Abstract
Nutrient sensing and coordination of metabolic pathways are crucial functions for all living cells, but details of the coordination under different environmental conditions remain elusive. We therefore undertook a systems biology approach to investigate the interactions between the Snf1 and the target of rapamycin complex 1 (TORC1) in Saccharomyces cerevisiae. We show that Snf1 regulates a much broader range of biological processes compared with TORC1 under both glucose- and ammonium-limited conditions. We also find that Snf1 has a role in upregulating the NADP+-dependent glutamate dehydrogenase (encoded by GDH3) under derepressing condition, and therefore may also have a role in ammonium assimilation and amino-acid biosynthesis, which can be considered as a convergence of Snf1 and TORC1 pathways. In addition to the accepted role of Snf1 in regulating fatty acid (FA) metabolism, we show that TORC1 also regulates FA metabolism, likely through modulating the peroxisome and β-oxidation. Finally, we conclude that direct interactions between Snf1 and TORC1 pathways are unlikely under nutrient-limited conditions and propose that TORC1 is repressed in a manner that is independent of Snf1.
Keywords: carbon metabolism, nitrogen metabolism, nutrient sensing, Snf1, TORC1
Introduction
Cells commonly face environmental changes such as varying availability of nutrients. Therefore, it is of crucial importance for them to adjust the metabolism accordingly. In this context, the nutrient sensing and related regulatory pathways are particularly important. Since there is a high degree of conservation in the functionality of these regulatory pathways in all eukaryotes, the budding yeast Saccharomyces cerevisiae serves as an excellent model and has been used for many studies on nutrient sensing and regulation in eukaryotic cells (Petranovic and Nielsen, 2008). S. cerevisiae senses extracellular nutrients and controls its metabolism through a complex regulatory network (Zaman et al, 2008). Key components in this regulatory network include the Snf1 kinase complex and the target of rapamycin complex 1 (TORC1). Snf1 complex belongs to a remarkably conserved serine/threonine kinase family called AMP-activated kinase (AMPK) that exists in all eukaryotes (Polge and Thomas, 2007). The Snf1 kinase was first identified as the key enzyme in releasing the glucose repression on glucose depletion (Celenza and Carlson, 1984), and later found to be involved in the regulation of transcription through post-translational modifications of histone H3 and Gcn5 (Lo et al, 2001; Liu et al, 2010) and interaction with RNA polymerase II holoenzyme (Kuchin et al, 2000). Upon activation by phosphorylation, Snf1 induces genes involved in gluconeogenesis, glyoxylate cycle and β-oxidation of fatty acids (FAs) by regulating a set of different transcription factors (TFs) (Soontorngun et al, 2007; Ratnakumar and Young, 2010) and represses lipid biosynthesis by inhibiting Acetyl-CoA carboxylase (Acc1) (Woods et al, 1994), the committed step of FA synthesis pathway. Besides the aforementioned processes, Snf1 is also involved in the general stress response, pseudohyphal growth, ageing and ion homeostasis (Alepuz et al, 1997; Kuchin et al, 2002; Lin et al, 2003; Portillo et al, 2005; Hong and Carlson, 2007; Ye et al, 2008)
TORC1 was first identified in the screening of yeast mutants against the antifungal reagent rapamycin. Similarly to Snf1, TORC1 is also highly conserved in eukaryotes (Schmelzle and Hall, 2000). TORC1 in S. cerevisiae (and some other unicellular eukaryotes such as Schizosaccharomyces pombe) consists of either Tor1 or Tor2, two homologous proteins, as well as other components, while in metazoans like flies, worms and mammals only one Tor protein can form the TORC1 (Inoki et al, 2005). However, unlike Tor1, Tor2 can also form the TOR complex 2 (TORC2), which is insensitive to rapamycin and has distinct structures and functions compared with TORC1 (Loewith et al, 2002; Jacinto et al, 2004; Wullschleger et al, 2005). TORC1 senses the availability and quality of the nutrients and regulates cell growth by antagonizing a spectrum of TFs in the cytoplasm. For example, TORC1 induces ribosome biogenesis through the TFs Sfp1 and Fhl1, in coordination with the protein kinase A (PKA) pathway, thus promotes protein translation and cell growth (Marion et al, 2004; Martin et al, 2004). TORC1 also negatively regulates those genes whose expression is induced by limitation of nitrogen sources through the transcription activators Gln3 and Gat1 (Beck and Hall, 1999), prevents amino-acid biosynthesis by modulating Gcn4 translation (Valenzuela et al, 2001) and represses stress responses through the TFs Msn2 and Msn4 (Monteiro and Netto, 2004; Petkova et al, 2010). It has been shown that TORC1 is also involved in many other processes such as autophagy, ageing and cell cycle (Kamada et al, 2000; Colomina et al, 2003; Medvedik et al, 2007).
Recent systematic approaches indicate some coordination between Snf1 and TORC1 signaling pathways under nutrient-excess conditions (Zaman et al, 2008; Smets et al, 2010). However, it is not clear if there is any interaction between them under nutrient limitation. Although AMPK was shown to directly inhibit mTORC1 (AMPK and mTORC1 are the orthologs of Snf1 and TORC1 in mammals, respectively), a similar interaction was not identified in yeast (Hardie, 2007). Instead, it was suggested that Snf1 may be involved in nitrogen metabolism through the regulation of the transcription activators Gln3 and Gcn4 (Bertram et al, 2002; Shirra et al, 2008), which are targets of TORC1 and positively regulates genes that are subject to nitrogen catabolic repression. It has also been shown that Snf1 is directly involved in nitrogen signaling and regulated by nitrogen availability (Orlova et al, 2006).
Thus far, our knowledge of nutrient sensing and related regulatory pathways is limited to studies conducted under nutrient-excess conditions (e.g., shake flask batch cultivation with excessive amount of carbon and nitrogen sources). The coordination of the signaling pathways under nutrient limitation remains largely obscure. To address this limitation, we took advantage of the chemostat cultivations, which permit correlating observations with the limiting nutrient (Daran-Lapujade et al, 2009). We focused on the cellular response to carbon (glucose) or nitrogen (ammonium) limitation in S. cerevisiae strains that lacked SNF1, TOR1 or both. We also assessed the role of Snf1 and TORC1 as kinases under these conditions using the state of the art phosphoproteomics technology, and their impact on gene expression with transcriptomics. We used the levels of selected metabolites, such as free amino acids and total FAs, as read-outs of the net contribution of these kinases, as amino-acid and FA metabolisms are known to be regulated by Snf1 and TORC1, respectively. By integrating these comprehensive data sets, we obtained substantial new insights into how Snf1 and TORC1 coordinate nutrient sensing and metabolic regulation.
Results and discussion
Cell physiology under nutrient-rich and -limited conditions
To evaluate the effects of deleting Snf1 and TORC1 on cell growth, we characterized the basic physiology of mutant strains snf1Δ, tor1Δ and snf1Δtor1Δ together with the reference strain CEN. PK113-7D (Supplementary Table S1) by growing them first in batch and then switched to chemostat, both using defined minimum medium with glucose as the sole carbon source. Chemostat cultures were used for several reasons. First, it is possible to correlate the observations with the limiting nutrient. Second, since the mutants have different maximum specific growth rates, chemostats offer a platform for comparing different strains at the same growth rate, thereby eliminating any growth-related effects (Fazio et al, 2008). Finally, since the mutant strain snf1Δ is unable to grow on non-fermentable carbon sources (such as ethanol and glycerol), glucose-limited chemostat is the only option to ensure Snf1 activity under comparable growth conditions. There are several good nitrogen sources for yeast, such as glutamine and ammonium (Zaman et al, 2008); however, glutamine can also be used as a carbon source and is therefore unsuitable for nutrient-limited cultures.
All mutant strains grew slower (by 12–22%) using glucose as the sole carbon source and ammonium as the sole nitrogen source, compared with the reference strain, indicating the contribution of Snf1 and TORC1 during exponential growth and deletion of either protein reduces cell growth on defined minimum medium (Table I). However, the observation of an equivalent reduction (about 20% lower) in the maximum specific growth rate for the snf1Δtor1Δ strain seems to contradict the hypothetic genetic interaction between Snf1 and TORC1 on these conditions, as one would expect a more severe phenotypic change in the double mutant strain if a genetic interaction was present (Boone et al, 2007). Deletion of Snf1 (snf1Δ and snf1Δtor1Δ) resulted in a substantial reduction (∼25%) in the biomass yield compared with the reference strain in glucose-limited chemostat cultures (Table I). On the contrary, we observed a small increase in biomass yield for tor1Δ under these conditions. The double mutant produced acetate and glycerol, even under completely respiratory conditions. Interestingly, we did not see any substantial difference in the biomass yield on N-limited condition.
Table 1. Cell physiology of all strains in batch and chemostat cultivations.
| Medium | Strain | μmax | Y SX | Y SAc |
|---|---|---|---|---|
| h−1 | g g−1 | Cmol Cmol−1 | ||
| All values are average±s.e.m. from at least three biological replicates. μmax, maximum specific growth rate on glucose in batch cultures; YSX, biomass yield on glucose in chemostat cultures; YSAc, acetate yield on glucose in chemostat cultures. μmax for each strain was determined based on the CO2 emission during the batch phase of the culture. | ||||
| C-limited | Reference | 0.37±0.01 | 0.515±0.007 | <0.002 |
| snf1Δ | 0.29±0.00 | 0.384±0.003 | 0.007±0.004 | |
| tor1Δ | 0.33±0.01 | 0.534±0.003 | <0.002 | |
| snf1Δtor1Δ | 0.30±0.00 | 0.382±0.002 | 0.028±0.003 | |
| N-limited | Reference | 0.097±0.002 | 0.006±0.000 | |
| snf1Δ | 0.102±0.000 | 0.008±0.000 | ||
| tor1Δ | 0.095±0.000 | 0.008±0.001 | ||
| snf1Δtor1Δ | 0.107±0.001 | 0.009±0.000 | ||
The effects of deleting Tor1 were relatively moderate, given that the TORC1 is the main regulator in cell growth and proliferation (Schmelzle and Hall, 2000). This could be due to the compensatory role of Tor2, which can also form TORC1. To evaluate this hypothesis, we examined the sensitivity of the reference and tor1Δ strains in the presence of rapamycin. While the reference strain could tolerate up to 2 nM of rapamycin in the growth media, there was no observable growth of the tor1Δ strain at any concentration of rapamycin (Supplementary Figure S1). The increased sensitivity to rapamycin caused by loss of Tor1, which was consistent with previous findings (Chan et al, 2000; Reinke et al, 2004; Xie et al, 2005), suggested that the deletion of TOR1 gene caused a substantial reduction in TORC1 signaling or complex activity and Tor2 could hence not fully compensate for the loss of Tor1 function. Since rapamycin inhibits the TORC1 by physically binding to the complex, these results clearly show that Tor1 is responsible for a majority of the TORC1 activity and the tor1Δ strain, therefore, represents a knockdown, but not necessarily a complete loss of function, of TORC1.
Global transcriptome changes due to loss of SNF1 but not TOR1
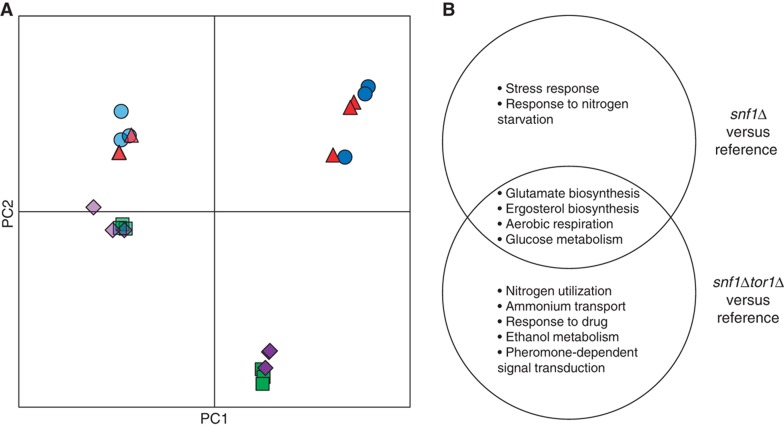
We used the Affymetrix DNA microarray platform to measure the expression level of all genes and evaluate the global effect caused by deletion of the SNF1 and TOR1 genes under nutrient-limited conditions. The transcriptome data were decomposed using principal component analysis (PCA). The first principal component (PC1), which accounted for about 40% of the total variation in the data (Supplementary Figure S2A), primarily distinguished the impact of nutrient limitation (Figure 1A), which was expected as the cells were respiring at C-limited condition while they were respiro-fermenting at N-limited condition. The second principal component (PC2) accounted for the impact of SNF1 deletion. It is also evident that the variance between snf1Δ and reference (represented by the distance between reference and snf1Δ in Figure 1A) is much larger at C-limited condition compared with N-limited condition, confirming that Snf1 has a bigger role on glucose-limited condition. Surprisingly, our data did not reveal any transcriptional role for the TOR1 gene, indicated by the overlapping of tor1Δ and the reference strain at all nutrient limitations (Figure 1). Furthermore, TOR1 deletion did not seem to have a large impact even in the snf1Δ background, as evident from the close proximity of the snf1Δtor1Δ with snf1Δ under all the conditions studied. This result suggests that Tor1 is dispensable under either of nutrient-limited conditions. It is not clear whether the dispensability arises due to its partial redundancy with Tor2 or due to the suppression of the Tor1 function under these conditions. To further examine the extent of the changes in each mutant strain, we performed pair-wise comparisons. On the C-limited condition, the expression of 519 and 603 genes was changed significantly (adjusted p<0.001) in snf1Δ and snf1Δtor1Δ, respectively, relative to the reference strain. Gene ontology (GO) terms analysis revealed that transcription of genes involved in nitrogen metabolism, ethanol metabolism and pheromone-dependent signal transduction appears to be specifically controlled by Tor1, other processes such as stress response and biosynthesis of ergosterol and glutamate were governed by Snf1 (Figure 1B).
Figure 1.
Deletion of SNF1 but not TOR1 caused global change in transcriptome. (A) Principal component analysis (PCA). Dark blue circles: reference on C-limited; dark green squares: snf1Δ on C-limited; dark red triangles: tor1Δ on C-limited; dark purple diamonds: snf1Δtor1Δ on C-limited; light blue circles: reference on N-limited; light green squares: snf1Δ on N-limited; light red triangles: tor1Δ on N-limited; light purple diamonds: snf1Δtor1Δ on N-limited. (B) The biological processes that were affected by deletion of SNF1 (snf1Δ and snf1Δtor1Δ) on C-limited condition.
The measurement of global gene expression provided clear insight into the metabolic differences between the strains. Increased acetate production (Table I) was in line with the lower expression of ACS1 (encoding acetyl-CoA synthetase) in the snf1Δ and snf1Δtor1Δ strains (Supplementary Figure S3). The expression of the other acetyl-CoA synthetase (encoded by ACS2) that is not subject to glucose repression increased slightly in the snf1Δ strain, which is consistent with previous findings (van den Berg et al, 1996). Two other genes in acetate metabolism, ADY2 (encodes an acetate transporter) and ALD4 (encodes a mitochondrial aldehyde dehydrogenase), showed similar expression patterns (Supplementary Figure S3). The change in gene expression for both ACS1 and ADY2 was more prominent in the snf1Δtor1Δ strain compared with the snf1Δ strain. However, the expression of these genes being unchanged in the tor1Δ strain indicates that the role of TORC1 in acetate metabolism relies on Snf1 activity, or in other words it seems like TORC1 has a role in the metabolism of alternative carbon sources through an active Snf1 kinase.
To identify transcriptional regulation of metabolism in response to deletion of SNF1 and/or TOR1, we overlaid the transcriptome onto a genome-scale metabolic model of S. cerevisiae. This method (Patil and Nielsen, 2005; Oliveira et al, 2008) allows identifying the so-called reporter features (metabolites, TFs, etc) around which significant transcriptional activity occurred and also subnetworks of coordinated transcriptional changes. On C-limited condition, deleting SNF1 (snf1Δ and snf1Δtor1Δ) resulted in an extensive transcriptional reprogramming around redox cofactors (NAD(P)+/NAD(P)H), Coenzyme A, several amino acids and α-ketoglutarate (Z-score >1.5) (Supplementary Table S2). These differences were identified primarily though global TFs such as Msn2/4, Cat8, Ino2/4, Oaf1/Pip2, and Hap1 that regulate stress response, aerobic respiration, as well as glucose and sterol metabolism (Supplementary Table S2). Although deleting TOR1 alone did not have any significant transcriptional response, deleting TOR1 in the snf1Δ background resulted in an altered expression for a small subset of genes (involved in several processes including stress response, tRNA methylation, protein targeting to vacuole, ammonium transport, intracellular protein transport), indicating that these processes might be co-regulated by Snf1 and TORC1. Overall transcriptome data from glucose limitation contradict the hypothesis that Snf1 inhibits TORC1.
TOR1 deletion had no distinct phosphorylation response
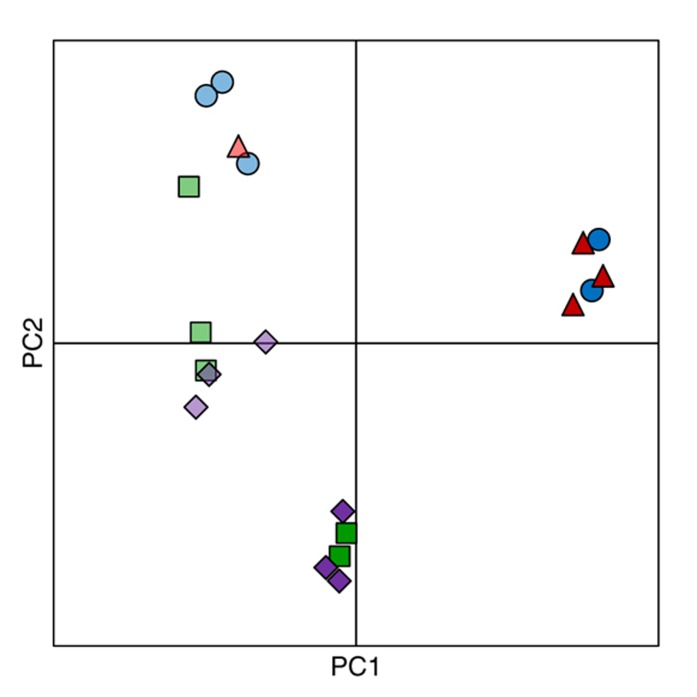
Since both Snf1 and TORC1 are kinase complexes and regulate several processes mainly through phosphorylation of their respective target proteins (Zaman et al, 2008; Smets et al, 2010), we measured the level of phosphorylation of various proteins for all strains under C- and N-limited conditions. As with the transcriptome data, the phosphoproteome data were also analyzed using PCA. The analysis revealed that the TOR1 deletion did not lead to a distinct phosphoproteome profile compared with the reference strain, irrespective of the nutrient limitation. Under all conditions studied, the phosphoproteome profile of tor1Δ always clustered with that of the reference and the phosphoproteome profile of snf1Δtor1Δ always clustered with that of snf1Δ (Figure 2). The reference and tor1Δ strains on C-limited were separated farthest from the other strains/conditions (Figure 2), indicating that the deletion of Snf1 has a dominant response irrespective of the limiting nutrient. Out of the 1714 phosphopeptides that were detected and identified, 399 and 206 peptides had significantly changed phosphorylation level in at least one mutant compared with the reference strain, on C- and N-limited conditions, respectively.
Figure 2.
Principal component analysis of phosphoproteome data for all strains on C- and N-limited conditions. Dark blue circles: reference on C-limited; dark green squares: snf1Δ on C-limited; dark red triangles: tor1Δ on C-limited; dark purple diamonds: snf1Δtor1Δ on C-limited; light blue circles: reference on N-limited; light green squares: snf1Δ on N-limited; light red triangles: tor1Δ on N-limited; light purple diamonds: snf1Δtor1Δ on N-limited.
We observed a clear Snf1-dependent pattern of phosphorylation (lower phosphorylation level in snf1Δ and snf1Δtor1Δ but not tor1Δ) for transcription repressor Cyc8 and its co-component Tup1 (Supplementary Figure S4). Since the Cyc8–Tup1 complex generally represses the transcription of many genes through different modes (Smith and Johnson, 2000), it may be responsible for upregulation of a subset of genes in the snf1Δ and snf1Δtor1Δ strains. This further supports that the transcriptome profile for snf1Δ and snf1Δtor1Δ was significantly changed directly due to the role of Snf1 in the regulation of transcription. This is supported by findings that several other proteins involved in regulation of transcription by histone modification (Bdf1, Eaf1, Leo1, Rph1 and Sin3) were also found to be differentially phosphorylated only in the snf1Δ and snf1Δtor1Δ strains. Significantly changed expression of ACS1 in snf1Δ and snf1Δtor1Δ might also contribute to changes in histone acetylation and global changes in transcription observed in these mutants (Takahashi et al, 2006). These results are in complete consistence with the important role of Snf1 in the regulation of gene transcription (Usaite et al, 2009).
Mammalian TORC1 (mTORC1) is repressed by AMPK (Dennis et al, 2001; Bolster et al, 2002; Inoki et al, 2003), and several lines of evidence suggest direct or indirect interaction between Snf1 and TORC1 (Bertram et al, 2002; Orlova et al, 2006) in yeast. However, in our analysis, we did not find any change in the phosphorylation level of TORC1 due to loss of Snf1 kinase or vice versa (Supplementary Figure S5). Only Tco89 (a non-essential component of TORC1) was identified as differentially phosphorylated in a Tor1-dependent manner. Despite the state-of-art methods used in identifying phosphoproteins, it is likely that some phosphopeptides have not been identified due to low abundance, inefficient purification, poor ionization, etc. Considering this limitation, the absence of all components of the Snf1 and TORC1 pathways in our phosphoproteome analyses compels us to conclude that Snf1 and TORC1 do not regulate the phosphorylation of each other under the conditions studied. It has been shown that phosphorylation of residual T210 on Snf1 is regulated by the nitrogen source or rapamycin through TORC1 (Orlova et al, 2006), which raised the possibility that TORC1 negatively regulates Snf1. However, both transcriptome and phosphoproteome data revealed a negligible role for Tor1 irrespective of Snf1 deletion, suggesting that TORC1 was mainly repressed at C-limited condition and this repression may be independent of Snf1. Based on these inferences, we propose that the Snf1 and TORC1 pathways only crosstalk via intermediate(s) under nutrient limitation. Such an intermediate would operate at the upstream of Snf1 and TORC1 and switches between the Snf1 and TORC1 activity (i.e., either Snf1 or TORC1, but not both, could be active under nutrient-limited conditions). One such intermediate could be the PKA/RAS pathway, which together with Snf1 can be perceived as a switch that senses the glucose concentration and regulates the cell metabolism accordingly (Zaman et al, 2009). The identification of significantly decreased phosphorylation of Bcy1, the regulatory subunit of PKA pathway, in the snf1Δ and snf1Δtor1Δ strains suggests a possible link between the Snf1 and PKA pathways (Supplementary Figure S5). PKA interacts with TORC1 in the regulation of protein translation and cell cycle (Martin et al, 2004; Wanke et al, 2005), and it may therefore bridge the Snf1 and TORC1 pathways. Since PKA and TORC1 are active in nutrient excess, while Snf1 is only fully active under glucose limitation or stress conditions, the media and growth conditions are essential for studying the regulatory pathways involved in nutrient sensing, because a shake flask cultivation using a rich medium (typically the YPD medium supplemented with 2% of glucose) is a completely different scenario from the chemostat culture fed with C- or N-limited medium.
Convergence of Snf1 and TORC1 onto amino-acid biosynthesis
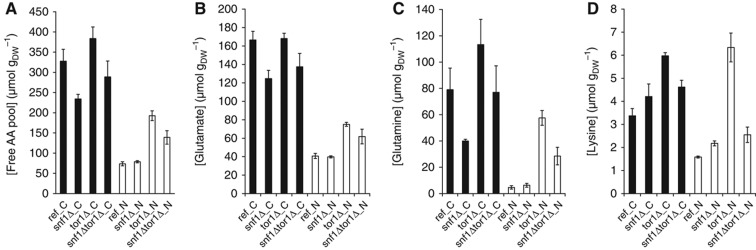
Neither the transcriptome nor the phosphoproteome data supported a direct link between Snf1 and TORC1 or the pathways they regulate. However, integration of these data using a metabolic model revealed extensive regulation around biosynthesis of amino acid and lipid (Supplementary Table S2). To investigate the regulation of amino-acid biosynthesis by Snf1 and TORC1, we quantified the intracellular level of 17 proteinogenic amino acids in all strains grown under both nutrient-limited conditions (Supplementary Table S3). Serine and arginine cannot be measured using the method applied and the cysteine level was below detection threshold. The free amino-acid pool under C-limited condition was 2- to 4.5-fold higher compared with that for N-limited condition for all the strains. During glucose limitation, the tor1Δ strain had about 17% higher level of free amino-acid pool, while the snf1Δ strain had a level about 29% lower compared with the reference strain (Figure 3A). During ammonium limitation, the strain snf1Δ had a similar level as the reference, while the tor1Δ and snf1Δtor1Δ strains had 170 and 93% higher total amino-acid level compared with the reference, respectively (Figure 3A). A substantial part of these differences was due to changes in glutamate and glutamine, which accounted for 60–75% of the total amino acid (Figure 3B and C).
Figure 3.
Intracellular levels of free amino acids. (A) Free amino-acid pool; (B) glutamate; (C) glutamine and (D) lysine. The error bars represent the s.e.m. from biological replicates from three chemostat cultures. Black—C-limited condition and white—N-limited condition.
To identify transcriptional regulation of amino-acid metabolism, we correlated the amino-acid levels with the expression of genes involved in their biosynthesis. Surprisingly, many genes responsible for the amino-acid biosynthesis were negatively correlated with the amino-acid levels (Figure 3; Supplementary Figure S6), indicating that the changes in the amino-acid abundance were not due to differential expression of corresponding amino-acid biosynthetic genes. We speculate that it may be due to that TORC1 senses a low level of glutamine (Figure 3B; Crespo et al, 2002) and consequently, the inhibition on Gcn4 is relieved (Valenzuela et al, 2001). Next, we studied the drain from the central carbon metabolism into the amino-acid biosynthesis. Since the expression of genes in TCA cycle is highly regulated by the nature and availability of carbon sources in an Snf1-dependent manner (Young et al, 2003), one may speculate if the lower level of Glx and other amino acids was due to the generally downregulated TCA cycle in the snf1Δ strain on C-limited and hence shortage of α-ketoglutarate, the direct precursor for amino acids of the glutamate family. However, a clear retrograde signaling response in the snf1Δ and snf1Δtor1Δ strains, reflected by both an induction of CIT2 (about 4-fold) and genes responsible for the early steps in the TCA cycle (CIT1, ACO1/ACO2, IDH1/IDH2) (Liu and Butow, 2006), confirmed that the amino-acid biosynthesis was not limited by the supply of α-ketoglutarate. Instead, a plausible explanation for the lower level of amino-acid pool in the snf1Δ and snf1Δtor1Δ strains would be that the expression of GDH3, which encodes one of the isoforms of glutamate dehydrogenase when cells are grown under derepressive conditions (DeLuna et al, 2001), was transcriptionally downregulated by >4-fold in the snf1Δ and snf1Δtor1Δ strains. The amino-acid biosynthesis was, therefore, likely limited by the inefficient conversion of α-ketoglutarate to glutamate, which is the main nitrogen source for cell growth. This phenomenon is similar to the one that was previously reported, where simply overexpression of GDH2 in the mutant strain gdh1Δ grown on batch culture (where the Gdh1 is the major isoform of glutamate dehydrogenase) changed the overall amino-acid pool significantly (Villas-Boas et al, 2005). On the other hand, the increased level for amino acids in tor1Δ could be attributed to an impaired balance between protein synthesis and degradation as a consequence of TOR1 deletion (Inoki et al, 2005).
We also found that amino-acid biosynthesis is regulated at the post-translational level. For example, homocitrate synthase (encoded by LYS20 and LYS21), which catalyzes the condensation of acetyl-CoA and α-ketoglutarate to form homocitrate, was found to be significantly more phosphorylated in the snf1Δ strain and to a lesser extent in the snf1Δtor1Δ strain. However, the phosphorylation of only LYS20 decreased in the tor1Δ strain (by ∼2-fold; Supplementary Figure S7). Since the intracellular lysine level significantly increased in all mutant strains (being highest in tor1Δ) at C-limited condition (Figure 3C), we could conclude that the homocitrate synthase isoenzymes (Lys20 and Lys21) are not only regulated through feedback inhibition by lysine, but could also be regulated through phosphorylation of these enzymes in an Snf1/TORC1-dependent manner. Collectively, we propose that Snf1 and TORC1 regulate the amino-acid biosynthesis via two independent mechanisms.
TORC1 may have a role in the regulation of FAs
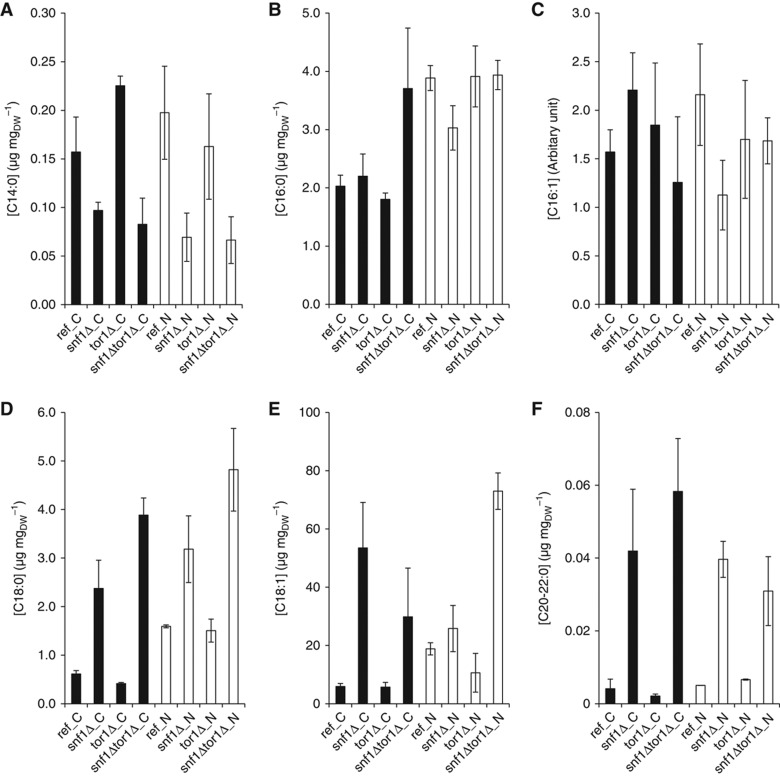
To unravel the role of Snf1 and TORC1 in the regulation of FA metabolism, we measured the relative abundance of FAs, including the free and ester form (e.g., in triacylglycerol), in the reference and mutant strains on both C- and N-limited conditions. Since Snf1 regulates FA biosynthesis by inhibiting acetyl-CoA carboxylase (Acc1) under derepressive conditions (Woods et al, 1994), the significant increase of total FA in the snf1Δ and snf1Δtor1Δ strains on C-limited condition was expectable (Figure 4). However, there was a significant variation in the FA species between different strains and the two growth conditions. The most abundant species was C18:1, where the largest differences between strains were observed (Figure 4E). The snf1Δ and snf1Δtor1Δ strains had higher levels of C18 (i.e., both C18:0 and C18:1) and longer FAs, on both C- and N-limited conditions, compared with the reference strain (Figure 4D–F), except for C14 where the result was contrary (Figure 4A). The snf1Δtor1Δ strain had higher amounts of C18 compared with the snf1Δ strain irrespective of the growth condition. The tor1Δ strain had higher C14 and C16 on N-limited condition, but the levels were lower on C-limited condition, compared with the reference strain.
Figure 4.
Abundance of fatty acids for all strains and two growth conditions. The abundance is based on the sum of FAs in the free as well as ester form. The error bars represent the s.e.m. from at least three replicates. (A) C14:0—myristic acid; (B) C16:0—palmitic acid; (C) C16:1—palmitoleic acid; (D) C18:0—stearic acid; (E) C18:1—oleic acid and (F) C20:0—arachidic acid and C22:0—behenic acid. Black—C-limited condition and white—N-limited condition.
However, this was only observed for C18 and longer FAs in these two strains (Figure 4D and E), while the abundance of C14 was reduced in the mutant strains in which SNF1 was deleted. There may be two mechanisms that can explain the different patterns between the FAs with different length. One possibility could be that while the inhibition of acetyl-CoA carboxylase by Snf1 was relieved, and the FA synthetase (encoded by FAS1 and FAS2) is constitutively functional and steadily converting short chain FAs to synthesize up to C16. Consistently, the elongase I (encoded by ELO1) that convert C12–16 to C18 was also found to be transcriptionally upregulated in snf1Δ and snf1Δtor1Δ; therefore, C16 was not accumulated in the strain snf1Δ and snf1Δtor1Δ (Figure 4B and C). It could also be that a lower peroxisome biogenesis due to the loss of Snf1 leads to a lower level of β-oxidation of the long chain FAs (Ratnakumar and Young, 2010), therefore not only the biosynthesis, but also the degradation of FAs is regulated by Snf1. We also observed that the deletion of TOR1 had some effect on the abundance of FAs, although to a lesser extent compared with those for SNF1 deletion (Figure 4D and E). The deletion of TOR1 in the snf1Δ background strengthened the changes caused by the deletion of SNF1 for C18:0, but rather dampened the changes for C18:1 (the most abundant FA species). The FA data support the hypothesis that Tor1 has a role in the regulation of FAs. However, TORC1 is unlikely involved in the regulation of acetyl-CoA carboxylase, and we suspect that the TORC1 may have a role in the regulation of peroxisome and β-oxidation of FAs. It is also interesting to notice that although deletion of TOR1 had not caused an evident change to the transcription and phosphorylation, many amino acids and FA species had changed significantly (Figures 3 and 4). This observation further supports the ideas that the intermediate metabolites are much more sensitive to mutations, while metabolic fluxes are rather robust (Cornish-Bowden and Cardenas, 2001; Raamsdonk et al, 2001).
Regulation of translation and cell growth
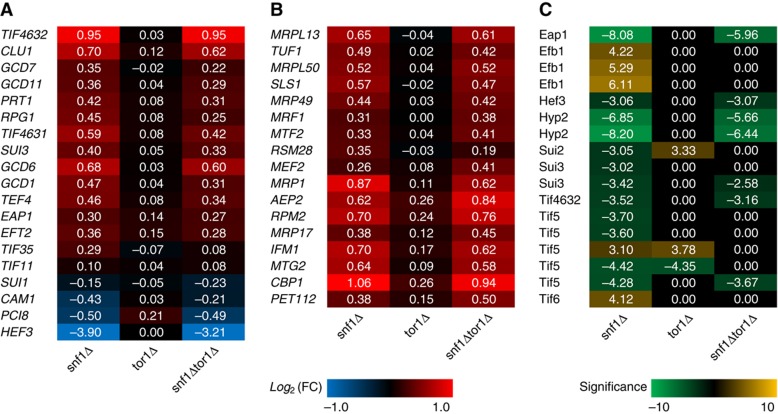
Since TORC1 promotes biosynthesis of ribosome and protein (Wullschleger et al, 2006), while Snf1 represses the energetically expensive processes such as biosynthesis of lipid and proteins (Hardie, 2007), we looked at the Snf1 and TORC1 regulation of protein translation, both at the transcriptome and at the phosphoproteome levels. Surprisingly, the deletion of SNF1 led to a significantly (despite <2-fold) increased expression of many genes involved in translation initiation or elongation, while the deletion of TOR1 alone did not cause any changes (Figure 5A). This held true even for the translation initiation or elongation factors (with only a few exceptions) that were found to be differentially phosphorylated in the mutant strains compared with the reference strain (Figure 5C). The observation that protein synthesis being primarily regulated by Snf1 instead of TORC1 seems to contradict the common knowledge that TORC1 is the main regulator for ribosomal translation (Inoki et al, 2005). However, taken the growth conditions (i.e., glucose limitation) into account it is actually consistent with the role of Snf1 as a global regulator of energy homeostasis and a repressor of anabolic processes (Hardie, 2007). The relative small changes in the snf1Δtor1Δ strain compared with the snf1Δ strain may advocate an inhibition of TORC1 under C-limited condition, as the TORC1 activity may require a high level of both ammonium and glucose (Figure 6). Interestingly, many genes involved in the mitochondrial ribosome and protein translation also had a similar pattern of expression where it was increased in the snf1Δ and snf1Δtor1Δ strains, and deletion of TOR1 had either no effect or similar effect with a lower magnitude (Figure 5B). Collectively, we conclude that the Snf1 has a major role in cell the mitochondrial proteome under C-limited condition.
Figure 5.
Regulation of genes and proteins involved in translation on C-limited condition. The red–blue heat map represents the relative changes of gene involved in (A) translation initiation and elongation and (B) mitochondrial ribosome and translation. (C) The yellow–green heat map represents the significance of phosphorylation changes of the proteins involved in translation. Positive numbers indicate higher while negative values indicate lower gene expression or protein phosphorylation in mutant strains compared with the reference on C-limited condition.
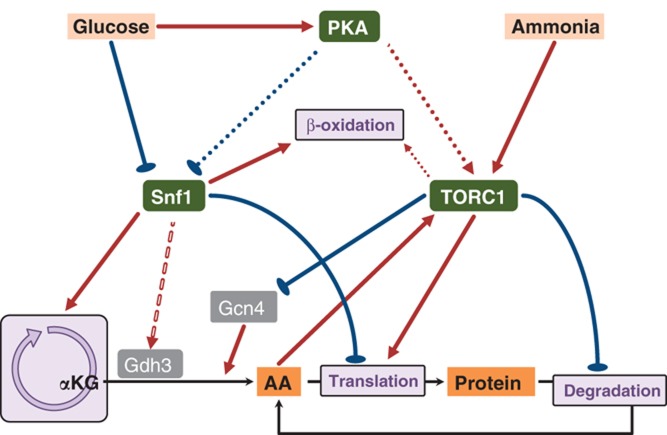
Figure 6.
Summary of the main regulatory network of Snf1 and TORC1.
Conclusion
Through integration of different omics data sets with metabolite profiles and strain physiology, we address the question of how Snf1 and TORC1, the two key regulators in the nutrient sensing pathways, coordinate metabolism with nutrient availability. The regulatory network is summarized in Figure 6. First, we showed that deletion of SNF1 caused bigger phenotypic changes compared with deletion of TOR1 grown on both nutrient-excess and -limited conditions and we demonstrate that it is likely due to that Snf1 kinase regulates a much broader range of biological processes such as global transcription, translation of protein, biogenesis of peroxisome and mitochondrion. The expression of NADP+-dependent glutamate dehydrogenase (Gdh3), which is upregulated under derepressing conditions (e.g., glucose limited), is regulated by Snf1, and the deletion of SNF1 likely results in an inefficient condensation of α-ketoglutarate and ammonium to form glutamate. Consequently, the synthesis of glutamine as well as the other amino acids is limited, resulting in a moderate induction of amino-acid biosynthetic genes through the TORC1/Gcn4 regulatory circuit (Figure 6). However, to elucidate the molecular mechanism by which Snf1 upregulates GDH3 gene requires extensive targeted studies such as the protein–protein/protein–DNA interaction assays. We also showed that besides Snf1, TORC1 may also have a role in the regulation of FAs, probably through modulating the peroxisome biogenesis and β-oxidation of FA, but via an unidentified mechanism than that of Snf1 pathway. Finally, we conclude that Snf1 and TORC1 do not seem to interact with each other directly under nutrient-limited conditions, although they have functional overlaps. We propose that TORC1 might be repressed by another regulator (or a signal molecule), which is activated (or raised) under nutrient-limited conditions, and this repression may not depend on the Snf1 activity. Furthermore, this unknown upstream regulator (or signal molecule) might also toggle switch between Snf1 and TORC1 activity to coordinate the cell growth and stress response under nutrient-rich and -limited conditions.
Materials and methods
Strains
The S. cerevisiae strains used in this study are the commonly used reference strain CEN.PK 113-7D (van Dijken et al, 2000) and its derivative strains (Supplementary Table S1). The tor1Δ strains (CEN.PK JZH-F1 and CEN.PK JZH-F2) were constructed by transforming the reference strains CEN.PK 113-7D and CEN.PK 113-1A (Matα) with a PCR amplified KanMX (from the strain BY4741) including ∼400 bp upstream and downstream of the TOR1 locus. The strain CEN.PK JZH-G1 snf1Δtor1Δ was constructed by crossing the strain CEN.PK 506-1C and CEN.PK JZH-F2, followed by dissection and screening as described previously (Zhang et al, 2010). The gene deletions were verified by PCR using primers outside the SNF1 and TOR1 loci and one primer inside the gene KanMX.
Chemostat cultivations
Chemostat cultures were grown at 30°C in 1.2 l bioreactors (DASGIP) with working volume of 0.5 l. The pH was controlled at 5.00±0.05 with 2 M KOH and the dissolved oxygen was kept above 30%. The dilution rate was adjusted to 0.10 h−1. For the C-limited cultures, one liter medium contained 10 g of glucose, 15 g of (NH4)2SO4, 3 g of KH2PO4, 1.5 g of MgSO4·7H2O, 1 ml of vitamin solution (Usaite et al, 2008), 1 ml of trace metal solution (Usaite et al, 2008), and 50 μl of Antiform 204 (Sigma-Aldrich, USA). For N-limited cultivation, the medium was the same as the one used in C-limited cultures except that the concentrations for (NH4)2SO4 and glucose were 1.0 and 60.0 g l−1, respectively. The CO2 emission (and residual O2) was monitored from the exhaust gas using the gas analyzer (DASGIP, Germany) and was used to determine the maximum specific growth rate during the batch growth phase. Samples for cell dry weight, extracellular and intracellular metabolites, transcriptome and proteome were taken from the cultures after steady state was achieved for about 50 h.
Transcriptome
The samples for transcriptome were taken as described previously (Zhang et al, 2010). The cells were mechanically disrupted using FastPrep homogenizer (MP Biomedicals) and total RNA was isolated using the RNeasy Mini Kit (QIAGEN). The quality of total RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) with RNA 6000 Nano LabChip kit (Agilent Technologies). The labeled RNA was synthesized using the GeneChip® 3′ IVT Express Kit (Affymetrix), which was then hybridized onto the GeneChip® Yeast Genome 2.0 Arrays (Affymetrix). Staining and washing of the hybridized arrays were carried out on the GeneChip® Fluidics Station 450 (Affymetrix) and scanned using the GeneChip® Scanner 3000 7G (Affymetrix). Affymetrix microarray data are available at GEO with the accession numbers GSE24421.
The transcriptome data were analyzed using Bioconductor in R. MAANOVA (MicroArray ANalysis Of VAriance) was performed to determine the genes whose expression level have significantly changed due to their genetic differences. PCA was applied to reduce the number of dimensions of the data set and simplify the data structure. Selected significant genes were clustered using a consensus clustering methods (Grotkjaer et al, 2006), and the GO terms for the genes in each cluster were analyzed using the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org/) to find the significant biological processes in each cluster (P<0.01). The Reporter Metabolite and Reporter Effector algorithms were applied to the transcriptome data to identify the ‘hot-spots’ in the metabolic or regulatory network, around which the significant changes have occurred (Patil and Nielsen, 2005; Oliveira et al, 2008).
Phosphoproteome
The samples collected from the chemostat cultures were rapidly quenched by adding trichloroacetic acid to a final concentration of 6.25%, incubated on ice for 10 min and spinned down by centrifuging (5000 r.p.m. at 4°C for 5 min). For each of the three replicates, 3 mg proteins were digested by trypsin (1:125 w/w) and cleaned by reverse phase chromatography. Phosphopeptides were enriched by titanium dioxide resin (1.25 mg GL Science resin for each sample) as previously described (Bodenmiller and Aebersold, 2010). The isolated phosphopeptides were analyzed by an LTQ-FT Ultra mass spectrometer (Thermo Scientific, Germany), interfaced with a nano-electrospray ion source. Chromatographic separation of peptides was performed on a Proxeon Easy-nLC II system (Odense, Denmark) using a 10.5 cm × 75 μm column packed with 3 μm Magic C18 material. Peptides were separated at a flow rate of 300 nl min−1 with a gradient increasing from 5 to 40% acetone. The five most intense ions detected in each MS1 scan were selected for fragmentation. The mass spectrometer data were searched against an SGD decoy database for yeast proteins using Sequest (Lundgren et al, 2009). OpenMS version 1.7 (Sturm et al, 2008) was used both to detect MS1 features and to align them between the different experimental conditions. By using a decoy database (Kall et al, 2008), a Peptide Prophet's probability threshold (0.9) was computed in order to achieve a false discovery rate <1%, and was used to filter OpenMS results. Phosphopeptides features with identical sequence and phosphorylation state but different charge were merged together. Only features which were detected at least twice in the three replicates were considered for statistical analysis by BAMarray version 3.0 (Ishwaran et al, 2006), which was used to compute the statistical significance of the regulated features. Two replicas for tor1Δ grown on N-limited condition were removed from statistical analysis due to their low data quality. The data can be downloaded from Tranche using the following link: https://proteomecommons.org/dataset.jsp?id=5JoVUbWQTC1tQWzvMlovAN8GJNgGqoWwZsdmcLwhgAjp4xJvlrJipf8V%2BbiCh2VjatUQaDbyCd%2F51j7%2B%2B5vI1EjfI9MAAAAAAAACUQ%3D%3D.
Free amino acids
The extraction of free amino acids was performed as described with modifications (Smits et al, 1998). First, 20 mg of freeze-dried cell pellets was suspended in 2.5 ml of cold methanol and 1 ml of chloroform, followed by addition of 4 ml of chloroform (−20°C) and 2 ml of Pipes-EDTA (3 mM each, pH 7.0). After shaking horizontally at 300 r.p.m. and −20°C for 45 min, the mixture was centrifuged at 3000 g and −10°C for 20 min, and the upper (aqueous) phase was collected. The free amino acids were concentrated and derivatized using the EZ:faast™ kit (Phenomenex) and quantified using GC-MS (Thermo Scientific) as described in the kit manual. The measurements are listed in Supplementary Table S3.
Fatty acids
The total FA was extracted and esterificated as described previously with modifications (Abdulkadir and Tsuchiya, 2008). First, about 15 mg of freeze-dried biomass was mixed with 5 μg of heptadecanoic acid (internal standard) in 625 μl of hexane and 250 μl of 14% BF3 in methanol. The head space of the tube was flushed with nitrogen gas to avoid oxidation and capped tightly before heated in a water bath (Grant OLS200, Cambridge, UK) at 100°C for 90 min with shaking at 70 r.p.m. After cooling to room temperature, 125 μl of hexane was added followed by addition of 250 μl distilled water. The tube was then shaken vigorously for 1 min and centrifuged for 3 min at 2500 r.p.m. (650 g). Finally, 750 μl of the upper phase, that is, hexane containing the FA methyl ester (FAME), was transferred into a gas chromatography-mass spectrometry (GC-MS) vial using a Pasteur pipette. The FAMEs were separated and quantified using Trace GC DSQII single quadrupole GC-MS (Thermo Scientific). Separation was performed with an Omegawax 250 (Supelco, Bellefonte, PA) column (30 m × 0.25 mm internal diameter, 0.25 μm film thickness). Helium was used as a carrier gas and the program was as follows. After the injection at 50°C, the oven temperature was raised to 180°C (20°C min−1), held for 1 min, raised to 210°C (3°C min−1), held for 5 min, raised to 215°C (1°C min−1), held for 3 min, raised to 221°C (1°C min−1), held for 5 min, raised to 230°C (3°C min−1), held for 5 min, raised to 250°C (3°C min−1), held for 2 min, and finally raised to 270°C (4°C min−1), held for 2 min. Mass transfer line and ion source were held at 250 and 200°C, respectively. FAME peaks were identified by searching their spectrum pattern against the NIST library. The FAME mixture (C14–22) standard (Sigma-Aldrich) and heptadecanoic acid (Sigma-Aldrich) serial diluted in hexane were injected in the same analysis to generate standard curves for the quantification. The measurements are listed in Supplementary Table S4.
Supplementary Material
Supplementary Figures S1–7, Supplementary Tables S1–4
Acknowledgments
This work was financially supported by the Chalmers Foundation, the Knut and Alice Wallenberg Foundation, the EU funded project UNICELLSYS and SystemsX.ch the Swiss Initiative in Systems Biology. We thank Dr Peter Kötter and Dr Matthias Rose (Frankfurt, Germany) for providing the strain BY4741.
Author contributions: JN conceived the study. RA and JN directed the research. JZ, SV, PC and RK performed the experiments. JZ, SV, PC, RK and GV analyzed the data. JZ wrote the manuscript. SV, PC, RK, GV, RA and JN edited and approved the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abdulkadir S, Tsuchiya M (2008) One-step method for quantitative and qualitative analysis of fatty acids in marine animal samples. J Exp Mar Biol Ecol 354: 1–8 [Google Scholar]
- Alepuz PM, Cunningham KW, Estruch F (1997) Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol Microbiol 26: 91–98 [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689. [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF (2002) Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol Cell Biol 22: 1246–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Aebersold R (2010) Quantitative analysis of protein phosphorylation on a system-wide scale by mass spectrometry-based proteomics. Methods Enzymol 470: 317–334 [DOI] [PubMed] [Google Scholar]
- Bolster DR, Crozier SJ, Kimball SR, Jefferson LS (2002) AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem 277: 23977–23980 [DOI] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ (2007) Exploring genetic interactions and networks with yeast. Nat Rev Genet 8: 437–449 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Carlson M (1984) Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol 4: 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TF, Carvalho J, Riles L, Zheng XF (2000) A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc Natl Acad Sci USA 97: 13227–13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomina N, Liu Y, Aldea M, Gari E (2003) TOR regulates the subcellular localization of Ime1, a transcriptional activator of meiotic development in budding yeast. Mol Cell Biol 23: 7415–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A, Cardenas ML (2001) Functional genomics. Silent genes given voice. Nature 409: 571–572 [DOI] [PubMed] [Google Scholar]
- Crespo JL, Powers T, Fowler B, Hall MN (2002) The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA 99: 6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daran-Lapujade P, Daran JM, van Maris AJ, de Winde JH, Pronk JT (2009) Chemostat-based micro-array analysis in baker′s yeast. Adv Microb Physiol 54: 257–311 [DOI] [PubMed] [Google Scholar]
- DeLuna A, Avendano A, Riego L, Gonzalez A (2001) NADP-glutamate dehydrogenase isoenzymes of Saccharomyces cerevisiae. Purification, kinetic properties, and physiological roles. J Biol Chem 276: 43775–43783 [DOI] [PubMed] [Google Scholar]
- Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G (2001) Mammalian TOR: a homeostatic ATP sensor. Science 294: 1102–1105 [DOI] [PubMed] [Google Scholar]
- Fazio A, Jewett MC, Daran-Lapujade P, Mustacchi R, Usaite R, Pronk JT, Workman CT, Nielsen J (2008) Transcription factor control of growth rate dependent genes in Saccharomyces cerevisiae: a three factor design. BMC Genomics 9: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotkjaer T, Winther O, Regenberg B, Nielsen J, Hansen LK (2006) Robust multi-scale clustering of large DNA microarray datasets with the consensus algorithm. Bioinformatics 22: 58–67 [DOI] [PubMed] [Google Scholar]
- Hardie DG (2007) AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8: 774–785 [DOI] [PubMed] [Google Scholar]
- Hong SP, Carlson M (2007) Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem 282: 16838–16845 [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Li Y, Guan KL (2005) Signaling by target of rapamycin proteins in cell growth control. Microbiol Mol Biol Rev 69: 79–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL (2003) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590 [DOI] [PubMed] [Google Scholar]
- Ishwaran H, Rao JS, Kogalur UB (2006) BAMarraytrade mark: Java software for Bayesian analysis of variance for microarray data. BMC Bioinformatics 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Kall L, Storey JD, MacCoss MJ, Noble WS (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res 7: 29–34 [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150: 1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin S, Treich I, Carlson M (2000) A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc Natl Acad Sci USA 97: 7916–7920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin S, Vyas VK, Carlson M (2002) Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol Cell Biol 22: 3994–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Manchester JK, Gordon JI (2003) Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J Biol Chem 278: 13390–13397 [DOI] [PubMed] [Google Scholar]
- Liu Y, Xu X, Kuo M-H (2010) Snf1p regulates Gcn5p transcriptional activity by antagonizing Spt3p. Genetics 184: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA (2006) Mitochondrial retrograde signaling. Annu Rev Genet 40: 159. [DOI] [PubMed] [Google Scholar]
- Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL (2001) Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293: 1142–1146 [DOI] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Lundgren DH, Martinez H, Wright ME, Han DK (2009) Protein identification using Sorcerer 2 and SEQUEST. Curr Protoc Bioinformatics Chapter 13: Unit 13.3 [DOI] [PubMed] [Google Scholar]
- Marion RM, Regev A, Segal E, Barash Y, Koller D (2004) Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA 101: 14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN (2004) TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969. [DOI] [PubMed] [Google Scholar]
- Medvedik O, Lamming DW, Kim KD, Sinclair DA (2007) MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol 5: e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro G, Netto LE (2004) Glucose repression of PRX1 expression is mediated by Tor1p and Ras2p through inhibition of Msn2/4p in Saccharomyces cerevisiae. FEMS Microbiol Lett 241: 221–228 [DOI] [PubMed] [Google Scholar]
- Oliveira AP, Patil KR, Nielsen J (2008) Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks. BMC Syst Biol 2: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M, Kanter E, Krakovich D, Kuchin S (2006) Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell 5: 1831–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil KR, Nielsen J (2005) Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA 102: 2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova MI, Pujol-Carrion N, Arroyo J, Garcia-Cantalejo J, Angeles de la Torre-Ruiz M (2010) Mtl1 is required to activate general stress response through Tor1 and Ras2 inhibition under conditions of glucose starvation and oxidative stress. J Biol Chem 285: 19521–19531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranovic D, Nielsen J (2008) Can yeast systems biology contribute to the understanding of human disease? Trends Biotechnol 26: 584–590 [DOI] [PubMed] [Google Scholar]
- Polge C, Thomas M (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–28 [DOI] [PubMed] [Google Scholar]
- Portillo F, Mulet JM, Serrano R (2005) A role for the non-phosphorylated form of yeast Snf1: tolerance to toxic cations and activation of potassium transport. FEBS Lett 579: 512–516 [DOI] [PubMed] [Google Scholar]
- Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, Westerhoff HV, van Dam K, Oliver SG (2001) A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol 19: 45–50 [DOI] [PubMed] [Google Scholar]
- Ratnakumar S, Young ET (2010) Snf1 dependence of peroxisomal gene expression is mediated by Adr1. J Biol Chem 285: 10703–10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A, Anderson S, McCaffery JM, Yates J 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T (2004) TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem 279: 14752–14762 [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN (2000) TOR, a central controller of cell growth. Cell 103: 253–262 [DOI] [PubMed] [Google Scholar]
- Shirra MK, McCartney RR, Zhang C, Shokat KM, Schmidt MC, Arndt KM (2008) A chemical genomics study identifies Snf1 as a repressor of GCN4 translation. J Biol Chem 283: 35889–35898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smets B, Ghillebert R, De Snijder P, Binda M, Swinnen E, De Virgilio C, Winderickx J (2010) Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr Genet 56: 1–32 [DOI] [PubMed] [Google Scholar]
- Smith RL, Johnson AD (2000) Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci 25: 325–330 [DOI] [PubMed] [Google Scholar]
- Smits HP, Cohen A, Buttler T, Nielsen J, Olsson L (1998) Cleanup and analysis of sugar phosphates in biological extracts by using solid-phase extraction and anion-exchange chromatography with pulsed amperometric detection. Anal Biochem 261: 36–42 [DOI] [PubMed] [Google Scholar]
- Soontorngun N, Larochelle M, Drouin S, Robert F, Turcotte B (2007) Regulation of gluconeogenesis in Saccharomyces cerevisiae is mediated by activator and repressor functions of Rds2. Mol Cell Biol 27: 7895–7905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm M, Bertsch A, Gropl C, Hildebrandt A, Hussong R, Lange E, Pfeifer N, Schulz-Trieglaff O, Zerck A, Reinert K, Kohlbacher O (2008) OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics 9: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD (2006) Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Mol Cell 23: 207–217 [DOI] [PubMed] [Google Scholar]
- Usaite R, Jewett MC, Oliveira AP, Yates JR 3rd, Olsson L, Nielsen J (2009) Reconstruction of the yeast Snf1 kinase regulatory network reveals its role as a global energy regulator. Mol Syst Biol 5: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usaite R, Nielsen J, Olsson L (2008) Physiological characterization of glucose repression in the strains with SNF1 and SNF4 genes deleted. J Biotechnol 133: 73–81 [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Aranda C, Gonzalez A (2001) TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J Bacteriol 183: 2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg MA, de Jong-Gubbels P, Kortland CJ, van Dijken JP, Pronk JT, Steensma HY (1996) The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem 271: 28953–28959 [DOI] [PubMed] [Google Scholar]
- van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin ML, Heijnen JJ, Hoare M, Lange HC, Madden EA, Niederberger P, Nielsen J, Parrou JL, Petit T, Porro D, Reuss M, van Riel N, Rizzi M, Steensma HY et al. (2000) An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol 26: 706–714 [DOI] [PubMed] [Google Scholar]
- Villas-Boas SG, Moxley JF, Akesson M, Stephanopoulos G, Nielsen J (2005) High-throughput metabolic state analysis: the missing link in integrated functional genomics of yeasts. Biochem J 388: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, De Virgilio C (2005) Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J 24: 4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D (1994) Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem 269: 19509–19515 [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Oppliger W, Hall MN (2005) Molecular organization of target of rapamycin complex 2. J Biol Chem 280: 30697–30704 [DOI] [PubMed] [Google Scholar]
- Xie MW, Jin F, Hwang H, Hwang S, Anand V, Duncan MC, Huang J (2005) Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method. Proc Natl Acad Sci USA 102: 7215–7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Elbing K, Hohmann S (2008) The pathway by which the yeast protein kinase Snf1p controls acquisition of sodium tolerance is different from that mediating glucose regulation. Microbiology 154: 2814–2826 [DOI] [PubMed] [Google Scholar]
- Young ET, Dombek KM, Tachibana C, Ideker T (2003) Multiple pathways are coregulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem 278: 26146. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR (2009) Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol 5: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR (2008) How Saccharomyces responds to nutrients. Annu Rev Genet 42: 27–81 [DOI] [PubMed] [Google Scholar]
- Zhang J, Olsson L, Nielsen J (2010) The beta-subunits of the Snf1 kinase in Saccharomyces cerevisiae, Gal83 and Sip2, but not Sip1, are redundant in glucose derepression and regulation of sterol biosynthesis. Mol Microbiol 77: 371–383 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–7, Supplementary Tables S1–4