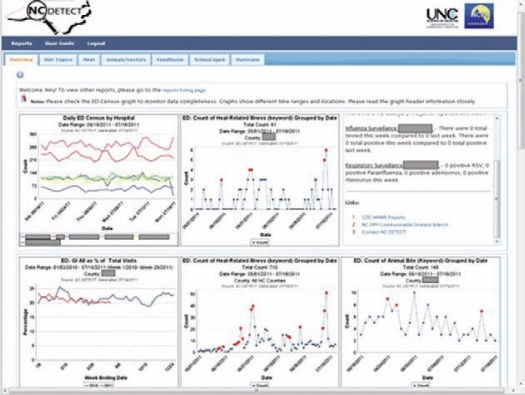
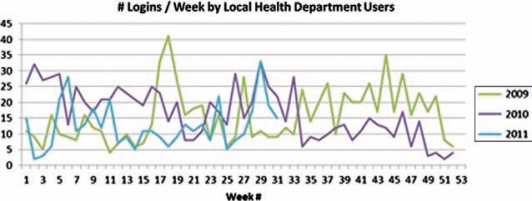
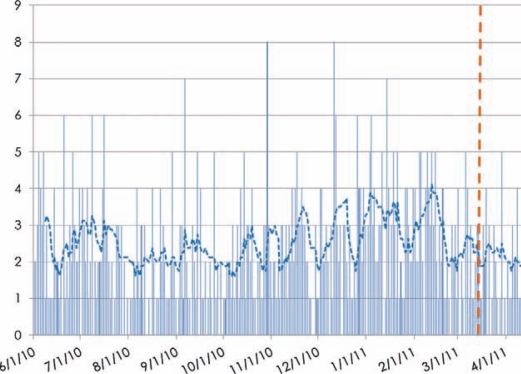
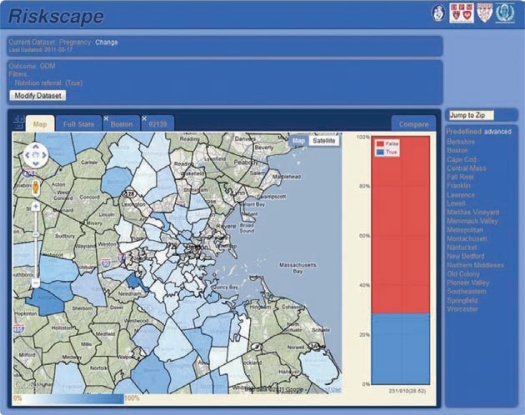
Objective
The objective of this study is to outline and demonstrate the robust methodology used by Geographic Utilization of Artificial Intelligence in Real-Time for Disease Identification and Alert Notification (GUARDIAN) surveillance system to generate and validate biological threat agent (BTA) profiles.
Introduction
Detection of BTAs is critical to the rapid initiation of treatment, infection control measures and public health emergency response plans. Due to the rarity of BTAs, standard methodology for developing syndrome definitions and measuring their validity is lacking.
Methods
BTA profile development consisted of the following steps.
Step 1: Literature scans for BTAs: articles found in a literature review on BTAs that met predefined criteria were reviewed by multiple researchers to independently extract BTA-related data including physical and clinical symptoms, epidemiology, incubation period, laboratory findings, radiological findings and diagnosis (confirmed, probable or suspected).
Step 2: Data analysis and transformation: articles were ran- domly divided, taking into account reported diagnosis and sample size, to generate detection (75% of articles), and testing (25% of articles) profiles. Statistical approaches such as combining frequencies, weighted mean, pooled variance, min of min and max of max were utilized for combining the data from articles to generate the profiles.
Step 3: Missing data analysis: based on generated statistical and clinical judgment, specific reasonable assumptions about the missing values for each element (i.e., always reported, never reported, representative and conditionally independent) were applied to the profile. Imputed case analysis (ICA) strategies (1) used these data assumptions to fill in missing data in each meta-analysis of the summary data.
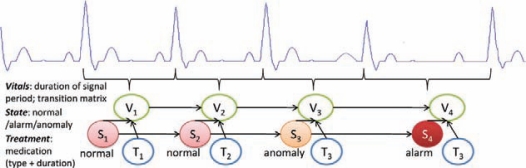
Step 4: Translation: the generated profiles and synthetic positive BTA cases were reviewed (via clinical filters and physician reviews) and programmed into GUARDIAN.
Step 5: Prior probability determination: using archived, historical patient data, the probabilities associated with each element of the profile were determined for the general (non-BTA) patient population.
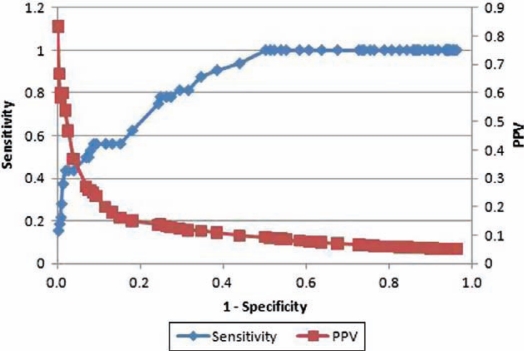
Step 6: Validation and testing: multiple mutually exclusive samples of ED cases along with synthetic/real positive BTA cases were utilized to perform 10-fold cross-validation as well as testing to generate statistical measures such as positive predicted value, negative predicted value, sensitivity, specificity, accuracy and receiver operating characteristic curve (ROC) for each BTA. To demonstrate the applicability and usability of BTA methodology, severe acute respiratory syndrome (SARS) was chosen since differentiating SARS symptoms from regular influenza is difficult and presents challenges for even robust surveillance systems.
Results
Literature scan yielded 34 articles with 4265 cases and 90 unique signs, symptoms and confirmatory features (frequency data = 68 and continuous data = 22) for SARS. After combining the data and assigning the assumptions, there were 18 representative, 63 conditionally independent and 9 confirmatory features.
Applying the BTA methodology for SARS, the positive predicted value, negative predicted value, sensitivity, specificity and accuracy based on 10-fold cross-validation were 55.7%, 94.6%, 74.8%, 88.1% and 85.8%, respectively. An ROC curve analysis revealed an area under the curve of 0.929. The main features contributing toward identifying the positive SARS cases were fever, chills/rigors, nonproductive cough, fatigue/malaise/lethargy and myalgias. The identified features were in agreement with clinicians’ judgment.
Conclusions
The GUARDIAN BTA profile development methodology provides a sound approach for creating disease profiles and a robust validation process even in a BTA (e.g., SARS) that may closely resemble regularly occurring diseases (e.g., influenza). The BTA profile development methodology has been successfully applied to other BTAs such as botulism, brucellosis and West Nile virus, with high sensitivity and specificity.
Keywords
Biological threat agents; real-time surveillance; surveillance methodologyReferences
References
-
1.Higgins J, White IR, Wood AM. Imputation methods for missing outcome data in meta-analysis of clinical trials. Clin Trials. 2008;5:225–39. doi: 10.1177/1740774508091600. [DOI] [PMC free article] [PubMed] [Google Scholar]