubMed] [Google Scholar]
- Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP (2010) MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38: D396–D400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Pace NR (2001) Environmental diversity of bacteria and archaea. Syst Biol 50: 470–478 [PubMed] [Google Scholar]
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW (2006) The evolution of mammalian gene families. PLoS One 1: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P, Huang H, Davis M, Calame K, Hood L (1980) An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell 19: 981–992 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH (2009) Adaptive evolution in zinc finger transcription factors. PLoS Genet 5: e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facciotti MT, Pang WL, Lo FY, Whitehead K, Koide T, Masumura K, Pan M, Kaur A, Larsen DJ, Reiss DJ, Hoang L, Kalisiak E, Northen T, Trauger SA, Siuzdak G, Baliga NS (2010) Large scale physiological readjustment during growth enables rapid, comprehensive and inexpensive systems analysis. BMC Syst Biol 4: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facciotti MT, Reiss DJ, Pan M, Kaur A, Vuthoori M, Bonneau R, Shannon P, Srivastava A, Donohoe SM, Hood LE, Baliga NS (2007) General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci USA 104: 4630–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AC, Amiri H, Andersson SG (2002) Genome deterioration: loss of repeated sequences and accumulation of junk DNA. Genetica 115: 1–12 [DOI] [PubMed] [Google Scholar]
- Freiman RN (2009) Specific variants of general transcription factors regulate germ cell development in diverse organisms. Biochim Biophys Acta 1789: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R (2010) Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet 11: 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz D, Paytubi S, Munro S, Lundgren M, Bernander R, White MF (2007) Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol 8: R220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor D, Pfeifer F (2005) In vivo analyses of constitutive and regulated promoters in halophilic archaea. Microbiology 151: 25–33 [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA (2003) Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57: 441–466 [DOI] [PubMed] [Google Scholar]
- Han MV, Zmasek CM (2009) phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 10: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He BZ, Holloway AK, Maerkl SJ, Kreitman M (2011) Does positive selection drive transcription factor binding site turnover? A test with Drosophila cis-regulatory modules. PLoS Genet 7: e1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janga SC, Perez-Rueda E (2009) Plasticity of transcriptional machinery in bacteria is increased by the repertoire of regulatory families. Comput Biol Chem 33: 261–268 [DOI] [PubMed] [Google Scholar]
- Jukes T, Cantor C (1969) Evolution of Protein Molecules. New York: Academic Press [Google Scholar]
- Juven-Gershon T, Kadonaga JT (2010) Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahm M, Hasenbrink G, Lichtenberg-Fraté H, Ludwig J, Kschischo M (2010) grofit: fitting biological growth curves with R. J Statist Softw 33: 1–21 [Google Scholar]
- Kaur A, Pan M, Meislin M, Facciotti MT, El-Gewely R, Baliga NS (2006) A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res 16: 841–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A, Van PT, Busch CR, Robinson CK, Pan M, Pang WL, Reiss DJ, DiRuggiero J, Baliga NS (2010) Coordination of frontline defense mechanisms under severe oxidative stress. Mol Syst Biol 6: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Ito T, Kamekura M, Nakayama M (2005) Ca(2+)-dependent cell aggregation of halophilic archaeon, Halobacterium salinarum. J Biosci Bioeng 100: 681–684 [DOI] [PubMed] [Google Scholar]
- Kjelleberg S, Givskov M (2007) The Biofilm Mode of Life: Mechanisms and Adaptations. Wymondham: Horizon Bioscience [Google Scholar]
- Koide T, Reiss DJ, Bare JC, Pang WL, Facciotti MT, Schmid AK, Pan M, Marzolf B, Van PT, Lo FY, Pratap A, Deutsch EW, Peterson A, Martin D, Baliga NS (2009) Prevalence of transcription promoters within archaeal operons and coding sequences. Mol Syst Biol 5: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM (2004) Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci USA 101: 3160–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka D, Hahn MW, Pollard KS (2010) Noncoding sequences near duplicated genes evolve rapidly. Genome Biol Evol 2: 518–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Pal C (2008) Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol Biol Evol 25: 559–567 [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P (2011) Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield CD (1998) Survival strategies for microorganisms in hypersaline environments and their relevance to life on early Mars. Meteorit Planet Sci 33: 813–819 [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB (1999) Combinatorial receptor codes for odors. Cell 96: 713–723 [DOI] [PubMed] [Google Scholar]
- Micorescu M, Grunberg S, Franke A, Cramer P, Thomm M, Bartlett M (2008) Archaeal transcription: function of an alternative transcription factor B from Pyrococcus furiosus. J Bacteriol 190: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Stubbs L (2010) Lineage-specific transcription factors and the evolution of gene regulatory networks. Brief Funct Genomics 9: 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. London, New York: Allen & Unwin; Springer-Verlag [Google Scholar]
- Paytubi S, White MF (2009) The crenarchaeal DNA damage-inducible transcription factor B paralogue TFB3 is a general activator of transcription. Mol Microbiol 72: 1487–1499 [DOI] [PubMed] [Google Scholar]
- Peeters E, Charlier D (2010) The Lrp family of transcription regulators in archaea. Archaea 2010: 750457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkonen M, Korhonen J, Laakso JT (2011) Increased survival during famine improves fitness of bacteria in a pulsed-resource environment. Evol Ecol Res 13: 1–18 [Google Scholar]
- Reiss DJ, Facciotti MT, Baliga NS (2008) Model-based deconvolution of genome-wide DNA binding. Bioinformatics 24: 396–403 [DOI] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A (2005) The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA 102: 9577–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F (1993) Introduction to saline environments. In The Biology of Halophilic Bacteria, Vreeland RH, Hochstein LI (eds), pp 1–23. Boca Raton, FL: CRC [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J (2006) TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Santoyo G, Romero D (2005) Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol Rev 29: 169–183 [DOI] [PubMed] [Google Scholar]
- Schmid AK, Reiss DJ, Kaur A, Pan M, King N, Van PT, Hohmann L, Martin DB, Baliga NS (2007) The anatomy of microbial cell state transitions in response to oxygen. Genome Res 17: 1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon PT, Reiss DJ, Bonneau R, Baliga NS (2006) The Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics 7: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Xia X (2003) Changes in growth parameters of Pseudomonas pseudoalcaligenes after ten months culturing at increasing temperature. FEMS Microbiol Ecol 45: 127–134 [DOI] [PubMed] [Google Scholar]
- Stone JR, Wray GA (2001) Rapid evolution of cis-regulatory sequences via local point mutations. Mol Biol Evol 18: 1764–1770 [DOI] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM (2004) Gene regulatory network growth by duplication. Nat Genet 36: 492–496 [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM (2006) The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178 [DOI] [PubMed] [Google Scholar]
- Thompson DK, Palmer JR, Daniels CJ (1999) Expression and heat-responsive regulation of a TFIIB homologue from the archaeon Haloferax volcanii. Mol Microbiol 33: 1081–1092 [DOI] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35: W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine DL (2007) Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol 5: 316–323 [DOI] [PubMed] [Google Scholar]
- Vasi F, Travisano M, Lenski RE (1994) Long-term experimental evolution in Escherichia-coli. 2. Changes in life-history traits during adaptation to a seasonal environment. Am Nat 144: 432–456 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information file containing supplementary Figures, Tables and Legends.
Supplementary Table in excel worksheet format showing copy number of TFBs across fully sequenced archaeal genomes.
Supplementary Table in excel worksheet format showing copy number of sigma factors across fully sequenced bacterial genomes.