Abstract
The generation of mathematical models of biological processes, the simulation of these processes under different conditions, and the comparison and integration of multiple data sets are explicit goals of systems biology that require the knowledge of the absolute quantity of the system's components. To date, systematic estimates of cellular protein concentrations have been exceptionally scarce. Here, we provide a quantitative description of the proteome of a commonly used human cell line in two functional states, interphase and mitosis. We show that these human cultured cells express at least ∼10 000 proteins and that the quantified proteins span a concentration range of seven orders of magnitude up to 20 000 000 copies per cell. We discuss how protein abundance is linked to function and evolution.
Keywords: mass spectrometry, protein abundance, proteomics
Introduction
The identification and determination of the concentrations of the molecules that constitute a cell is important and technically challenging. It is important, because progress in systems, cell, and structural biology depends on knowledge of cellular protein quantities. For example, the generation of mathematical models that describe and simulate the behavior of a cell's energy consumption, signaling networks and other systems depend on the knowledge of protein copy numbers per cell. Similarly, atomic models of large cellular structures such as kinetochores, centrosomes, nuclear pores, and others depend on the knowledge of their respective subunit stoichiometries. Furthermore, experimental biology increasingly depends on the comparison and integration of quantitative data sets, a task that depends on comparable, i.e., absolute values.
Until now, absolute protein concentration estimates for large fractions of the expressed proteome have been extremely scarce and existent for very view species. Among these, Saccharomyces cerevisiae is the species with the most extensively studied proteome. In a pioneering study, the cellular concentration of 3868 proteins was derived from genetically altered cells via epitope tagging and quantification of the detected tag (Ghaemmaghami et al, 2003). However, the technique used is costly in terms of time and resources, not generally portable from yeast to other species and bears the risk of perturbing the proteome by the presence of the tagged proteins. Mass spectrometry (MS)-based methods can overcome these difficulties and were recently used to determine protein copy numbers per cell for a significant fraction of the proteome of two bacterial species, namely Mycoplasma pneumonia and Leptospira interrogans (Malmstrom et al, 2009; Maier et al, 2011) and yeast (de Godoy et al, 2008). However, due to technical limitations, the measurement of large-scale absolute protein abundances in higher eukaryotes remained challenging. Protein copy numbers of about 6000 mouse proteins (Schwanhausser et al, 2011) and about 1000 human proteins have previously been reported (Vogel et al, 2010).
We have previously described a quantitative tandem MS strategy to estimate the cellular concentration of a substantial fraction of the proteome of microbial species. We applied it to the human pathogen Leptospira interrogans to estimate the concentration of the majority of expressed proteins in 25 different cellular states (Malmstrom et al, 2009; Schmidt et al, 2011). This method was established for the low to medium complexity proteomes such as single cellular species. It is not directly scalable to the more complex proteomes of multicellular species, particularly those of mammals. In this study, we have determined the cellular concentration of the majority of the proteins expressed by the commonly used human tissue culture cell line U2OS. To cope with the enormous complexity of these samples on the peptide level, we made use of (i) extensive peptide fractionation to reduce sample complexity per fraction, (ii) integration of quantification data per peptide and protein across multiple peptide fractions, and (iii) directing MS data acquisition for in-depth proteome coverage. We demonstrate that U2OS cells express at least ∼10 000 proteins. For ∼7300 of these proteins, we also estimated their cellular concentrations to generate the most extensive quantitative data set on a human cell to date. It was previously shown that cellular core functions are conducted by relatively stable proteins (Schwanhausser et al, 2011). We demonstrate that cellular core functions are often carried out by relatively few proteins, which are present at very high abundance. In contrast, regulatory functions are often orchestrated by large protein families existing in variable but predominantly low abundance in the cell. The fraction of the proteome devoted to such functions is expanded in higher organisms. This finding is underlined by the observation that protein domain duplication is negatively correlated with protein abundance.
Results
At first, we generated an extensive proteome map of the U2OS (human osteosarcoma) cell line. We trypsinized lysates from cells grown in log phase and analyzed them by bottom-up proteomics. LC-MS/MS systems are, at the presently achievable dynamic range and scan speed, incapable of covering a whole, unfractionated proteome digest. We, therefore, used peptide isoelectric focusing (Malmstrom et al, 2006) via off-gel electrophoresis (OGE) to generate peptide fractions of reduced complexity (Horth et al, 2006), shotgun MS together with charge state fractionation to establish an initial map. We then used directed MS (Jaffe et al, 2008; Schmidt et al, 2008) together with charge state and gas phase fractionation (Yi et al, 2002; Scherl et al, 2008) to complement and refine the proteome map (see Supplementary information for detail). To exclude the possibility of an inflated protein false discovery rate (FDR) due to error propagation from peptide to protein level inference, we used the Mayu software tool that determines the protein FDR in large data sets as a function of the peptide FDR (Reiter et al, 2009). Overall, 174 066 peptide-spectrum matches (PSMs) were identified at a FDR of 1% (Figure 1). From the identified peptides, we inferred 10 006 proteins (Supplementary Table S1, raw data available at https://proteomecommons.org), which is to our knowledge the by far most comprehensive proteome map of a mammalian cell line, with earlier studies reaching, e.g. 5399 proteins in U2OS (Lundberg et al, 2010) and 2859 proteins in HeLa cells (Wisniewski et al, 2009). Previous studies of U2OS cells that used other proteomic approaches discovered even fewer proteins (n=237) (Niforou et al, 2008). To assess the comprehensiveness of our approach, we asked whether mRNAs that are highly expressed in U2OS cells remain undetected on the protein level. Based on RPKM values (reads per kilobase of exon model per million mapped reads) provided by the aforementioned study of U2OS cells (Lundberg et al, 2010), we detected proteins corresponding to ∼84% of the most abundant quartile of mRNAs. Although it remains unknown whether all mRNAs are translated into proteins, gene ontology (GO) analysis revealed that ∼33% of the mRNAs unidentified on the protein level encode transmembrane proteins, suggesting that these proteins are less accessible for our proteomic approach (discussed below). To determine whether additional fragment ion spectra from additional LS-MS/MS runs would have further increased proteome coverage, we used proteome coverage prediction software tools we previously developed (Claassen et al, 2009). Our analysis revealed that under the experimental conditions chosen saturation of proteome coverage was already reached from the available data (Figure 1).
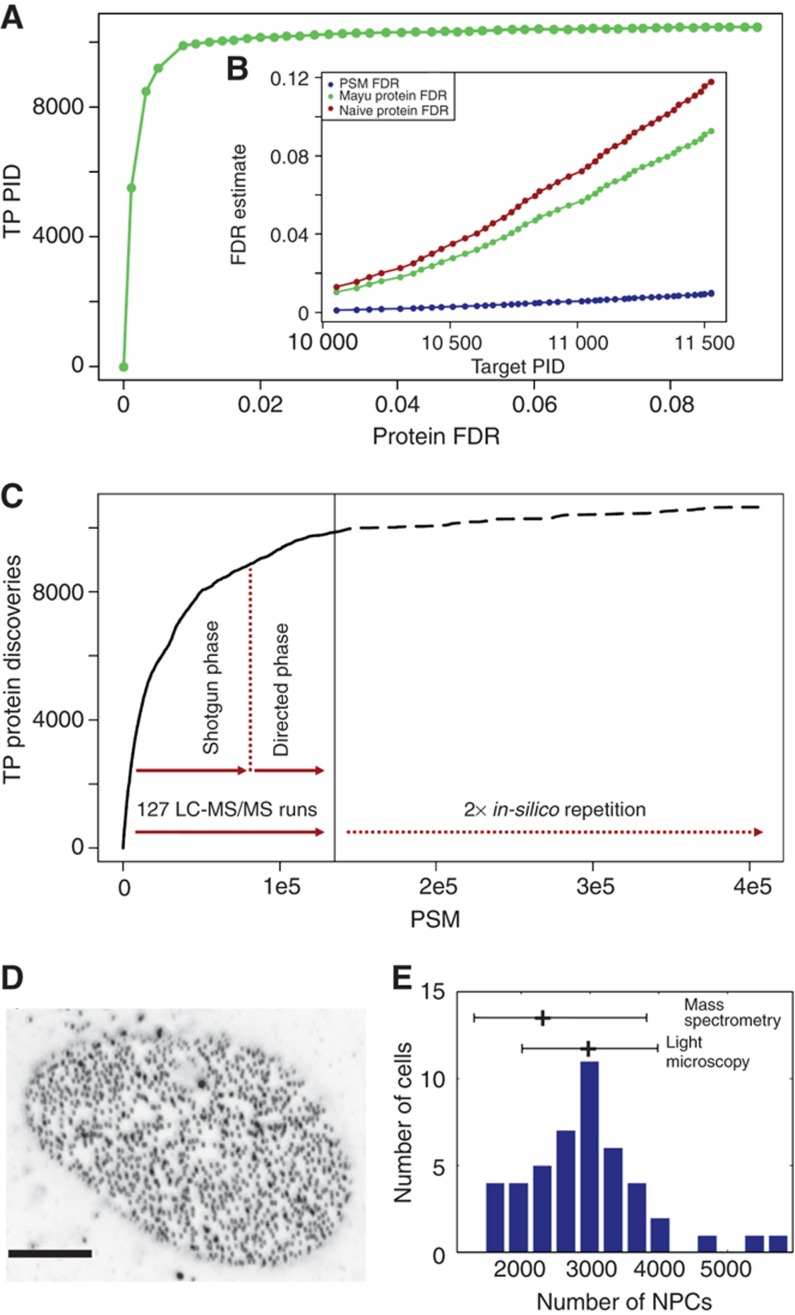
Figure 1.
Protein and PSM FDRs for the ‘U2OS data set’ and independent validation of protein copy numbers. (A) Number of expected true positive protein identifications (TP PIDs) for varying protein FDRs. Number of PIDs stagnates at 2% protein FDR (∼0.2% PSM FDR). Stringent PSM filter preserves true PIDs. (B) FDR estimates for different entities as a function of the number of total, i.e., true and false PIDs (target PID). PSM FDR (blue), Mayu protein FDR (green) and the (frequently used and yet) too pessimistic naive protein FDR (Reiter et al, 2009; see Supplementary information for detail) estimate (brown). (C) Proteome coverage prediction (dashed) for repetition of experiments that gave rise to the ‘U2OS data set’ (solid). Number of acquired confident PSMs is plotted against the number of true positive protein discoveries (TP PIDs). Effective saturation coverage reached at level of TP PIDs for given experimental set-up. (D) Confocal section of U2OS cell with punctuate pattern of NPCs stained with monoclonal antibody mAb414 (scale bar 5 μm). (E) Distribution of number of NPCs in U2OS cells as determined by quantification of images from 46 cells as shown in (D). The mean value of 3000 NPCs per cell and the standard deviation of 1000 is displayed and put into relation to the number of NPCs per cell measured by MS and the corresponding precision of the MS method (mean fold error <2; Supplementary Figure S1).
To quantify the MS detectable proteins of the U2OS proteome, we selected 144 high-intensity proteotypic peptides (PTPs) (Kuster et al, 2005) covering 84 human proteins from the proteome map described above (Supplementary Table S1). These peptides were synthesized as heavy isotope labeled, absolutely quantified reference peptides and defined amounts were spiked into trypsin-digested extracts from 1exp7 U2OS cells growing in log phase. The peptide samples were processed and analyzed as described above except that data-dependent precursor ion selection was used exclusively (see Supplementary information for detail). Within the combined data from the 16 OGE fractions set, we confidently detected and quantified 70 reference and endogenous peptides pairs corresponding to 53 proteins in a concentration range from 4.5exp3 to 2.5exp6 copy numbers per cell. We used the same data to calculate three different protein abundance scores as previously described (Ishihama et al, 2005; Silva et al, 2006; Malmstrom et al, 2009) and validated their precision by statistical analysis (Supplementary Figure S1; see Supplementary information for detail). Based on this analysis, we decided to use the share of spectrum identification index (SSID) normalized by protein molecular weight to estimate the abundance of >7300 human proteins (Supplementary Table S1). Since we cannot assess the precision of the quantitative estimates outside of the dynamic range covered by heavy labeled reference peptides, we masked proteins <500 copies per cell and >20 000 000 copies per cell, respectively, assuming a correlation similar to the validated concentration range within the respective range of protein copies. To validate our abundance scale with an independent method, we used high-resolution confocal fluorescence microscopy to count the number of nuclear pore complexes (NPCs) in U2OS cells (Figure 1D). In contrast to MS that measures average signals from the combined lysates of many cells, this method can provide a distribution of the number of NPC's across many individual cells. The number of NPCs determined by light microscopy was in very good agreement with the copy numbers of the relevant proteins determined by MS (Figure 1E). Although this validation method relies only onto a single measurable value, namely NPC copies per cell, it demonstrates that we neither systematically overestimate nor underestimate protein abundance in the MS-derived quantitative scale (see Supplementary information for detail). Further, the variance measured across individual cells was in a similar range as the estimated precision of the MS method. Thus, we conclude that we successfully determined the copy numbers per cell for 73% of U2OS proteins that are detectable with the MS method used, with an estimated mean error of about two-fold.
To investigate whether changes in protein copy numbers across different biological states can be observed, we repeated the experiment-described above with cells arrested in M phase using nocodazole (Supplementary Figure S2) and quantified 6800 proteins. Proteins detected with copy number variations as compared with a sample from non-synchronized cells were significantly enriched for biological processes carrying out mitotic functions (Supplementary Figure S3; Supplementary Table S2). To investigate whether the proteins with higher copy number during M phase are essential for cell division, we compared our data set with mitotic gene silencing phenotypes identified by a recent genome-wide study (Neumann et al, 2010). The frequency of mitotic phenotypes discovered by RNAi screening that were associated with proteins displaying a higher copy number during mitosis was considerably higher than in a control set (Supplementary Figure S3; Supplementary Table S2). Nevertheless, a considerable fraction of proteins with increased copy number in mitosis did not show a strong phenotype despite being known to have a role in mitosis (e.g., kinetochore components or mitotic kinases). This finding indicates that mammalian cells can often cope with the effects of gene silencing without displaying an obvious phenotype. This may be because related proteins, e.g. isoforms, can compensate for the function of the targeted gene or because compensatory effects at the network level can attenuate phenotypes (Bodenmiller et al, 2010).
High- and low-abundant proteins have specific cellular functions
We next analyzed the relationship between protein abundance and function. At first, we used GOs (Ashburner et al, 2000; Huang da et al, 2009) to compare the number of genes and expressed gene products devoted to specific functions on the level of the genome, qualitative proteome (i.e., number of different proteins associated with a function) and absolutely quantified proteome, respectively (Figure 2A). Such analyses reveal the fraction of total protein mass that the cell devotes to specific biological functions. Processes such as transcription, translation, protein, and nucleic acid metabolism and transport make up considerable fractions of the total proteome. Others, such as cell adhesion, communication, and signaling are underrepresented on the quantitative as compared with the qualitative proteome and genome level, indicating that the corresponding gene products are expressed at relatively low copy numbers. The opposite is true for translation and cytoskeleton, for example.
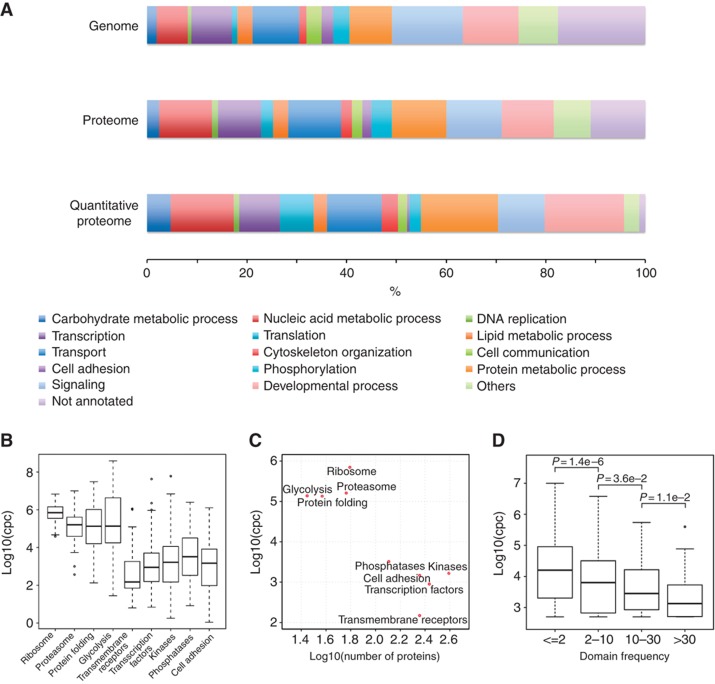
Figure 2.
Abundance levels of functional protein categories. (A) The fraction of predicted gene models or detected gene products, respectively, functioning in specific biological processes is shown in percent on the genome, qualitative, and quantitative proteome levels. Processes such as signaling and cell adhesion are underrepresented in the quantitative proteome, while processes such as protein and nucleic acid metabolism, development, cytoskeleton, translation, and carbohydrate metabolism are overrepresented in the total protein amount of U2OS cells. GO was used to categorize genes and gene products; protein copy numbers within each group were considered in case of the quantitative proteome. (B) The abundance of proteins of different molecular functions and functioning together in distinct protein complexes or biological processes varies over several orders of magnitude. (C) The median protein abundance of the functional groups shown in (B) shows a moderate inverse correlation with the number of proteins per group. (D) The frequency of protein domains in the human genome negatively correlates with their median abundance. P-values were calculated using an one-sided Wilcoxon rank sum test.
To test whether the proteins functioning in specific cellular components, functional pathways or protein complexes have distinct cellular abundance patterns, we grouped all absolutely quantified proteins into four abundance groups: high (>100 000 copies), moderate (5000–100 000 copies), low (500–5000 copies), and very low-abundant proteins (<500 copies) and tested if specific ontology or pathway terms were significantly enriched within these groups (Supplementary Table S3). We discovered first that no significant enrichment of functional categories was detectable for the low-abundant protein class (500–5000 copies per cell). Metabolic processes require proteins expressed over the entire range of protein abundance: glycolysis and purine metabolism depend mostly on high-abundant proteins. However, proteins functioning in lipid, fatty acid, steroid, and phospholipid metabolism were frequently of very low abundance. Second, we found that cellular processes associated with protein synthesis and turn over, namely translation, protein folding, splicing, and degradation as well as RNA processing, are mostly conducted by high-abundant proteins. However, proteins involved in protein sorting and localization, e.g. nuclear transport, are moderately abundant, while proteins involved in catalyzing post-translational modifications such as glycosylation, phosphorylation, and ubiquitination are mostly conducted by very low-abundant proteins. Third, we found that proteins involved in functions such as signaling, cell communication, and regulation of cellular processes are often of very low abundance. Functional groups falling into this category are, e.g., cell adhesion, transcription factors, kinases, or calcium signaling.
The abundance distribution of the components of several protein complexes and functional groups is shown in Figure 2B (see also Supplementary Table S3). The ribosome and proteasome are the most abundant protein complexes and display a narrow component distribution, indicating that they are stably associated. Proteins functioning together in biological processes but in multiple protein complexes display a broader distribution but can still be associated with specific abundance ranges. These include the functions ‘protein folding’ and ‘glycolysis’ as well as kinases and phosphatases. Interestingly, functional protein abundance classes seem to split up into two major categories (Figure 2C): cellular core functions with a smaller number of genes that are expressed at high copy numbers and regulatory cell functions that are orchestrated by families composed of a large number of genes each expressed at low copy numbers (discussed below). Inspired by this finding, we further investigated the link between protein abundance and gene frequency using the superfamiliy classification of protein domains (Gough et al, 2001). This classification is based on a database of 1106 Hidden Markov Models that assign >75% of the quantified human proteome to at least one superfamily, and provides a more comprehensive and complementary view to the functional annotation used in Figure 2A. We observed a negative correlation between the frequency of protein domains and their median copy number per cell (Figure 2D; Supplementary Figure S4A; Supplementary Table S5), which implicates that abundant proteins are generally composed of domains that occur at lower frequency in the human genome and, consequently, likely to be under purifying selection. Vice versa, domains that underwent expansion during evolution are more likely to be present in low-abundant proteins. The same trend can also be observed when gene duplication events are considered instead of domain expansion. In this case, proteins that have a higher number of paralogs tend to be expressed at lower copy number (Supplementary Figure S4B).
Discussion
In this study, we determined the so far most extensively measured human cell proteome. We identified >10 000 proteins expressed in the commonly used human tissue culture cell line U2OS and demonstrate that protein discovery has reached saturation under the experimental conditions used, i.e., that further measurements of the same type would not be expected to identify additional proteins. We furthermore describe a large-scale estimate of protein abundances in a human cell. We and others have previously shown that the dynamic range of protein concentrations spans more than three orders of magnitude in the bacterium L. interrogans (Malmstrom et al, 2009) and five orders of magnitude in yeast (Ghaemmaghami et al, 2003; de Godoy et al, 2008; Picotti et al, 2009). In the present study, we demonstrate that the protein copy numbers of a human cell span at least seven orders of magnitude. This range is similar to that determined in mouse cells (Schwanhausser et al, 2011). This finding is furthermore in good agreement with the volume of the relevant cell types, namely ∼0.2 μm3 in L. interrogans (Beck et al, 2009), and about ∼30 μm3 in S. cerevisiae and ∼4000 μm3 in U2OS, (assuming spherical shape and 4 and 20 μm diameter for yeast and U2OS, respectively).
Interestingly, the bacterium L. interrogans expresses a relatively small number of in very high copy proteins, e.g. proteins of the translation and protein folding system, metabolic enzymes as well as components of the cell wall. Those proteins make up the majority of the total protein mass (Malmstrom et al, 2009) and a considerable fraction of the cytoplasmic volume (Beck et al, 2009), while proteins functioning in signaling, protein transport, or regulatory pathways, e.g. transcription factors, comprise a minority of the quantitative proteome. To investigate whether the same holds true for eukaryotes, we systematically compared the four available data sets mentioned above (Figure 3). We arbitrarily grouped all functional categories into three major classes: (i) cellular core functions containing carbohydrate, nucleobase, nucleoside, nucleotide, nucleic acid metabolic processes, lipid and other metabolic processes as well as transcription, translation, DNA replication, transport, and other core functions; (ii) regulatory functions, namely cytoskeleton organization, cell adhesion, cell division, phosphorylation, protein metabolic processes, signaling, developmental process, cell communication, and other regulatory functions; and (iii) others. The bacterium L. interrogans devotes most of its protein mass (∼75%) to core and <25% to regulatory functions. In contrast, less than half of the analyzed protein mass of U2OS fulfills core functions, and 51% carries out regulatory functions. In particular, the total fraction of protein devoted to cytoskeleton organization, protein metabolic processes and signaling is largely expanded in U2OS cells, while other processes with the exception of central metabolic processes are largely reduced. A very similar picture emerged for mouse cells. Yeast, at a first glance, does not seem to follow this trend. However, it devotes only one third of the total protein mass to metabolism, while the corresponding number is >50% in L. interrogans. As a single cell eukaryote, yeast expands a significant fraction of its protein mass (∼30%) on translation and protein sorting. Taken together, this analysis indicates that the fraction of total protein mass devoted to regulatory functions is largely expanded in higher eukaryotes.
Figure 3.
Comparative analysis of protein abundance. Pie charts representing the annotated quantitative proteome of human U2OS cells, mouse NIH3T3 cells, S. cerevisiae and L. interrogans taking protein copy numbers per functional category into account. Functional categories are classified into three major groups: cellular core, regulatory functions and others. Protein abundance data sets were taken from this study and references (Ghaemmaghami et al, 2003; Malmstrom et al, 2009; Schwanhausser et al, 2011).
In multicellular species, domain families fulfilling regulatory functions have been more frequently subject to gene expansion than domains fulfilling core functions (Vogel and Chothia, 2006; Ori et al, 2011). We therefore investigated, using the quantitative data generated in this study, how this effect is linked to protein abundance. We and others showed that protein abundance is linked to function, namely that high-abundant proteins are often responsible for core functions, such as energy metabolism and translation, while regulatory functions such as protein phosphorylation and transcriptional regulation are often carried out by low-abundant proteins (Figure 2B and C; Supplementary Table S3; Schwanhausser et al, 2011). There are several lines of evidence suggesting that protein abundance is also linked to evolvability. It has been previously shown that highly expressed proteins evolve more slowly than proteins expressed at lower levels, i.e., they display a reduced protein divergence on the sequence level (Pal et al, 2001; Subramanian and Kumar, 2004), while low-abundant proteins display decreased sequence conservation across organisms (Schrimpf et al, 2009). It was further shown that protein families displaying lower abundance variability across species less often underwent gene duplication and that abundance variability scales inversely with protein expression (Weiss et al, 2010). These findings indirectly suggest a link between protein abundance and gene duplicability. Our data support this hypothesis. We show a negative correlation between the frequency of domain families in the human genome and their median copy number per cell (Figure 2D; Supplementary Figure S4A; Supplementary Table S5). We also show that proteins, which have a higher number of paralogs, tend to be expressed at lower copy number (Supplementary Figure S4B). These findings underline the view that duplications of genes encoding for proteins expressed at high level are maintained under purifying selection, likely because of energy constraints (Lane and Martin, 2010) or higher risk of protein aggregation and toxicity (Drummond et al, 2005). Interestingly, a recent study that compares the relative expression level of gene products of three human cell lines on proteome and transcriptome level showed that proteins involved in regulatory functions more often vary in their expression levels as compared with core functions (Lundberg et al, 2010). One might thus speculate that the large fraction of the human proteome expressed at low copy number and involved in regulatory function was the main source of biological innovation during evolution. This hypothesis is supported by the following lines of evidence: (i) domain families occurring in low-abundant proteins are significantly more correlated with increase in organism complexity than the ones present in highly expressed proteins (P=7.8e−9, one-sided Wilcoxon rank sum test; Supplementary Figure S4C; Supplementary Table S5). (ii) The abundance of proteins involved in core functions is more strongly conserved across species than for proteins involved in regulatory functions (Schrimpf et al, 2009). (iii) The fraction of the proteome devoted to regulatory functions significantly expanded during the course of evolution (Figure 3).
Regulatory, often low-abundant proteins are key players in mediating the integration of external stimuli with the cell's internal state and they control fundamental biological processes such as cell proliferation, migration, and cell differentiation. It was recently shown for mouse cells that low-abundant proteins and mRNAs are less stable than high-abundant ones (Schwanhausser et al, 2011). Therefore, expression at low copy numbers might provide an efficient way of dynamic regulation by translation and rapid turnover. Vice versa, cellular core functions might be more efficiently regulated by other means than degradation.
Current limitations of protein abundance indices determined from MS data are the availability of PTPs accounting for the multitude of isoforms within protein families and a bias toward proteins that produce fewer well-ionizing peptides. In particular, GO analysis reveals an underrepresentation of transmembrane proteins in the identified proteome (Supplementary Table S4). Such an effect has been observed before (Schrimpf et al, 2009) and is likely a result of the reduced accessibility of membrane proteins for MS analysis, although we had used an MS compatible detergent during sample preparation. This finding is further underlined by fact that a significant fraction of high-abundant mRNAs not discovered on the protein level encodes for membrane proteins. Otherwise, the distribution of functional categories on the genome and proteome level is quite similar, suggesting high proteome coverage and that the assumption of an even extractability of proteins holds true for the majority of proteins but not for membrane proteins. We demonstrate the feasibility of establishing protein abundance scales in very complex proteomes with precision that is likely sufficient to allow the analysis of biological systems by means of computational modeling. The method used in this study is principally applicable to the majority of all cell types and might be useful to study a multitude of cellular states and organisms in the future.
Materials and methods
A detailed description of all Materials and methods is provided in Supplementary information.
Cell culture
U2OS cells were grown in DMEM medium supplemented with 10% bovine serum, harvested by trypsinization, washed twice in phosphate-buffered saline (PBS), resuspended in 0.5 ml PBS, and lysed by adding 0.5 ml of 10 mM Tris pH 7.5, 10 M urea, 0.1% Rapigest. Synchronization was carried out by adding nocodazole to a final concentration of 330 nM for 18 h.
Mass spectrometry
Proteins were reduced with 10 mM TCEP for 20 min at 37°C and alkylated with 10 mM iodoacetamide and digested with trypsin (1/100, w/w). AQUA peptides were spiked into the sample at this stage, if applicable. The peptides were cleaned up by C18 reversed-phase spin columns according to the manufacturer's instructions (Harvard Apparatus). The peptides were separated on pH 3–10 IPG strips (GE Healthcare) with a 3100 OFFGEL fractionator (Agilent) according to the manufacturer's instructions. The set-up of the μRPLC-MS system was as described previously (Schmidt et al, 2008). Depending on the sample complexity, each fraction was analyzed 3–4 times in shotgun and 2–5 times in directed (inclusion list) mode. Directed LC-MS measurements of features and reference peptides were performed according to Schmidt et al (2008) using a rolling inclusion list if the number of masses exceeded 500. Thereby, large feature lists were automatically split into smaller lists covering a certain mass range and charge to enable more specific directed MS analysis (Scherl et al, 2008). Samples for absolute protein quantification were analyzed using an Easy-nLC/Orbitrap-Velos (both from ThermoScientific, Bremen, Germany) LC-MS system with the following modified parameters: peptides were separated using a linear gradient from 92% solvent A (98% water, 2% acetonitrile, 0.15% formic acid) and 8% solvent B (98% acetonitrile, 2% water, 0.15% formic acid) to 40% solvent B over 120 min. Each survey scan acquired in the Orbitrap at 60 000 FWHM was followed by MS/MS scans of the 20 most intense precursor ions in the linear ion trap. MS/MS spectra were searched using the SEQUEST algorithm (Yates et al, 1995) against a decoy database (consisting of forward and reverse protein sequences). The database search results were further processed using the PeptideProphet (Keller et al, 2002) and ProteinProphet (Keller et al, 2005) program. PSM FDRs have been estimated by means of the target-decoy strategy (Elias and Gygi, 2007). Protein FDRs have been estimated by the generalized target-decoy strategy Mayu (Reiter et al, 2009). Proteome coverage prediction has been performed as described in Claassen et al (2011).
Data deposition
The three MS raw data sets corresponding to the proteome mapping and both quantification experiments (of synchronized and non-synchronized cells) were deposited at Proteome Commons (https://proteomecommons.org) with the following hash codes:
3Wj0424JA2DCVkBnfqm45v+UfMZOHgf3p2PTwUe83RwjqQvtr4mQnloYvUSrMHCYBz+krDIXmz50spF2TNYGw3/8jZIAAAAAAAAB9w==;
x8hmYUs40bOspaY+EMuyUtDkyiw+xgyjSynVK/ggQXhl+bbDV5QbiAMakzsSKonz/XszxEEUThtmn6cIS/STS1Y0n2QAAAAAAAAB3g==;
Gm5TsXK3crQV70MqiIIH+/uaKyioNCFWi+Ri7fpLq+W1ga5OQA0dTe2u0LMvN+ty7uuRsA1o3WTWb79Bc/XqYK7v9D0AAAAAAAACBA==.
Absolute abundance estimation
AQUA peptides were grouped into three abundance classes based on spectral counts and spiked into human peptide mixtures directly after digestion, at a final concentration of 0.5, 5, or 50 pmol/μl, respectively. Heavy and light ratios between spiked AQUA and endogenous peptides were calculated using XPRESS as implemented into the trans-proteomic pipeline (Keller et al, 2005). The exponentially modified protein abundance index (emPAI) was calculated as previously described (Ishihama et al, 2005) and based on the peptide statistics calculated by PeptideProphet (Keller et al, 2002). To the SSID the ‘percent share of spectrum ID’ output of ProteinProphet (Keller et al, 2005) was normalized by dividing through the proteins molecular weight. The median of the extracted precursor ions (XICs) of the three best flying peptides per protein (Top3) index (Silva et al, 2006) was calculated as described previously (Malmstrom et al, 2009). The arbitrary protein abundance indices were calibrated to absolute protein copy numbers per cell their precision validated by bootstrapping analysis as described earlier (Malmstrom et al, 2009). To estimate the average number of NPCs per cell, we performed immunofluorescence labeling, high-resolution confocal microscopy and computational image analysis. In brief, U2OS cells fixed and permeabilized and NPCs were then stained with mab414 (Covance) and secondary goat anti-mouse IgG coupled to Alexa Fluor 488 (Invitrogen). Confocal z-stacks (xyz pixel size 44/44/380 nm) through the entire nucleus of 46 randomly chosen cells were acquired using a Zeiss LSM 710 confocal microscope. In each cell, the NPC density was quantified with an in-house developed intensity peak identification macro in Image J.
Functional analysis
The functional annotation of the U2OS proteome was performed using a custom designed GO slim annotation. The comparative analysis of quantitative proteomes across species was performed using published data set for L. interrogans (Malmstrom et al, 2009), S. cerevisiae (Ghaemmaghami et al, 2003), and mouse (Schwanhausser et al, 2011), and the data set presented in this study for human. Domain distribution across quantified proteins was investigated using the Superfamily v1.75 annotation (Uniprot 2011_07 assignment; Gough et al, 2001). RNA abundance data, specifically reads per kilobase of exon model per million mapped reads (RPKM) values of U2OS cells were taken from a previously published data set (Lundberg et al, 2010).
Supplementary Material
Supplementary figures S1-5, Supplementary tables S1-7
Acknowledgments
Jean-Karim Heriché and Thomas Walter (EMBL) are acknowledged for help in preparing Supplementary Figure S3. This project was funded in part by ETH Zurich, SystemsX.ch, the Swiss initiative for systems biology (project PhosphonetX) and by the European Union Seventh Framework Program PROSPECTS (Proteomics Specification in Space and Time Grant HEALTH-F4-2008-201648). RA is supported by the European Research Council (Grant # ERC-2008-AdG 233226). JM was supported by a fellowship from the Swedish Society of Medical Research (SSMF), MB was supported by a long-term fellowship of the European Molecular Biology Organization and a Marie Curie fellowship of the European Commission, AS was supported by the Competence Center for Systems Physiology and Metabolic Diseases. AO is supported by a postdoctoral fellowship from the Alexander von Humboldt Foundation.
Author contributions: MB and AS designed and performed experiments, generated and processed samples, processed and validated data, and wrote the manuscript; JM designed and performed experiments, processed and validated data; MC processed and validated data; AO processed and validated data and wrote the manuscript; AS designed and performed experiments, processed and validated data; FH designed and performed experiments; OR designed experiments, processed and validated data; JE validated data and wrote the manuscript; and RA managed the project, validated data, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Malmstrom JA, Lange V, Schmidt A, Deutsch EW, Aebersold R (2009) Visual proteomics of the human pathogen Leptospira interrogans. Nat Methods 6: 817–U855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, Brusniak MY, Roschitzki B, Zhang C, Shokat KM, Schlapbach R, Colman-Lerner A, Nolan GP, Nesvizhskii AI, Peter M, Loewith R et al. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal 3: rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassen M, Aebersold R, Buhmann JM (2009) Proteome coverage prediction with infinite Markov models. Bioinformatics 25: i154–i160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassen M, Aebersold R, Buhmann JM (2011) Proteome coverage prediction for integrated proteomics datasets. J Comput Biol 18: 283–293 [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455: 1251–1254 [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH (2005) Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA 102: 14338–14343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313: 903–919 [DOI] [PubMed] [Google Scholar]
- Horth P, Miller CA, Preckel T, Wenz C (2006) Efficient fractionation and improved protein identification by peptide OFFGEL electrophoresis. Mol Cell Proteomics 5: 1968–1974 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Jaffe JD, Keshishian H, Chang B, Addona TA, Gillette MA, Carr SA (2008) Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics 7: 1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Eng J, Zhang N, Li XJ, Aebersold R (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol 1: 2005.0017; DOI: 10.1038/msb4100024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kuster B, Schirle M, Mallick P, Aebersold R (2005) Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol 6: 577–583 [DOI] [PubMed] [Google Scholar]
- Lane N, Martin W (2010) The energetics of genome complexity. Nature 467: 929–934 [DOI] [PubMed] [Google Scholar]
- Lundberg E, Fagerberg L, Klevebring D, Matic I, Geiger T, Cox J, Algenas C, Lundeberg J, Mann M, Uhlen M (2010) Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol 6: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T, Schmidt A, Guell M, Kuhner S, Gavin AC, Aebersold R, Serrano L (2011) Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol 7: 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom J, Lee H, Nesvizhskii AI, Shteynberg D, Mohanty S, Brunner E, Ye M, Weber G, Eckerskorn C, Aebersold R (2006) Optimized peptide separation and identification for mass spectrometry based proteomics via free-flow electrophoresis. J Proteome Res 5: 2241–2249 [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wunsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R et al. (2010) Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464: 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niforou KM, Anagnostopoulos AK, Vougas K, Kittas C, Gorgoulis VG, Tsangaris GT (2008) The proteome profile of the human osteosarcoma U2OS cell line. Cancer Genomics Proteomics 5: 63–78 [PubMed] [Google Scholar]
- Ori A, Wilkinson MC, Fernig DG (2011) A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J Biol Chem 286: 19892–19904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD (2001) Highly expressed genes in yeast evolve slowly. Genetics 158: 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R (2009) Full dynamic range proteome analysis of S.cerevisiae by targeted proteomics. Cell 138: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R (2009) Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics 8: 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A, Shaffer SA, Taylor GK, Kulasekara HD, Miller SI, Goodlett DR (2008) Genome-specific gas-phase fractionation strategy for improved shotgun proteomic profiling of proteotypic peptides. Anal Chem 80: 1182–1191 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Beck M, Malmstrom J, Lam H, Claassen M, Campbell D, Aebersold R (2011) Absolute quantification of microbial proteomes at different states by directed mass spectrometry. Mol Syst Biol 7: 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Gehlenborg N, Bodenmiller B, Mueller LN, Campbell D, Mueller M, Aebersold R, Domon B (2008) An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol Cell Proteomics 7: 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrimpf SP, Weiss M, Reiter L, Ahrens CH, Jovanovic M, Malmstrom J, Brunner E, Mohanty S, Lercher MJ, Hunziker PE, Aebersold R, von Mering C, Hengartner MO (2009) Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol 7: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 5: 144–156 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S (2004) Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 168: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Abreu Rde S, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Chothia C (2006) Protein family expansions and biological complexity. PLoS Comput Biol 2: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M, Schrimpf S, Hengartner MO, Lercher MJ, von Mering C (2010) Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics 10: 1297–1306 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Yates JR III, Eng JK, McCormack AL (1995) Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem 67: 3202–3210 [DOI] [PubMed] [Google Scholar]
- Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, Goodlett DR (2002) Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis 23: 3205–3216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1-5, Supplementary tables S1-7