Abstract
We manufactured a novel type of lipid-coated superparamagnetic nanoparticles that allow for a rapid isolation of plasma membranes (PMs), enabling high-resolution proteomic, glycomic and lipidomic analyses of the cell surface. We used this technology to characterize the effects of presenilin knockout on the PM composition of mouse embryonic fibroblasts. We found that many proteins are selectively downregulated at the cell surface of presenilin knockout cells concomitant with lowered surface levels of cholesterol and certain sphingomyelin species, indicating defects in specific endosomal transport routes to and/or from the cell surface. Snapshots of N-glycoproteomics and cell surface glycan profiling further underscored the power and versatility of this novel methodology. Since PM proteins provide many pathologically relevant biomarkers representing two-thirds of the currently used drug targets, this novel technology has great potential for biomedical and pharmaceutical applications.
Keywords: eukaryotic cell systems; glycomics; lipidomics; presenilin, proteomics
Introduction
Contemporary cell biology is increasingly challenged toward the global and comparative analysis of the molecular phenotype of the cell under investigation. Whereas powerful techniques exist for the quantification of genome-wide changes in gene expression, these approaches provide only indirect insights in the global protein, lipid and sugar composition of the cell. Global biochemical compositional analysis remains therefore challenging. Despite major technological improvements, mass spectrometry (MS) analysis of total tissue or cell extracts is biased towards the identification of the more abundant proteins (Wisniewski et al, 2009). This problem can be partially circumvented by isolating individual subcellular compartments, which allows to identify also the more rare proteins, resulting in a better understanding of their functional roles in their specific micro-environment (Huber et al, 2003; Foster et al, 2006). The most reliable results have been obtained with organelles for which good purification protocols are available, including cilia, exosomes, mitochondria and synaptic vesicles (Pisitkun et al, 2004; Forner et al, 2006; Ghesquière et al, 2006; Takamori et al, 2006; Mayer et al, 2008; Deery et al, 2009). A major challenging compartment is the plasma membrane (PM), and by extension, its dynamic interplay with the endocytic/recycling system. These compartments are highly dynamic in nature with overlapping physical features such as buoyant densities, rendering it far more difficult to isolate them to the quality required for ‘organellar omics.’ Being the physical boundary of the cell, the PM acts as an interface between the cell and its environment for inter- and extracellular communication, intracellular transport, stress responses, cell–pathogen interaction and downstream signaling (Tan et al, 2008). Furthermore, these processes can induce rapid and selective changes in the overall composition through internalization and endocytic recycling, adding significantly to the dynamic nature of the PM. Likewise, the membrane composition can be affected in many diseases, giving rise to the expression of proteins that can act as markers in, for example, specific cancers and inflammatory diseases (Josic and Clifton, 2007). Therefore, resolving the structural complexity of the PM is an important challenge.
In this study, we developed a procedure to isolate PMs under physiological conditions based on novel phospholipids-coated superparamagnetic nanoparticles (SPMNPs; Ø 10–15 nm). This technology does not involve affinity tags (biotin, ligands or antibodies) nor invasive approaches like silica coating, is compatible with MS-based proteomic and lipidomic analysis and can be used for the analysis of post-translational modifications (e.g. N-glycoproteomics, N-glycan profiling) of cell surface proteins.
We used this novel technology here to begin to address a long-standing question in the field of Alzheimer's disease (AD), that is to what extent the presenilins (PSENs), apart from their function as catalytic subunits of the γ-secretase complex (De Strooper and Annaert, 2010), are also involved in membrane or protein trafficking events. Genetic knockout of PSEN does indeed not only block the proteolytic processing of APP (De Strooper et al, 1998), Notch (De Strooper et al, 1999), cadherins (Marambaud et al, 2002) and many other substrates, but additionally causes alterations in the subcellular localization and turnover of proteins like caveolin-1 (Wood et al, 2005), telencephalin (Esselens et al, 2004) and β1-integrin (Zou et al, 2008). PSEN-deficient cells have also increased levels of cholesterol and sphingomyelin (SM) (Grimm et al, 2005) and present with defects in lysosomal fusion (Esselens et al, 2004). This raises the question whether this diverse array of alterations can be traced to selective defects in the transport of proteins or membranes to and from the cell surface. As a first step, we analyze here the effect of PSEN deficiency on the global biomolecular composition of the PM of mouse embryonic fibroblasts (MEFs). We indeed identify qualitative and quantitative changes of the cell surface proteome and lipid composition, likely resulting from transport defects. We analyzed also the cell surface N-glycosylation profile, which appeared less affected. Overall, this novel SPMNP-based isolation procedure constitutes a timely and much sought-after tool for subsequent generation of protein and lipid inventories (and their modifications) of PMs of potentially different cell types.
Results
Particle synthesis and magnetic isolation
SPMNPs are synthesized by thermal decomposition resulting in oleic acid coated nanoparticles of 9.4±3.0 nm size with superparamagnetic properties (Supplementary Figure S1a and b, left panel). Subsequent coating with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE)-PEG-NH2 increases their hydrodynamic diameter to 16.3±5.0 nm (Supplementary Figure S1a and b, right panel), without compromising the superparamagnetic properties (Supplementary Figure S1c). Even after several weeks, these particles remained dispersed in the water phase in a water-organic solvent mixture (Supplementary Figure S1a′). To characterize their behavior in cell culture, we used fluorescently labeled variants in which carboxyfluorescein and the NH2-functionalized PEG-linked phospholipid conjugates were mixed in 1:4 ratios during ligand addition. When briefly applied to cells at 4 °C, these SPMNPs adhered strongly to the cell surface with no detectable internalization as judged from confocal and transmission EM (TEM) analysis (Figure 1B (confocal) and Figure 1C (TEM)). Zeta-potential measurements confirmed the positive charge of SPMNPs, suggesting that they have strong electrostatic interactions with the negatively charged PMs (Supplementary Figure S1d).
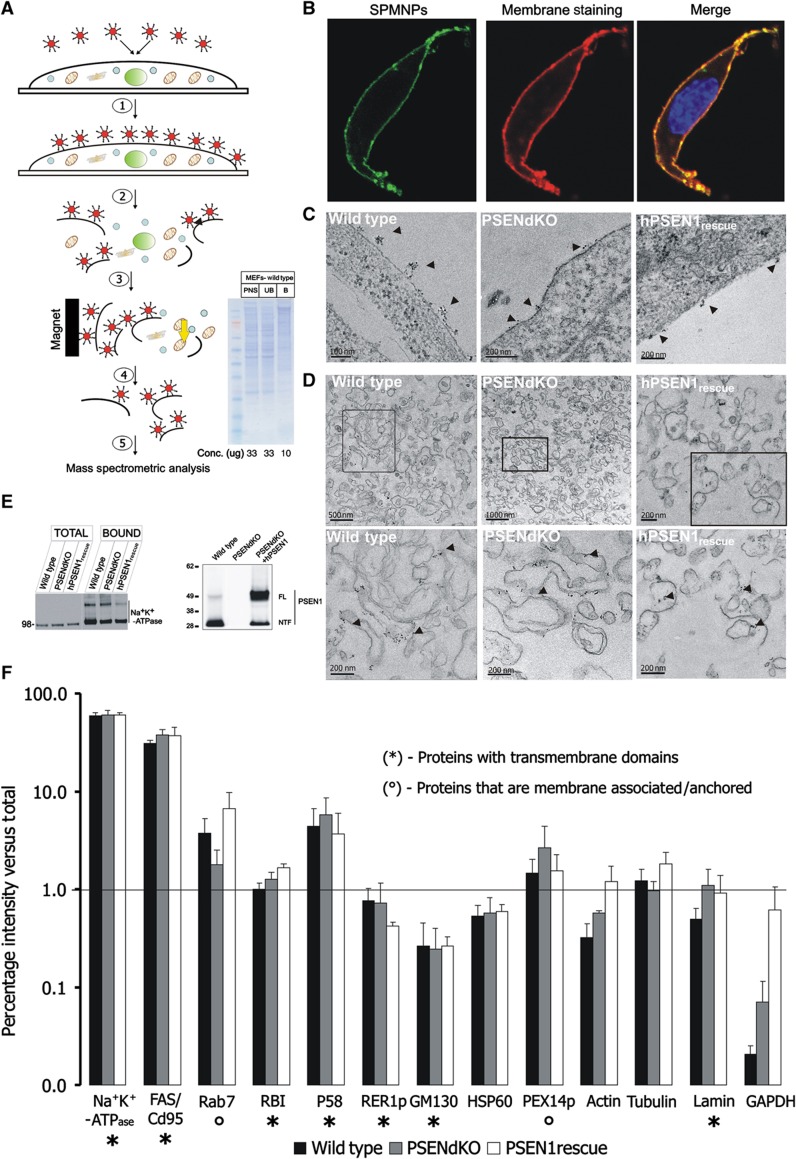
Figure 1.
Schematic diagram and quality control of the PM isolation procedure using SPMNPs. (A) Stepwise outline of the SPMNP-based affinity purification of PMs: (1) SPMNPs are incubated with cell monolayers at 4 °C for 15 min; (2) after washing, cells are harvested, homogenized and centrifuged (200g, 10 min) to yield a PNS, (3) PMs are magnetically retained on an LS column, (4) followed by elution and concentration by ultracentrifugation and finally (5) protein/lipid are extracted for subsequent analysis. (Inset) Coommassie Brilliant Blue staining of PNS, unbound (UB) and bound (B) fractions obtained from WT MEFs (first lane: SeeBlue plus2 rainbow protein marker (Invitrogen)). The distinct protein profile in the bound fraction underscores the high enrichment for PMs. (B) Confocal laser scanning microscopy images of WT MEFs incubated with fluorescently modified NH2-lipid end-group SPMNPs and Alexafluor-647 conjugated cholera toxin subunit B (CTB) (15 min at 4 °C). (C) TEM of WT, PSENdKO and hPSEN1rescue MEFs showing SPMNPs adhering to the cell surface (arrowheads). (D) TEM of an isolated PM fraction of WT, PSENdKO and hPSEN1rescue MEFs showing uniform long PM sheets decorated with SPMNPs, that were virtually devoid of other subcellular organelles. (E) (Left) Representative western blot analysis showing the strong enrichment for the PM marker Na+K+-ATPase between total cell extracts (TOTAL) and isolated PMs (BOUND) for all three investigated cell lines. (Right) Western blot analysis demonstrating endogenous PSEN1 in WT and exogenous human PSEN1 in PSENdKO-rescued MEFs. (F) Quantitative western blot analysis of the indicated organelle marker proteins (x axis) in isolated PM fractions as a percentage of the total cell lysate (y axis, presented in logarithmic scale) for WT, PSENdKO and hPSEN1rescue MEFs (mean±s.e.m., n=3). Na+K+-ATPase and FAS/CD95 were used as PM-localized integral membrane proteins. Overall, very-to-extreme low levels of contaminations were detected for all subcellular compartments as judged from the many marker proteins. RBI represents ER while the membrane proteins p58 and RER1p reside in intermediate to cis-Golgi compartments. GM130 is a Golgi-associated protein while lamin, HSP60 and the membrane-associated PEX14p denote nuclear envelope, mitochondria and peroxisomes respectively. Rab7 represents late endosomes. Cytosolic and cytoskeletal marker proteins are GAPDH and actin/tubulin. Proteins were resolved in a precast 4–12% SDS–PAGE gel. (*) Represents integral membrane proteins and (°) represents membrane-associated/-anchored proteins. Line denotes 1% contamination level on the logarithmic y axis.
We exploited this feature to magnetically isolate PMs (Figure 1A). Following a brief incubation (4 °C, 20 min) (1), SPMNP-coated cells are homogenized using a cell cracker (2). The total homogenate is centrifuged (200 g, 10 min) to remove major contaminating membranes like cell debris and nuclei. The resulting post-nuclear supernatant (PNS) is run over an LS column placed within a strong magnetic field (0.5 T) to retain SPMNP-coated PMs and to separate them ‘on line’ from remaining non-coated contaminating membranes (3). After extensive washing, PMs are eluted by withdrawal of the magnet (4), concentrated by ultracentrifugation and processed for analysis of proteins, lipids and glycoproteins (5). The whole procedure, from applying SPMNPs to the isolated PM fraction can be accomplished in ∼2 h. This short isolation protocol significantly reduces the time that free or uncoated parts of SPMNPs may non-specifically adhere to membranes thereby minimizing potential contaminations and contributing to a higher efficacy to obtain pure PM fractions (see below).
We next applied this approach to isolate PMs from cultured wild-type (WT) and PSENdKO MEFs as well as from PSENdKO MEFs stably rescued with human PSEN1 (hPSEN1rescue) (Figure 1E). For all cell lines, western blot analysis of magnetically isolated PMs showed very low levels of major contaminating organelles compared with total cell lysates. On average and for all three cell lines, recoveries for marker proteins of the endoplasmic reticulum (Ribophorin, RBI), intermediate compartment (p58), cytoskeleton (tubulin) and peroxisomes (PEX14p) were around or below 4%, while for mitochondria (HSP60), nuclear envelope (lamin A/C), cytosol (GAPDH, actin) and intermediate compartment/cis-Golgi (RER1p, GM130) even below 1% (Figure 1F, mean±s.e.m., n=3). On the contrary, two bona fide integral membrane proteins of the PM, FAS and Na+K+-ATPase, were recovered up to 40 and 60% compared with total cell lysates in agreement with SPMNPs associating essentially with the PM facing the culture medium and indicating a very high and identical degree of recovery of the PM from all three cell lines. This is further corroborated by the >20-fold enrichment of the Na+K+-ATPase in PM isolates obtained from the different cell lines as well as by the distinct protein composition as revealed by SDS–PAGE followed by Coommassie blue staining of WT PMs (Figure 1A, inset). We systematically recovered higher levels (between 4 and 7% in the different cell lines) of the late endosomal Rab7 GTPase. However, when we performed in WT MEFs cell surface biotinylation followed by internalization (20 min, 37 °C) and removal of remaining cell surface-bound biotin (using sodium 2-mercaptoethanesulfonate to reduce the biotin-S-S-NHS), subsequent SPMNP isolation of PMs recovered only 1.23±0.90% (n=3) of biotin. This experiment indicates that the contamination from endosomal compartments is extremely low (Supplementary Figure S2a and b) and suggests that the Rab7 ‘contamination’ in our isolation procedure may originate from Rab7 being transiently associated with the cell surface, as previously documented, for example, in the case of cell injury (Andrews, 2002) or in exosomal secretion (Simpson et al, 2008). To estimate the purity, we next measured the percentage of total immunoreactivity of the PM marker Na+K+-ATPase relative to the sum of the immunoreactivities of all compartments analyzed (each represented by a single organelle-specific marker protein, that is, Na+K+-ATPase-PM, RBI-roughER, RER1p-intermediate compartment, GM130-Golgi-associated protein, internalized biotin endosomes; lamin-nucleus, HSP60-mitochondria, PEX14p-peroxisomes; GAPDH-cytosol, actin and tubulin-cytoskeletal; see also Figure 1F). This resulted in purities of 92.2±1.4% (WT), 88.7±4.2% (PSENdKO) and 90.0±3.6% (hPSEN1rescue) (mean±s.e.m., n=3) and allows us to conclude that with our new method, purities can be obtained in the range of 85–95%. This is finally supported by TEM of the PM isolates for all three cell lines confirming the presence of large membranous sheets covered with SPMNPs and the apparent lack of other subcellular compartments (Figure 1D).
As our approach obviates the need of detergents and the fact that we isolate intact PM sheets, we retain full active protein complexes in their in situ lipid environment. γ-Secretase consists of PSEN complexed with nicastrin, APH-1 and PEN-2 and liberates through intramembrane proteolysis amyloid β peptides from APP C-terminal fragments (APP-C99) (De Strooper and Annaert, 2010). Quantification of γ-secretase activity in PNS and PM fractions resulted in an 8.0±2.6-fold (n=3) fold enrichment of surface-associated activity (Supplementary Figure S2c).
Qualitative and quantitative PM proteomics
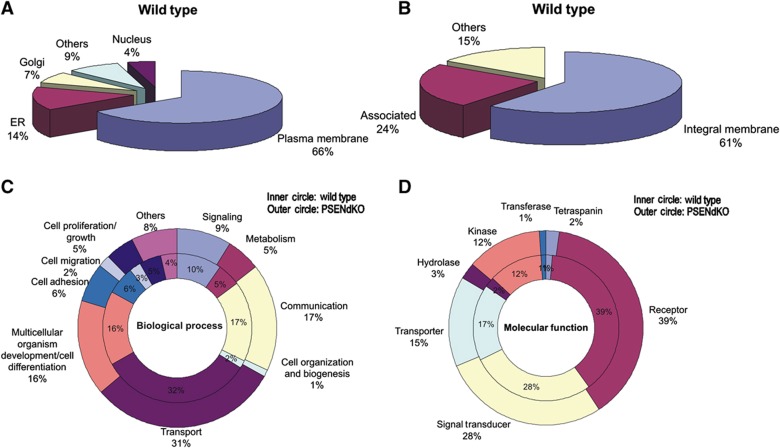
We obtained high quality and quantity of purified PMs from MEFs (±80–140 μg protein out of eight 10 cm dishes) that allows MS-based proteomics. Following trypsin digestion of PM isolates, extracted peptides were identified by LC-MS/MS. A single analysis gave rise to over 10 000 peptides identifying over 1000 membrane proteins (using Gene Ontology) in each cell type (n=3) (Figure 2A and Pride Database Project No: 15826). In WT MEFS, 66% of the identified proteins are authentic PM proteins, with the remaining assigned to the ER (14%), Golgi (7%), nucleus (4%) or ‘not known’ (9%). These numbers demonstrate the exceptionally high enrichment for cell surface proteins in one single purification step. The estimate of 66% is likely an underestimation of the technique as many of the proteins assigned to non-PM compartments are also partially present at the cell surface, either in transition or because wrongly allocated to only one compartment. Examples of ER-annotated proteins are nicastrin, PSEN1, caveolin-2, stromal interaction molecule 1 and endophilin, all of which have been in other studies located at the cell surface or in functional complexes with cell surface proteins (Mora et al, 1999; Herreman et al, 2003; Chyung et al, 2005; Cheung and Ip, 2009; Carrasco and Meyer, 2010).
Figure 2.
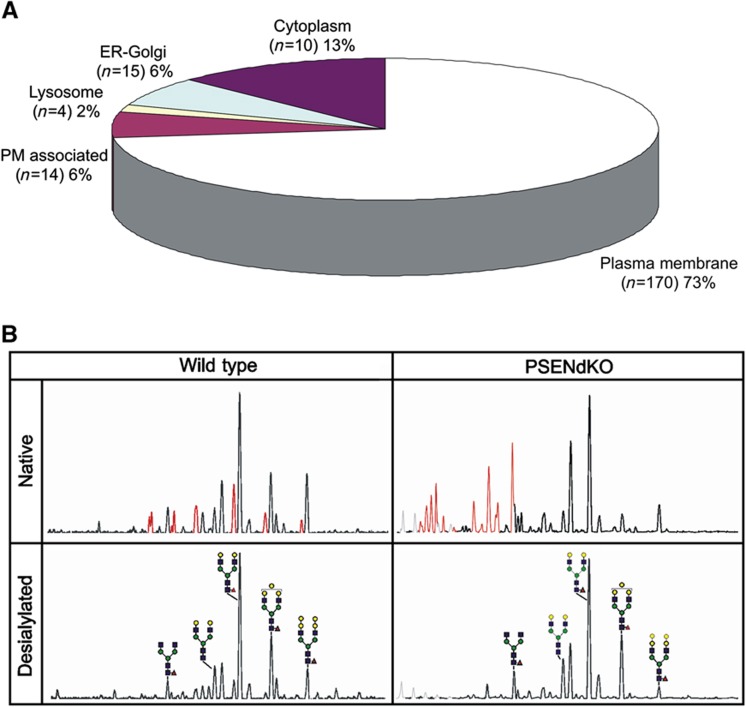
Qualitative MS/MS-based PM proteomics. (A) Pie chart showing that almost 70% of the proteins identified in the isolated PM fraction of WT MEFs (of a total of ∼1000 identified in one magnetic isolation procedure) are expected to be PM proteins, based on Gene Ontology annotations. About 14% are annotated to the ER, 7% to Golgi and 4% nuclear. (B) Subclassification of PM proteins with 61% being integral membrane proteins and 24% associated. (C, D) Comparative classification of PM proteomes of WT (inner circle) and PSENdKO (outer circle) MEFs does not show gross alterations in the biological processes and molecular function profiles, respectively.
About two-third of the PM proteins are integral (Figure 2B), 60% of which are multitransmembrane proteins, consistent with literature (Durr et al, 2004; Zhao et al, 2004). Of the remaining proteins, 24% are membrane-associated (inner peripheral/cytoskeletal-associated) and 15% are not characterized. Related to γ-secretase, we did not identify Aph-1 and PEN-2 likely because they have minimal hydrophilic domains, thereby generating insufficient numbers of MS-detectable tryptic peptides. No nicastrin was identified in PSENdKO PMs in agreement with its failure to mature and traffic beyond Golgi in the absence of PSENs (Leem et al, 2002). Lastly, only subtle differences between cell lines where found when membrane proteins were categorized based on biological processes or molecular function (Figure 2C and D).
In a second series of experiments, we compared quantitatively the membrane composition of PSEN-deficient cells versus WT. As a control, we stably expressed human PSEN1 (hPSEN1rescue) in PSEN knockout cells, to keep the cellular genetic background completely identical.
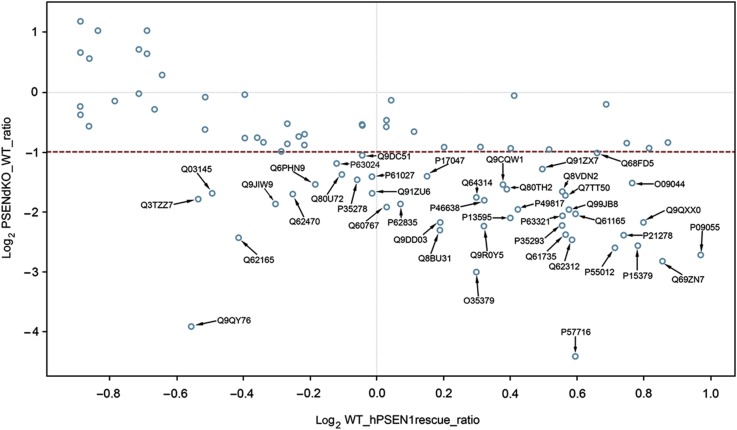
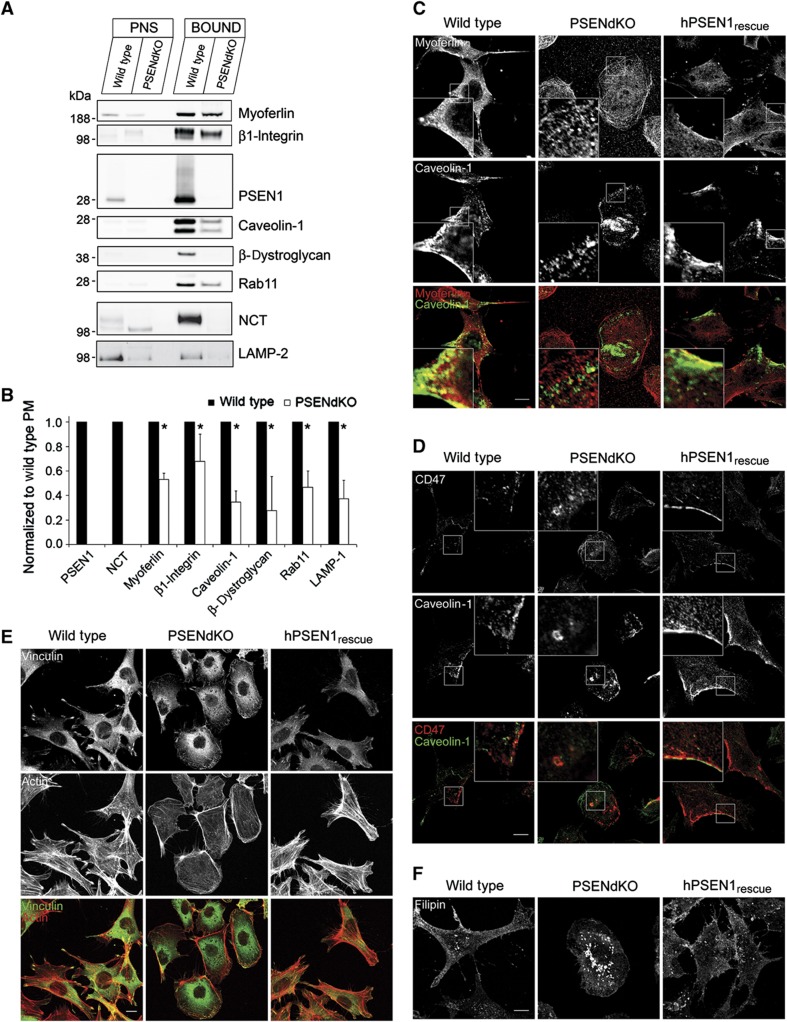
For these experiments, we post-metabolically labeled endoproteinase Lys-C-digested PMs using either light 12C3 or heavy 13C3-N-propionylation prior to LC-MS/MS analysis (Ghesquière et al, 2009, 2011). In each analysis, the ratios of the abundances of light labeled WT peptides over that of heavy labeled peptides (PSENdKO or hPSEN1rescue) were quantified (Supplementary Figure S3a and b and Pride Database Project No: 15828). The broad ratio distribution when comparing WT versus PSENdKO PMs indicates that a considerable number of proteins are differentially expressed at the cell surface between both cell lines. The unimodal ratio distribution between WT and hPSEN1rescue PMs (Supplementary Figure S3b) underscores that the changes in surface protein expression detected by these experiments are indeed caused by PSEN deficiency. We further selected the PM proteins that were not different in WT and hPSEN1rescue MEFs but were changed in the PSENdKO analysis (resp. x axis and y axis in Figure 3). This scatter plot of 61 shared proteins reveals a group of 20 unregulated (open circles) and 41 downregulated proteins (filled circles) in PSENdKO PMs. Among these we find, besides nicastrin, many proteins linked to migration/adhesion, integrin signaling as well as proteins associated with cholesterol-rich domains such as lipid rafts (e.g. CD44, CD47) and caveolae (e.g. caveolin-1, dystroglycan, myoferlin) (Table I). Furthermore, we discerned categories identifying proteins involved in PM fusion (e.g. SNAREs) and endosomal redistribution, notably several Rab GTPases. Quantitative western blotting of several of these proteins confirmed that they were significantly downregulated in PSENdKO PMs (Figure 4A and B; mean±s.e.m. (n=3), *P>0.05). Immunofluorescence experiments confirmed also alterations emerging from the proteomic analysis. For instance, while caveolin-1 localizes to the PM region in the trailing edge of migrating WT and hPSEN1rescue MEFs, this was not apparent in PSENdKO MEFs (Figure 4C and D). Other proteins, shown to be altered in PSENdKO cells like myoferlin, CD47 and β1-integrin, were similarly shown to be less expressed at the surface of PSENdKO and/or colocalized intracellularly with caveolin-1 in PSENdKO only. Finally, the lower surface expression of proteins like β1-integrin and caveolin-1 suggests alterations in cellular adhesion. In agreement with this, immunostaining for actin/vinculin confirmed that PSENdKO MEFs exhibited a more round morphology and a clearly altered focal adhesion pattern (Figure 4E) as observed previously (Waschbüsch et al, 2009).
Figure 3.
Quantitative proteomics using shotgun approach on WT, PSENdKO and PSEN1rescue MEFs. Scatter plot of the PM protein in Log2 ratio's of PSENdKO versus WT (y axis) and WT versus PSEN1rescue (x axis) generated using Tableau software. For the x axis, only ratio's in the interval (−1, 1) between WT versus PSEN1rescue are taken (hence not significantly regulated between WT and PSEN1rescue PM). Analysis of the Log2 ratio's of these proteins in PSENdKO versus WT (y axis) identified 41 proteins that were significantly higher expressed in WT PMs (or downregulated in PSENdKO PMs) (ratio>2, marked blue circles with accession number as opposed to unregulated proteins (unmarked blue circles)). Identified proteins are listed in Table I along with their known or proposed functions.
Table 1. List of 41 differentially regulated proteins between WT/PSENdKO and WT/PSEN1rescue along with their known or proposed functions.
| Annotation | Description | WT/rescue ratio | WT/dKO ratio | Function/location | |
|---|---|---|---|---|---|
| Proteins are grouped according to their functions in cell adhesion/migration (A), association with caveolae (B), or functionally involved in transport regulation from endosomal recycling compartments to the surface, such as SNAREs (C), small GTPases (D), signaling molecules (E), and transporters (F). | |||||
| Q64314 | CD34 (hematopoietic progenitor cell antigen CD34) | 1.23 | 3.38 | Cell adhesion | |
| P15379 | CD44 | 1.72 | 5.91 | Cell adhesion/integrin signaling/raft associated | |
| Q61735 | CD47 | 1.48 | 5.2 | Cell adhesion/integrin signaling/raft associated | |
| Q62470 | ITA3 (integrin α-3) | 0.84 | 3.25 | Cell adhesion | |
| P09055 | Integrin β | 1.96 | 6.59 | Cell adhesion/negative regulation of cell migration | A |
| Q91ZU6 | BPA1 (bullous pemphigoid antigen 1) | 0.99 | 3.23 | Cell adhesion/calcium ion binding | |
| P13595 | NCAM1 (neural cell adhesion molecule 1) | 1.32 | 4.28 | Cell adhesion/GPI | |
| Q80TH2 | LAP2 | 1.31 | 3.06 | Cell junction | |
| Q7TT50 | MRCKB (serine/threonine-protein kinase MRCK β) | 1.48 | 3.31 | Cell junction | |
| P49817 | Caveolin-1 | 1.34 | 3.88 | Caveolae associated | |
| Q03145 | EPHA2 (ephrin type-A receptor 2) | 0.71 | 3.23 | Caveolae associated | B |
| Q62165 | Dystroglycan | 0.75 | 5.38 | Caveolae associated/calcium release | |
| Q69ZN7 | Myoferlin | 1.81 | 7.07 | Caveolae/lipid raft associated | |
| Q91ZX7 | LRP1 | 1.41 | 2.44 | Endocytosis/cholesterol homeostasis | |
| Q60767 | Lymphocyte antigen 75 | 1.02 | 3.77 | Endocytosis | |
| Q99JB8 | PACN3 (protein kinase C and casein kinase II protein 3) | 1.49 | 3.9 | Endocytosis | |
| P63024 | VAMP3 | 0.92 | 2.29 | Endosomal SNARE/required for integrin transport | |
| Q9QY76 | VAPB (vesicle-associated membrane protein-B) | 0.68 | 15.09 | Ligand for Eph receptor | C |
| O09044 | SNP23 (synaptosomal-associated protein 23) | 1.7 | 2.86 | Plasma membrane SNARE/involved in migration | |
| Q9CQW1 | YKT6 (synaptobrevin) | 1.3 | 2.92 | Plasma membrane SNARE | |
| Q3TZZ7 | ESYT2 (extended synaptotagmin-2) | 0.69 | 3.45 | Exocytosis | |
| P17047 | LAMP-2 | 1.11 | 2.65 | Late endosome associated | |
| Q9DC51 | GNAI3 (guanine nucleotide-binding protein subunit-α) | 0.97 | 2.07 | GTPase/vesicular trafficking | |
| P35278 | RAB5 | 0.96 | 2.76 | Early endosome fusion | |
| P61027 | RAB10 | 0.99 | 2.66 | Endosome recycling to PM | |
| P46638 | Rab11B | 1.25 | 3.5 | Endosome recycling to PM | |
| Q9DD03 | RAB13 (Ras-related protein Rab13) | 1.14 | 4.52 | Endosomal sorting | |
| P35293 | RAB18 (Ras-related protein Rab18) | 1.47 | 4.69 | Endosomal sorting | |
| Q6PHN9 | RAB35 (Ras-related protein Rab35) | 0.88 | 2.91 | Endosome recycling to PM | D |
| P63321 | RALA (Ras-related protein Ral-A) | 1.47 | 4.19 | GTPase/vesicular trafficking | |
| Q8BU31 | RAP2C (Ras-related protein Rap-2c) | 1.14 | 4.95 | GTPase/vesicular trafficking | |
| Q9JIW9 | Ras-related protein Ral-B/RABIB | 0.81 | 3.65 | GTPase/vesicular trafficking | |
| P62835 | Ras-related protein Rap-1A/RAB1A | 1.05 | 3.65 | GTPase/vesicular trafficking | |
| Q80U72 | SCRIB (protein scribble homolog OS) | 0.93 | 2.6 | GTPase/cell polarity and migration | |
| Q9QXX0 | JAG1 (protein jagged-1) | 1.74 | 4.51 | Notch signaling | |
| Q62312 | TGFR2 | 1.5 | 5.52 | TGF signaling | E |
| P57716 | Nicastrin | 1.51 | 21.23 | Component of γ-secretase complex | |
| Q8VDN2 | AT1A1 (sodium/potassium-transporting ATPase α-1) | 1.47 | 3.16 | Sodium/potassium ion transport | |
| P55012 | S12A2 (solute carrier family 12 member 2) | 1.64 | 6.07 | Sodium/potassium ion transport | F |
| Q61165 | SL9A1 (sodium/hydrogen exchanger 1) | 1.51 | 4.08 | Sodium/potassium ion transport | |
Figure 4.
Validation of quantitative proteomics data using western blot analysis and indirect immunofluorescence microscopy. (A, B) Western blot analysis of PNS and PM fractions for various proteins that were significantly downregulated in PSENdKO MEFs compared with WT/PSEN1rescue MEFs as obtained by quantitative proteome analysis following shotgun approach. (A) Proteins (12 μg protein/lane) were resolved on 4–12% precast SDS–PAGE gels, transferred to nitrocellulose and immunoprobed for the indicated proteins isolated from PM using the SPMNPs-based isolation method. NCT and LAMP-2 were immunoprobed after PM protein isolation using cell surface biotinylation. (B) Quantification of the ratio of the indicated proteins in PSENdKO PM fractions normalized to WT. No mature NCT was detected at the surface of PSENdKO MEFs (mean ratio±s.e.m., n=3, *P<0.05). (C–E) Altered cell morphology and localization of adhesion and migration proteins in PSENdKO MEFs. WT, PSENdKO and hPSEN1rescue MEFs were fixed and immunostained for caveolin-1 and myoferlin (C); CD47 and caveolin-1 (D); and F-actin (using phalloidin conjugated to Alexafluor-488) and vinculin (E). (F) Filipin staining reveals the intracellular accumulation of cholesterol in PSENdKO MEFs in contrast to a prominent cell surface localization in WT and hPSEN1rescue MEFs. Scale bar=10 μm in all panels.
PM glycoproteomics/N-glycan profiling
We performed a global N-glycoproteome analysis on isolated PMs using a COFRADIC protocol. 533 N-glycosylation sites in WT PMs were identified in 347 glycoproteins of which about 70% were annotated as PM proteins, with 75% of all glycosylaton sites belonging to membrane proteins (Pride Database Project No: 15827 and Figure 5A). Further analysis using the gene ontology glycosylation qualifiers revealed 17.2% known, 28.4% potential and 54.3% non-reported sites.
Figure 5.
FACE-based glycan profiling and N-glycoproteomics. (A) Subcellular localization classification of glycoproteins identified from SPMNPs-based PM isolation. For known and characterized proteins, protein subcellular localization was classified based from the SWISS-PROT database. For N-glycosylation, sites from hypothetical proteins and peptide sequences were identified using gene ontology. (B) N-glycan profile (before and after sialidase treatment) of PM fractions derived from WT and PSENdKO MEFs. Sialylated N-glycans are represented in red and gray peaks originate from the presence of Nonidet P-40 detergent. [ : galactose,
: galactose,  : N-acetylglucosamine,
: N-acetylglucosamine,  : mannose,
: mannose,  : fucose; symbols are those suggested by the Consortium for Functional Glycomics (http://glycomics.scripps.edu/CFGnomenclature.pdf)].
: fucose; symbols are those suggested by the Consortium for Functional Glycomics (http://glycomics.scripps.edu/CFGnomenclature.pdf)].
N-glycan profiling was next performed using the DNA-Sequencer Aided fluorophore-assisted carbohydrate electrophoresis (DSA-FACE) method. Only the PM fraction gave a clean N-glycan profile in contrast to PNS where fluorescent non-glycan compounds severely interfered with the analysis (data not shown). Interestingly, the PM fraction was strongly depleted for immature and enriched for mature/complex N-glycans further underscoring the high purity of the isolated membranes. Secondly, we identified the most prevalent murine glycan structures present on WT and PSENdKO PMs, that is, mainly α(1,6)-fucosylated, biantennary N-glycans showing differential modification with α(1,3)-galactose or sialic acid at the β-linked galactose residues. In the PSENdKO MEFs, the level of sialylation was higher, accompanied with a lower extent of α(1,3)-galactose modification, indicating that our method allows for the detection of alterations in N-glycan profile (Figure 5B).
Quantitative PM lipidomics
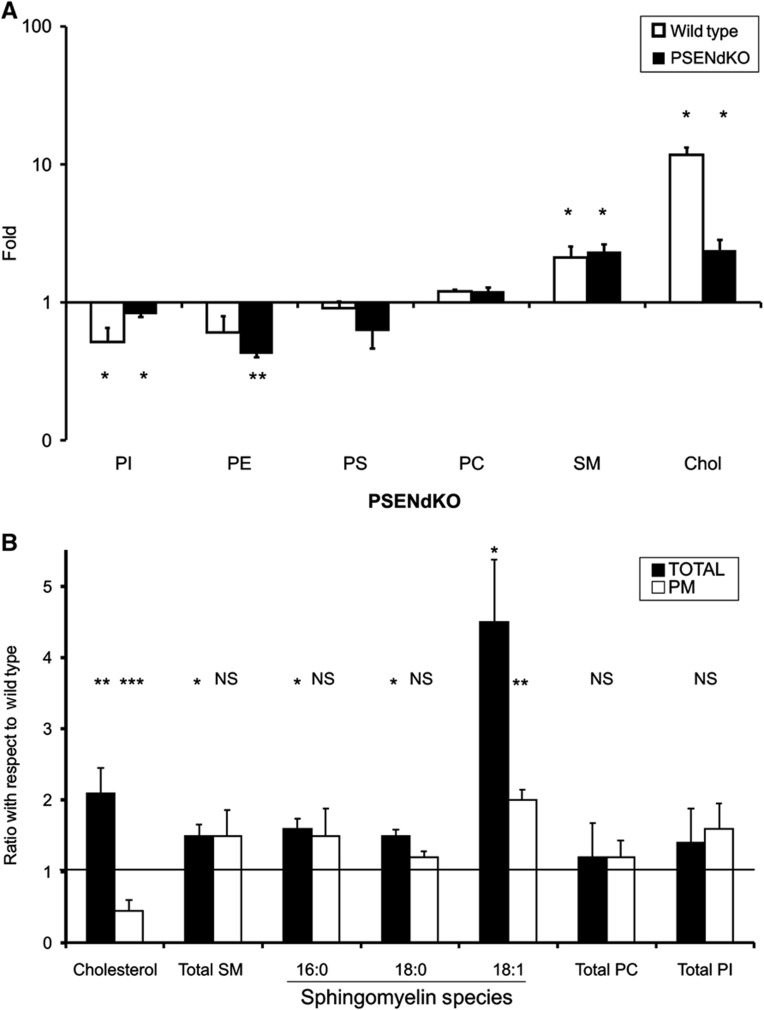
We next explored the potential of the technique for the study of the lipid composition of the cell surface. We extracted lipids from total cells and isolated PMs (see Materials and methods) and analyzed them using nanoelectrospray ionization (ESI)-MS, except for cholesterol, which was measured by fluorometry. Distinct lipid species were used as internal standards and data are presented as mol% of lipid species relative to the total amount of lipids quantified. In WT PMs, cholesterol and SMs were significantly increased compared with the total cell extract (7.8±3.0 mol% (PM) compared with 3.98±0.40 mol% (total) for SMs; P<0.05) while phosphatidylinositol (PI) levels were decreased (from 8.7±1.1 mol% (total) to 4.3±1.8 mol% (PM); P<0.05) (Figure 6A; Supplementary Table SI). Phosphatidylethanolamine (PE) was also decreased, although not reaching statistical significance (Figure 6A; Supplementary Table SI). Similar changes were obtained for PSENdKO PMs, to a lesser extent for PI but significant for PE (see below and Figure 6B). High SM and cholesterol, and low PI and PE are characteristic for cell surface lipid composition (Renkonen et al, 1972; Kalvodova et al, 2009). We also found moderate changes for other glycerophospholipids that were not significant. To illustrate the power of the technique, we analyzed in more detail the combined acyl chain lengths of phosphatidylcholine (PC) species and noticed a significant increase in the PM compared with total cell extract for species with 34 carbons and a decrease for fatty acid moieties of ⩾36 atoms (Supplementary Figure S4a and b). In the PMs, these PC species have more zero or one degree of unsaturation (both acyl chains combined) while polyunsaturation (⩾2 unsaturations in both chains combined) is less abundant, as reported earlier (Kalvodova et al, 2009) (Supplementary Figure S4c and d). In the case of phosphatidylserine (PS), a significant decrease in polyunsaturation of ⩾4° (in both chains combined) was observed (Supplementary Figure S5).
Figure 6.
Quantitative lipidomics. (A) Enrichment plot of PM versus total cell extracts of WT and PSENdKO MEFs for cholesterol (Chol) and the following major lipid families: PI, PE, PC, PS and SM. (B) Lipid ratio levels in total cell extracts and in PM fractions of PSENdKO versus WT MEFs, respectively. Increases are essentially observed in total cell extracts of PSENdKO, but not in their PM fraction. Cholesterol levels were determined by the amplex cholesterol assay. Total SM, SM species, PC and PI were determined by ESI-MS (mean±s.e.m., n=3; *P<0.05; **P<0.01; ***P<0.001; NS, not significant with respect to fold increase for (A) and with respect to WT for (B).
Surprisingly, cholesterol and SM levels (mainly d18:1/16:0 (1.6±0.1, n=3, P<0.05), d18:1/18:0 (1.5±0.1, n=2, P<0.05) and d18:1/18:1 (4.5±0.8, n=3, P<0.05)) were significantly increased in the total cell extracts of PSENdKO compared with WT MEFs and this trend was not recapitulated in isolated PMs of PSENdKO (Figure 6B; Supplementary Table SII) (Grimm et al, 2005). In fact, while SM levels were not different between WT and PSENdKO PMs, the latter harbor significantly lower levels of cholesterol concentrations compared with WT PMs. This was further validated by filipin staining, indeed revealing an excessive intracellular accumulation of cholesterol in PSENdKO but not WT MEFs (where cholesterol is more surface localized) and this was rescued again in hPSEN1rescue MEFs (Figure 4F). Finally, we generated heat maps (using log2 ratios of PSENdKO/WT in total cell extract and PM) to investigate subtle variations in lipid species (Supplementary Figure S6; Supplementary Table SIII). This revealed a higher abundance of polyunsaturated glycerophospholipid species in PSENdKO PMs compared with total cell extract. In the case of SM species, no enrichments were found, instead we detected decreased ratios of saturated and monounsaturated species. Thus, these alterations in the lipid composition of PSENdKO PMs underscore the proteomics finding and also point towards an underlying transport defect in PSENdKO, something that could only be revealed by the comparative and comprehensive analysis of the biomolecular composition of isolated PMs.
Discussion
We here introduce an SPMNP-based isolation procedure allowing multiplexed analysis of the PM with respect to lipids, proteins and N-glycosylation, one of the most common co/post-translational modifications. The technology is exceptionally gentle, as it is based on a non-covalent association of phospholipid-coated SPMNPs with the PM lipid bilayer. This approach obviates the use of detergents and classic affinity labeling procedures with biotin or antibodies directed against surface proteins, the latter strategy being in many cases limited by antibody availability (Zhang et al, 2003; Watarai et al, 2005; Wollscheid et al, 2009). The method is clearly superior to the often used colloidal silica coating (Durr et al, 2004), as it avoids multistep surface chemistry on living cells. Such chemistry inherently affects membrane fluidity, nanodomain dynamics and compromises PM-associated enzymatic activities. As we demonstrate here, PMs can be isolated in a preparative way and in an unsurpassed short experimental time window. The ease of the procedure opens the way to profile the overall PM composition of any given cell type allowing to evaluate conclusively PM changes in differentiated or diseased conditions. The SPMNP-loaded isolated PMs are, as demonstrated, compatible (i.e. no interference from SPMNPs themselves) with protein/lipid extraction procedures and enzymatic digests required for (N-glyco)-proteomics, lipidome analysis as well as N-glycan profiling. We thus present a very significant methodological advance that enables exploration of the overall PM composition in the most comprehensive way possible.
Comparative quantitative proteome analysis of the PM of WT, PSENdKO and hPSEN1rescue MEFs revealed significant changes in a subset of proteins. The changes at the cell surface were PSEN dependent, as they became rescued in hPSEN1rescue PMs. Absence of PSENs resulted in significant downregulation of 41 proteins, including many proteins functionally involved in migration/adhesion, either as surface proteins or within the process of endosomal redistribution and fusion with the PM (SNAREs and RABs). Also evident is the cell surface depletion of proteins known to associate or scaffold on cholesterol-rich microdomains or lipid rafts. These include several GPI-anchored proteins like NCAM1 and dystroglycan and associated proteins like flotillin-2. Another protein, myoferlin, scaffolds on rafts as well as caveolae (Bernatchez et al, 2009) and β1-integrin recycling is caveolae dependent (Wang et al, 2010). Lipidome profiling recapitulated major hallmarks of PM composition including high cholesterol and SM levels and more saturated lipids (Kalvodova et al, 2009; Zech et al, 2009). Besides SM, these lipids turned out to be relatively depleted from the PM of PSENdKO. This was unexpected as previous publications documented accumulations of cholesterol and SM in total cell extracts of PSENdKO MEFs (Grimm et al, 2005). Therefore, these increases had to be intracellular in nature, as we demonstrated (Figure 4F). Secondly, in membrane modeling systems, lateral segregation is aided by the interaction of cholesterol with hydrocarbon chains of saturated lipids, while associations with polyunsaturated lipids are disfavored, excluding them from cholesterol-rich domains (Kahya and Schwille, 2006; García-Sáez et al, 2007; Lingwood and Simons, 2010). In agreement with a relative depletion of cholesterol, PMs of PSENdKO are relatively enriched in polyunsaturated lipid species (Supplementary Figure S6). We noticed also a concomitant and significant downregulation of many raft-/caveolae-associated proteins. These findings overall suggest a selective but severe imbalance in the sorting of cholesterol-rich microdomains, resulting in the intracellular accumulation of such lipids and associated (membrane) proteins in PSENdKO MEFs and causing a significant remodeling of their PMs. While these observations do not resolve the mechanism by which PSEN affects these changes, they demonstrate at least the amazing impact of PSEN deficiency on the cell membrane composition to an extent that was not realized before. We are currently addressing whether the observed PM remodeling is caused by PSEN-related defects in selective endosomal routing/recycling as endosomal/lysosomal dysfunction is among the earliest features in AD neuronal pathology (Nixon, 2005).
Integrating lipidomics as well as N-glycoprotein analysis and N-glycan profiling using the same isolation strategy that allowed the analysis of the protein composition of the PM is an unprecedented, distinctive advantage of this novel technique allowing the interpretation of congruent changes in lipid and protein compositions. The identification of novel glycosylation sites in surface-localized proteins is important and can provide experimental evidence for topology predictions (Wollscheid et al, 2009). Moreover, glycosylation of proteins and lipids has a significant impact on several biological processes including immunity and signaling (Dennis et al, 2009). However, progress in this field suffered from the fact that no technology was available to reliably profile the glycans that are specifically present at the outer surface of the cells. Until now, lectin-based microarrays were used for this purpose, but they yield only limited glycan structural information (Vanderschaeghe et al, 2010). Recent approaches to capture cell surface glycoproteins use carbohydrate oxidation to selectively label the carbohydrates (Wollscheid et al, 2009). While suitable for the identification of N-glycosylation sites, these approaches inherently destroy at least part of the carbohydrate structure, which makes it complicated or impossible to obtain full structural information on the cell surface glycan composition, a problem that can be now overcome using our approach.
Our technology allows for the first time to consider the generation of integral PM ‘fingerprints’ of likely any cell type. Thus identifying the biomolecular composition of PMs with respect to proteins, lipids and their major modifications may accelerate the systematic set-up of more comprehensive PM inventories. It is conceivable now to profile the changes of the cell surface during, for example, stem cell differentiation (Gronthos et al, 2003), neuronal polarization as well as cancer cells becoming metastatic. The identification of PM protein and lipid alterations, as they occur as a consequence of disease, is of paramount importance in experimental medicine. Such alterations may provide novel biomarkers for diagnosis or targets for drug discovery.
Materials and methods
Chemicals, antibodies and cell lines
For SPMNP synthesis, LC-grade reagents for iron (III) acetylacetonate, 1,2-hexadecanediol, oleic acid and benzyl ether were purchased from Sigma-Aldrich, while Oleylamine was from Accor, Chloroform from Merck and ethanol from Honeywell. DSPE-PEG(2000)-NH2 and DSPE-PEG(2000)-CF were from Avanti Polar Lipids and MACS LS column from Miltenyi Biotec. The following monoclonal antibodies (mabs) were purchased: anti-actin, -Flag (M2) and -acetylated tubulin (Sigma), anti-GM130 and -HSP60 (BDBiosciences), anti-Na+K+-ATPase (Novus-Biologicals), anti-GAPDH (Millipore), anti-BIP (Sanbio), anti-RER1p (19432) (Spasic et al, 2007), polyclonal antibodies (pabs) to murine Rer1p (19432) were produced in rabbits using the 17 carboxyterminal amino acids coupled to KLH (Pierce) as immunogen. Pabs against p58 and RBI were provided by R Schekman (Berkeley) and against RAB7 by P Chavrier (CNRS, Paris).
Studies were performed in MEFs WT, PSENdKO and PSEN1rescue using retroviral transduction and followed by puromycin selection (Nyabi et al, 2003) and further subcloning. MEFs were cultured in DMEM/F-12 (Invitrogen) supplemented with 10% FBS (Invitrogen).
Synthesis and functionalization of SPMNPs
Fe3O4 SPMNPs were synthesized using the thermal decomposition method as reported (Sun and Zeng, 2002). In a typical synthesis for 8 nm Fe3O4 nanoparticles, iron (III) acetylacetonate (2 mmol), 1,2-hexadecanediol (10 mmol), oleic acid (6 mmol), oleyl amine (6 mmol) and benzyl ether (20 ml) were magnetically stirred under N2 flow, heated to 200 °C for 2 h and then refluxed at 300 °C for 1 h. The black-colored mixture (SPMNPs) was cooled to room temperature, further precipitated by the addition of ethanol and then magnetically separated using a rare earth magnet. Finally, SPMNPs were dispersed in hexane and centrifuged (5000 r.p.m., 10 min) to remove aggregates. The SPMNPs concentration and size were determined using thermal gravimetric analysis (TGA) and dynamic light scattering (DLS), respectively. Phospholipids-SPMNPs were synthesized by adopting the ligand addition procedure described for water-soluble quantum dots (Dubertret et al, 2002). In a typical experiment, 5 mg of SPMNPs were dissolved in 1 ml of chloroform with DSPE(2000)-PEG-NH2 (10 mg) and vortexed for 4 h followed by the removal of chloroform by evaporation. The residual solid was dried by N2 flow for 5 min, and 1 ml of deionized water was added immediately. After 5 min of vigorous stirring, the uniform dark-colored water-soluble SPMNP solution was centrifuged for 10 min at 5000 r.p.m. to remove the aggregates. The supernatant was further purified on a Miltenyi MACS LS column in the presence of a magnetic field. Removal from the magnetic field allowed collection of the bound lipid-coated SPMNPs; these were next pelleted, resuspended in 1 ml of PBS solution and stored at 4 °C until use. To generate fluorescently labeled SPMNPs, 1:4 compositions (CF and NH2) were used in the functionalization step.
Quality control of SPMNPs
Transmission EM
SPMNP suspensions were adhered onto a carbon-coated copper grid, dried and imaged on a 300-kV Philips CM30 instrument equipped with a field emission gun electron source. The statistical size distribution was determined on at least 50 particles visualized on TEM images.
Thermal gravimetric analysis
SPMNP concentration measurements were performed on a TA instrument Q5000 IR under N2 atmosphere. Briefly, 100 μl aliquots of SPMNPs were heated to 80 °C at the rate of 10 °C/min and kept at 80 °C for 30 min to remove all solvent/dH20. Next, the sample was heated to 800 °C at 20 °C/min to determine the nanoparticle concentration.
DLS and zeta-potential measurement
The hydrodynamic diameters and zeta-potential measurement of SPMNPs were measured using Zetasizer Nano-ZS DLS system (Malvern Instruments Ltd., UK) and reported using DTS application software 7.1. All sizes reported here were based on number average, which was obtained using a non-negative least squares analysis based algorithm. Zeta-potential measurements of SPMNPs in water were measured between pH 2 and 11 (adjusted using 10 mM HCl and 10 mM NaOH).
Magnetic characterization of SPMNPs
Magnetization measurements were made using a standard alternating gradient field magnetometer (AGFM Model 2900; Princeton Instruments, NJ). All measurements were made at room temperature with the magnetic field applied in the film plane.
Isolation and purification of PM
MEFs were grown to near confluency in eight times 10-cm culture dishes, placed on ice and washed once with ice-cold DMEM followed by three washes with ice-cold PBS. Next, MEFs were incubated with SPMNPs (for development and synthesis of SPMNPs, see Supplementary Information) diluted in PBS (2 mg/ml; 20 min, at 4 °C on a slow rocking platform). After incubation, cells were washed once to remove unattached SPMNPs, harvested in PBS and centrifuged (1000 r.p.m., 10 min). Cell pellets were resuspended in 250 mM sucrose (supplemented with 10 mM Hepes, 1 mM EDTA (pH 7.4) and protease inhibitors (Roche) and homogenized using a ball-bearing cell cracker (20 passages, clearance 10 μm, Isobiotec, Germany). After low-speed centrifugation (200 g, 10 min), the PNS was loaded on a PBS-equilibrated LS column placed inside a strong magnetic field (SuperMACSII, Miltenyi Biotec). This LS column is packed with a hydrophilically coated matrix that strongly enhances the magnetic field and thereby ensures a more efficient retainment of the SPMNPs (and adhering PMs) on the column. After loading, the column was extensively and sequentially washed with ice-cold homogenization buffer, high salt 1 M KCl and high pH 0.1 M Na2CO3 buffers. The PM fraction was eluted from the column by removal of the magnetic field. Pelleted PM fractions (55 000 r.p.m., 1 h) were resuspended in 200 μl homogenization buffer and subjected to further analysis. Total proteins were measured using a Bradford assay (Bio-Rad) and denatured in sample buffer prior to separation on precast 4–12% gradient Bis-Tris gels (Invitrogen). Western blot detection was done using ECL (Western Lightning, Perkin-Elmer), imaged on a Fuji MiniLAS3000 and quantified using Aida software (Raytest, Germany). For further MS-based protein/lipid or glycan analysis, the SPMNPs in the membrane fraction did not interfere with any of these subsequent processing steps (enzymatic digestions, solvent extractions, etc) or during the analysis.
Biotin internalization assay
To evaluate potential contaminations arising from endosomal compartments in PM fractions, we performed cell surface biotinylation using cleavable NHS-SS-biotin (0.5 mg/ml; Pierce Chemical Co.) as described (Spasic et al, 2007). After labeling, cells were incubated at 37 °C for 20 min to induce biotin internalization (Zwang and Yarden, 2006). After removing the remaining cell surface-bound biotin using 100 mM 2-sodium-2-mercaptoethanesulfonate (Sigma) as a reducing agent (15 min at 4 °C), PMs were isolated using SPMNPs, as described in the Materials and methods section.
Cell-free γ-secretase activity assay
PM and PNS fractions were extracted in CHAPS and mixed with recombinant APP-C99-FLAG affinity isolated from transiently transfected Aph-1 triple knockout MEFs as described (Spasic et al, 2007). Newly produced APP intracellular domain (AICD) was separated on 10% precasted gels (NuPAGE) in MES buffer and analyzed for western blotting.
Transmission EM
After magnetic labeling, cells were washed twice with PBS−/− and subsequently fixed using 2.5% glutaraldehyde in 0.1 M Na-cacodylate buffer for 30 min. Fixed cells were scraped in 1% gelatin and centrifuged. The cell pellets were washed in buffer, post-fixed in 2% osmiumtetroxide (1 h), rinsed with dH2O and dehydrated in a graded ethanol series (50–100%). Finally, after embedding in Agar, ultrathin sections of 50 nm were examined and micrographs were taken in a JEOL JEM2100. Epon embedding and ultrastructural analysis of isolated PMs was done as described (Ikin et al, 1996).
Confocal imaging
Following incubation with fluorescently labeled SPMNPs, cells were washed in PBS−/−, fixed with 4% paraformaldehyde and mounted in Moviol. Fluorescence was captured on confocal microscope (Radiance2100, Zeiss) connected to an upright NikonE800 microscope and using an oil-immersion plan Apo60x/1.40 NA objective. Image processing was done using Lasersharp2000 (Zeiss) and Photoshop (Adobe, CA).
PM proteomics
MS/MS analysis of PM and PNS
PM and PNS were fully dried in a vacuum concentrator and redissolved in 200 μl of 100 mM TEAB (tri-ethylammoniumbicarbonate; Sigma-Aldrich, Steinheim, Germany), pH 7.8. Samples were heated for 10 min at 95 °C and immediately put on ice for another 15 min. Trypsin (Promega, WI, USA) was added in a 1/100 (w/w) ratio and digestion took place overnight at 37 °C.
Disulfides were reduced in 10 mM (final concentration) triscarboxyethylphosphine (Pierce, Rockford, IL, USA) and free thiols were alkylated using 20 mM iodoacetamide (Sigma-Aldrich) for 15 min at 37 °C. Next, the sample was acidified with 20 μl of 5% acetic acid, and 100 μl of the acidified mixture was injected onto a RP-HPLC column (2.1 mm internal diameter × 150 mm (length) 300SB-C18 column, Zorbax®, Agilent, Waldbronn, Germany) using an Agilent 1100 Series HPLC system. Following a 10-min wash with HPLC solvent A (10 mM ammonium acetate in water/acetonitrile, 98/2 (v/v), water (LC-MS grade, Biosolve, Valkenswaard, The Netherlands) and acetonitrile (HPLC grade, Baker, Deventer, The Netherlands)), a linear gradient to 100% solvent B (10 mM ammonium acetate in water/acetonitrile, 30/70 (v/v)) was applied over 100 min. Using Agilent's electronic flow controller, a constant flow of 80 μl/min was used. Peptides eluting between 20 and 80 min were collected in 60 fractions of 1 min each (80 μl) in a 96-well plate (Agilent). To reduce the total number of LC-MS/MS runs, collected fractions that were separated by 15 min were pooled, vacuum dried, redissolved in 100 μl 2% acetonitrile and stored at −25 °C until further analysis.
A measure of 10 μl of the isolated peptides was sampled by LC-MS/MS using an Ultimate 3000 HPLC system (Dionex, Amsterdam, The Netherlands) in-line connected to a LTQ Orbitrap XL mass spectrometer (Thermo Electron, Bremen, Germany). Peptides were first trapped on a trapping column (PepMap™ C18 column, 0.3 mm I.D. × 5 mm (Dionex)) and following back-flushing from the trapping column, the sample was loaded on a 75-μm I.D. × 150 mm reverse-phase column (PepMap C18, Dionex). Peptides were eluted with a linear gradient of 1.8% solvent B′ (0.05% formic acid in water/acetonitrile (2/8, v/v)) increase per minute at a constant flow rate of 300 nl/min.
Lipid extraction and analysis
Phospholipids were extracted and analyzed by ESI-MS/MS as published (Rysman et al, 2010). Briefly, the PM fractions and PNSs were mixed with 0.9 ml of 1 N HCl: methanol 1:8 (v/v). Subsequently, CHCl3 (0.8 ml), 200 μg/ml of the anti-oxidant 2,6-di-tert-butyl-4-methylphenol (Sigma) and lipid standards (PC (26:0), PC (28:0), PC (40:0), SM (d18:1/12:0), PE (28:0), PI (12:0/13:0), PS (28:0)) were added. Lipid standards were added at 7.5 nmol/mg protein, 3.75 nmol/mg protein, 3.75 nmol/mg protein, 0.43 nmol/mg protein and 0.15 nmol/mg protein for PC, SM, PE, PI and PS, respectively. Finally, the organic fractions were collected by centrifugation (17 300 g for 5 min), evaporated and reconstituted in CH3OH:CHCl3:NH4OH (90:10:1.25, v/v/v) before MS analysis. Phospholipids were analyzed by nanoelectrospray ionization tandem MS (ESI-MS/MS) on a hybrid quadrupole linear ion trap MS (4000 QTRAP; Applied Biosystems, Foster City, CA) equipped with a robotic nanoflow ion source (Advion Biosciences) operated in the infusion mode. Quantification of individual molecular species with respect to lipid standards was carried out by multiple reaction monitoring (MRM) with collision energies of 45, 40, 35, −40 and −55 eV for PC, SM, PE, PS and PI, respectively. Individual MRM signals (100 ms dwell time) were typically averaged over a period of 2 min. Data were calculated according to Liebisch et al (2004) and were expressed as fold change relative to the control samples (WT or total). Heatmaps were build using the Heatmap Builder software (Clifton Watt, Stanford University, USA).
Cholesterol levels were determined using the Amplex-Red cholesterol assay (Molecular Probes).
PM glycomics
FACE-based N-glycan profiling was performed as published (Callewaert et al, 2001; Laroy et al, 2006). After centrifugation (13 000 r.p.m., 5 min), the PM fractions were resuspended in 50 μl ultrapure water (final protein concentration ranged from 0.25 to 0.5 μg/μl) and an equal amount of the corresponding PNS fraction was diluted to 50 μl with ultrapure water. To each sample, 5 μl of 5.0% SDS and 0.4 M dithiothreitol in water was added and incubated for 10 min at 100 °C, followed by 5 min at 4 °C. Subsequently, 7 μl of 0.5 M sodium phosphate pH 7.5 buffer and 7 μl of 10% Nonidet P-40 in water were added to each sample. Then, the samples were incubated at 37 °C for 3 h after the addition of 2 μl Peptide N-glycosidase F (2 New England Biolabs units), followed by an extra microliter of enzyme preceding an overnight incubation at 37 °C. In all, 10 μl of each sample was evaporated to dryness (vacuum) and 5 μl of a 1:1 mixture (v/v) of 50 mM APTS (Molecular Probes, USA) in 1.2 M citric acid and 1 M NaCNBH3 in DMSO was added. After overnight labeling at 37 °C, the samples were subjected to size-exclusion chromatography using Sephadex G-10 resin (Laroy et al, 2006). Then, the samples were dried and reconstituted in 100 μl of 60% acetonitrile plus 0.05% TFA. They were loaded on NuTip P100 carbon tips (Glygen), conditioned with 100 μl of 60% acetonitrile plus 0.05% TFA (3 × ) and with 100 μl of 0.05% TFA in water (3 × ), by 20 aspirate-expel cycles using a micropipette. Subsequently, the NuTips were washed five times in 100 μl of 0.05% TFA in water and the glycans were then eluted two times with 50 μl of 60% acetonitrile plus 0.05% TFA. Finally, the obtained samples were dried and reconstituted in 10 μl of ultrapure water and diluted at least 150 times prior to their injection in the capillaries of an ABI 3130 DNA sequencer (Applied Biosystems) for 80 s at 1200 V. To remove the sialic acid residues from the N-glycans, 1 μl of 20 mM sodium acetate pH 5.0 buffer containing 40 mU of Arthrobacter ureafaciens α(2-3,6,8)-sialidase (Glyko) was added to 2 μl of sample, incubated overnight at 37 °C and quenched to 10 μl with ultrapure water prior to their analysis on a sequencer ( × 40 dilution). To determine the level of sialylation, the amount of sialylated N-glycan branches was normalized to the total amount of N-glycan branches present in the N-glycome profile. The N-glycan structure of each peak was assigned by enzymatic digestions with (combinations of) exoglycosidases (e.g. α(1–3)- and β(1–4)-galactosidase, α(1–2,3,4,6)-fucosidase and β(1–2,3,4,6)-N-acetylhexosaminidase) that cleave specific sugar residues. APTS-labeled dextran was used as internal standard in every sample to correct for injection differences due to different salt concentrations.
Statistical analysis
All quantified data are represented as an average of at least three independent experiments (mean±s.e.m.). Statistical significance was determined by two-tailed Student's t-test and set at *P⩽0.05; **P⩽0.01; and ***P⩽0.001.
Proteomics database online
Proteomics data from the experiments can be found on the PRIDE database (http://www.ebi.ac.uk/pride/) using login ‘review43406’ and password ‘n3mSbXfK.’ Project number 15826 contains all the MS data from the shotgun non-differential experiment, project 15827 contains the MS data from the N-glycosylation experiments and finally project 15828 contains data from the differential experiments involving WT, PSENdKO and PSEN1rescue cells.
Supplementary Material
Supplementary Figures S1–6, Supplementary Tables SI–II
Lipidomics Data
Lipidomics Data Set-II
Acknowledgments
This work was financially supported by grants from the KULeuven (GOA 2011–2015 to WA and JS; GOA08/16 to EW; CREA/07/023 to WA and Methusalem (BDS)) and UGent (GOA 2005–2010 to NC), the Research Foundation Flanders (FWO) (G.0663.09 and G.0.708.10 to WA and G.0541.08 to NC), Stichting Alzheimer Onderzoek (SAO-FRMA/2010), the Alzheimer Association (IIRG-08-91535) and Hercules Foundation (AKUL/09/037) to WA, the federal government (IAP P6/43 to WA, IAP P6/28 to KG and EW). KG acknowledges support by research grants from the Concerted Research Actions (project BOF07/GOA/012) from the UGent and EW from two impulse financing initiatives (http://biomacs.kuleuven.be; http://www.prometa.kuleuven.be). AK holds a fellowship of the VIB-International PhD program. KC is holder of an IWT fellowship. BG is a Postdoctoral Research Fellow and ER a research assistant of the Fund for Scientific Research (FWO)—Flanders (Belgium).
Author contributions: DBTGR, GB and WA planned the project. DBTGR developed the SPMNPs and conducted the major part of the experimental work. BG and KG carried out the proteomics. RD, ER, EW and JS conducted the lipidomics. DV and NC performed N-glycan analysis. AKT, KC, PB contributed with confocal and EM analysis. DBTGR and WA wrote the paper with input from BDS, GB, ER and DV. All authors contributed in the discussion/analysis of the data and contributed to manuscript editing.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andrews NW (2002) Lysosomes and the plasma membrane: trypanosomes reveal a secret relationship. J Cell Biol 158: 389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatchez PN, Sharma A, Kodaman P, Sessa WC (2009) Myoferlin is critical for endocytosis in endothelial cells. Am J Physiol Cell Physiol 297: C484–C492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert N, Geysens S, Molemans F, Contreras R (2001) Ultrasensitive profiling and sequencing of N-linked oligosaccharides using standard DNA-sequencing equipment. Glycobiology 11: 275–281 [DOI] [PubMed] [Google Scholar]
- Carrasco S, Meyer T (2010) STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem 80: 973–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ZH, Ip NY (2009) Endophilin B1: guarding the gate to destruction. Commun Integr Biol 2: 130–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyung JH, Raper DM, Selkoe DJ (2005) Gamma-secretase exists on the plasma membrane as an intact complex that accepts substrates and effects intramembrane cleavage. J Biol Chem 280: 4383–4392 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W (2010) Novel research horizons for presenilins and gamma-secretases in cell biology and disease. Annu Rev Cell Dev Biol 26: 235–260 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R (1999) A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398: 518–522 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F (1998) Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391: 387–390 [DOI] [PubMed] [Google Scholar]
- Deery MJ, Maywood ES, Chesham JE, Sládek M, Karp NA, Green EW, Charles PD, Reddy AB, Kyriacou CP, Lilley KS, Hastings MH (2009) Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol 19: 2031–2036 [DOI] [PubMed] [Google Scholar]
- Dennis JW, Lau KS, Demetriou M, Nabi IR (2009) Adaptive regulation at the cell surface by N-glycosylation. Traffic 10: 1569–1578 [DOI] [PubMed] [Google Scholar]
- Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A (2002) In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science 298: 1759–1762 [DOI] [PubMed] [Google Scholar]
- Durr E, Yu J, Krasinska KM, Carver LA, Yates JR, Testa JE, Oh P, Schnitzer JE (2004) Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat Biotechnol 22: 985–992 [DOI] [PubMed] [Google Scholar]
- Esselens C, Oorschot V, Baert V, Raemaekers T, Spittaels K, Serneels L, Zheng H, Saftig P, De Strooper B, Klumperman J, Annaert W (2004) Presenilin 1 mediates the turnover of telencephalin in hippocampal neurons via an autophagic degradative pathway. J Cell Biol 166: 1041–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner F, Foster LJ, Campanaro S, Valle G, Mann M (2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics 5: 608–619 [DOI] [PubMed] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M (2006) A mammalian organelle map by protein correlation profiling. Cell 125: 187–199 [DOI] [PubMed] [Google Scholar]
- García-Sáez AJ, Chiantia S, Schwille P (2007) Effect of line tension on the lateral organization of lipid membranes. J Biol Chem 282: 33537–33544 [DOI] [PubMed] [Google Scholar]
- Ghesquière B, Colaert N, Helsens K, Dejager L, Vanhaute C, Verleysen K, Kas K, Timmerman E, Goethals M, Libert C, Vandekerckhove J, Gevaert K (2009) In vitro and in vivo protein-bound tyrosine nitration characterized by diagonal chromatography. Mol Cell Proteomics 8: 2642–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghesquière B, Jonckheere V, Colaert N, Van Durme J, Timmerman E, Goethals M, Schymkowitz J, Rousseau F, Vandekerckhove J, Gevaert K (2011) Redox proteomics of protein-bound methionine oxidation. Mol Cell Proteomics 10: M110.006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghesquière B, Van Damme J, Martens L, Vandekerckhove J, Gevaert K (2006) Proteome-wide characterization of N-glycosylation events by diagonal chromatography. J Proteome Res 5: 2438–2447 [DOI] [PubMed] [Google Scholar]
- Grimm MO, Grimm HS, Pätzold AJ, Zinser EG, Halonen R, Duering M, Tschäpe JA, De Strooper B, Müller U, Shen J, Hartmann T (2005) Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat Cell Biol 7: 1118–1123 [DOI] [PubMed] [Google Scholar]
- Gronthos S, Zannettino AC, Hay SJ, Shi S, Graves SE, Kortesidis A, Simmons PJ (2003) Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J Cell Sci 116 (Part 9): 1827–1835 [DOI] [PubMed] [Google Scholar]
- Herreman A, Van Gassen G, Bentahir M, Nyabi O, Craessaerts K, Mueller U, Annaert W, De Strooper B (2003) Gamma-secretase activity requires the presenilin-dependent trafficking of nicastrin through the Golgi apparatus but not its complex glycosylation. J Cell Sci 116 (Part 6): 1127–1136 [DOI] [PubMed] [Google Scholar]
- Huber LA, Pfaller K, Vietor I (2003) Organelle proteomics: implications for subcellular fractionation in proteomics. Circ Res 92: 962–968 [DOI] [PubMed] [Google Scholar]
- Ikin AF, Annaert WG, Takei K, De Camilli P, Jahn R, Greengard P, Buxbaum JD (1996) Alzheimer amyloid protein precursor is localized in nerve terminal preparations to Rab5-containing vesicular organelles distinct from those implicated in the synaptic vesicle pathway. J Biol Chem 271: 31783–31786 [DOI] [PubMed] [Google Scholar]
- Josic D, Clifton JG (2007) Mammalian plasma membrane proteomics. Proteomics 7: 3010–3029 [DOI] [PubMed] [Google Scholar]
- Kahya N, Schwille P (2006) How phospholipid-cholesterol interactions modulate lipid lateral diffusion, as revealed by fluorescence correlation spectroscopy. J Fluoresc 16: 671–678 [DOI] [PubMed] [Google Scholar]
- Kalvodova L, Sampaio JL, Cordo S, Ejsing CS, Shevchenko A, Simons K (2009) The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J Virol 83: 7996–8003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroy W, Contreras R, Callewaert N (2006) Glycome mapping on DNA sequencing equipment. Nat Protoc 1: 397–405 [DOI] [PubMed] [Google Scholar]
- Leem JY, Vijayan S, Han P, Cai D, Machura M, Lopes KO, Veselits ML, Xu H, Thinakaran G (2002) Presenilin 1 is required for maturation and cell surface accumulation of nicastrin. J Biol Chem 277: 19236–19240 [DOI] [PubMed] [Google Scholar]
- Liebisch G, Lieser B, Rathenberg J, Drobnik W, Schmitz G (2004) High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim Biophys Acta 1686: 108–117 [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K (2010) Lipid rafts as a membrane-organizing principle. Science 327: 46–50 [DOI] [PubMed] [Google Scholar]
- Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, Baki L, Wen P, Efthimiopoulos S, Shao Z, Wisniewski T, Robakis NK (2002) A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J 21: 1948–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Ungerer N, Klimmeck D, Warnken U, Schnölzer M, Frings S, Möhrlen F (2008) Proteomic analysis of a membrane preparation from rat olfactory sensory cilia. Chem Senses 33: 145–162 [DOI] [PubMed] [Google Scholar]
- Mora R, Bonilha VL, Marmorstein A, Scherer PE, Brown D, Lisanti MP, Rodriguez-Boulan E (1999) Caveolin-2 localizes to the golgi complex but redistributes to plasma membrane, caveolae, and rafts when co-expressed with caveolin-1. J Biol Chem 274: 25708–25717 [DOI] [PubMed] [Google Scholar]
- Nixon RA (2005) Endosome function and dysfunction in Alzheimer's disease and other neurodegenerative diseases. Neurobiol Aging 26: 373–382 [DOI] [PubMed] [Google Scholar]
- Nyabi O, Bentahir M, Horré K, Herreman A, Gottardi-Littell N, Van Broeckhoven C, Merchiers P, Spittaels K, Annaert W, De Strooper B (2003) Presenilins mutated at Asp-257 or Asp-385 restore Pen-2 expression and Nicastrin glycosylation but remain catalytically inactive in the absence of wild type Presenilin. J Biol Chem 278: 43430–43436 [DOI] [PubMed] [Google Scholar]
- Pisitkun T, Shen RF, Knepper MA (2004) Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA 101: 13368–13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkonen O, Gahmberg CG, Simons K, Kääriäinen L (1972) The lipids of the plasma membranes and endoplasmic reticulum from cultured baby hamster kidney cells. Biochim Biophys Acta 255: 66–78 [DOI] [PubMed] [Google Scholar]
- Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D, Daniëls VW, Machiels J, Vanderhoydonc F, Smans K, Waelkens E, Verhoeven G, Swinnen JV (2010) De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res 70: 8117–8126 [DOI] [PubMed] [Google Scholar]
- Simpson RJ, Jensen SS, Lim JW (2008) Proteomic profiling of exosomes: current perspectives. Proteomics 8: 4083–4099 [DOI] [PubMed] [Google Scholar]
- Spasic D, Raemaekers T, Dillen K, Declerck I, Baert V, Serneels L, Füllekrug J, Annaert W (2007) Rer1p competes with APH-1 for binding to nicastrin and regulates gamma-secretase complex assembly in the early secretory pathway. J Cell Biol 176: 629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zeng H (2002) Size-controlled synthesis of magnetite nanoparticles. J Am Chem Soc 124: 8204–8205 [DOI] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, Müller SA, Rammner B, Gräter F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmüller H, Heuser J et al. (2006) Molecular anatomy of a trafficking organelle. Cell 127: 831–846 [DOI] [PubMed] [Google Scholar]
- Tan S, Tan HT, Chung MC (2008) Membrane proteins and membrane proteomics. Proteomics 8: 3924–3932 [DOI] [PubMed] [Google Scholar]
- Vanderschaeghe D, Festjens N, Delanghe J, Callewaert N (2010) Glycome profiling using modern glycomics technology: technical aspects and applications. Biol Chem 391: 149–161 [DOI] [PubMed] [Google Scholar]
- Wang C, Yoo Y, Fan H, Kim E, Guan KL, Guan JL (2010) Regulation of integrin beta 1 recycling to lipid rafts by Rab1a to promote cell migration. J Biol Chem 285: 29398–29405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschbüsch D, Born S, Niediek V, Kirchgessner N, Tamboli IY, Walter J, Merkel R, Hoffmann B (2009) Presenilin 1 affects focal adhesion site formation and cell force generation via c-Src transcriptional and posttranslational regulation. J Biol Chem 284: 10138–10149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watarai H, Hinohara A, Nagafune J, Nakayama T, Taniguchi M, Yamaguchi Y (2005) Plasma membrane-focused proteomics: dramatic changes in surface expression during the maturation of human dendritic cells. Proteomics 5: 4001–4011 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Wollscheid B, Bausch-Fluck D, Henderson C, O'Brien R, Bibel M, Schiess R, Aebersold R, Watts JD (2009) Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol 27: 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DR, Nye JS, Lamb NJ, Fernandez A, Kitzmann M (2005) Intracellular retention of caveolin 1 in presenilin-deficient cells. J Biol Chem 280: 6663–6668 [DOI] [PubMed] [Google Scholar]
- Zech T, Ejsing CS, Gaus K, de Wet B, Shevchenko A, Simons K, Harder T (2009) Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J 28: 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhou G, Zhao Y, White MA, Zhao Y (2003) Affinity enrichment of plasma membrane for proteomics analysis. Electrophoresis 24: 2855–2863 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang W, Kho Y, Zhao Y (2004) Proteomic analysis of integral plasma membrane proteins. Anal Chem 76: 1817–1823 [DOI] [PubMed] [Google Scholar]
- Zou K, Hosono T, Nakamura T, Shiraishi H, Maeda T, Komano H, Yanagisawa K, Michikawa M (2008) Novel role of presenilins in maturation and transport of integrin beta 1. Biochemistry 47: 3370–3378 [DOI] [PubMed] [Google Scholar]
- Zwang Y, Yarden Y (2006) p38 MAP kinase mediates stress-induced internalization of EGFR: implications for cancer chemotherapy. EMBO J 25: 4195–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–6, Supplementary Tables SI–II
Lipidomics Data
Lipidomics Data Set-II