Abstract
DNA interstrand cross-links (ICLs) covalently link both strands of the DNA duplex, impeding cellular processes like DNA replication. Homologous recombination (HR) is considered to be a major pathway for the repair of ICLs in mammalian cells as mutants for HR components are highly sensitive to DNA-damaging agents that cause ICLs. This chapter describes GFP assays to measure HR following site-specific ICL formation with psoralen through DNA triplex technology. This approach can be used to determine the genetic requirements for ICL-induced HR in relation to those involved in HR repair of other DNA lesions such as double-strand breaks.
Keywords: Homologous recombination, interstrand cross-link repair, triplex-forming oligonucleotide, GFP reporters
1. Introduction
DNA interstrand cross-links (ICLs) are toxic to dividing cells because they impede DNA replication and other cellular processes, and as a result, agents that cause ICLs such as cisplatin are frequently used in cancer chemotherapy (1). In addition to causing lethal damage, ICLs can induce mutations and gross chromosomal rearrangements. Multiple pathways have been implicated in ICL repair, including nucleotide excision repair, translesion synthesis, and homologous recombination (HR) (2). In mammalian cells, a role for HR in ICL repair is postulated based on the extreme sensitivity of cells deficient in HR components, such as BRCA1 (3, 4) and BRCA2 (5, 6), to various agents that cause ICLs. Cells deficient in Fanconi anemia pathway components are also sensitive to ICL agents (7, 8) and show defects in HR, especially HR coupled to DNA replication (9, 10).
Given the many open questions about the relationship between ICL repair and HR, we developed an approach to assay ICL-induced HR in mammalian cells based on the DR-GFP reporter which has previously been developed to detect HR induced by another type of lesion, a DNA double-strand break (DSB) (Fig. 16.1a) (11). DR-GFP is composed of two differentially mutated green fluorescent protein (GFP) genes oriented as direct repeats (hence, “DR”): the upstream repeat contains the recognition site for the rare-cutting I-SceI endonuclease and the downstream repeat is a 5′ and 3′ truncated GFP fragment. Transient expression of I-SceI leads to a DSB in the upstream GFP gene; HR to repair the DSB results in GFP+ cells which are quantified by flow cytometry (12). This assay has been widely used to identify proteins required for HR repair, such as BRCA1 and BRCA2, and to determine which pathways suppress HR repair, using both candidate gene approaches (13) and whole genome screens (14). While developed for chromosomal DSB repair assays, the DR-GFP reporter can also be used to assay repair in plasmids.
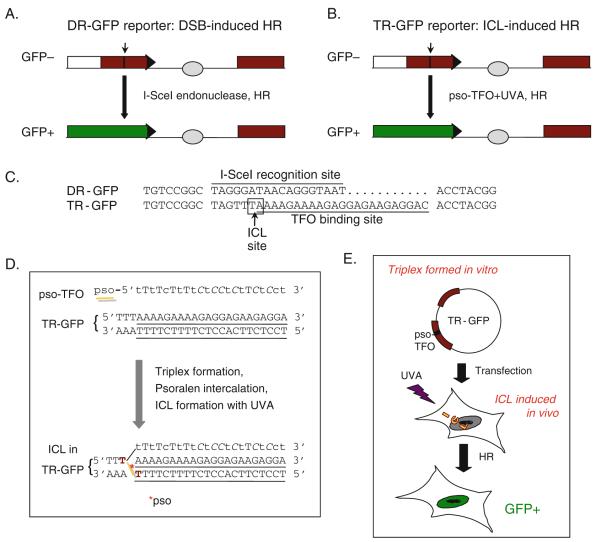
Fig. 16.1.
Design of HR assays. a DR-GFP reporter measures DSB-induced HR (11, 12, 18). This reporter consists of two defective GFP genes, the first of which contains an I-SceI endonuclease site (arrow) such that cells are GFP−. Cellular expression of I-SceI leads to a DSB which can be repaired by HR using the downstream wild-type GFP sequence as a template, resulting in GFP+ cells. Two different DR-GFP plasmids have been created: pDR-OriP-GFP has an EBV OriP placed between the GFP repeats (light gray circle) to allow the plasmid to replicate in the presence of EBNA1; in pDR-GFP-hprt, the reporter is flanked by Hprt genomic sequences for targeting the reporter to the mouse Hprt locus (not shown). Short black bar box, GFP repeat; white box and horizontal arrow head, non-repetitive parts of the GFP gene; long black bar, GFP+ gene. b TR-GFP reporter measures ICL-induced HR (10). A TFO binding site (arrow) replaces the I-SceI site of the DR-GFP reporter. After triplex formation at the TFO binding site with a psoralen-conjugated TFO (pso-TFO:TR-GFP), followed by UVA irradiation (ICL formation), ICL-induced HR repair restores an intact GFP gene, giving rise to GFP+ cells. Similar to DR-GFP, two TR-GFP plasmids have been constructed, pTR-GFP-hprt and TR-OriP-GFP. c Sequences of the I-SceI recognition site in DR-GFP and the TFO binding site in TR-GFP. The ICL site – a TA/AT sequence suitable for psoralen intercalation and ICL formation – is boxed. The TAG of the I-SceI site and the same triplet in TR-GFP are stop codons, truncating the GFP protein. Eight base pairs of flanking sequences in DR/TR-GFP are shown on both sides of the damage sites. d Details of ICL formation. After triplex formation between the pso-TFO and the TR-GFP, the 5′-psoralen moiety of pso-TFO intercalates at the TA/AT site. UVA irradiation cross-links psoralen (ICL, gray line) to the two Ts (bold) of the double-stranded DNA of TR-GFP. See the legend for Fig. 16.2a for more details about the pso-TFO. e Scheme for the TR-GFP assay. The pso-TFO:TR-GFP triplex is formed in vitro and transfected into cells which are then treated with UVA to form the ICL. Cells are incubated for 48 h and GFP+ cells arising by HR are quantified by flow cytometry.
To elucidate the role of HR in the repair of ICLs, we modified DR-GFP to contain a specific site for ICL formation, creating the TR-GFP reporter (TR, triplex, and repeats of GFP; Fig. 16.1b, c) (10). The modification was accomplished by replacing the I-SceI site with a sequence that can bind a triplex-forming oligonucleotide (TFO) conjugated with psoralen at its 5′-end (pso-TFO) (15). Following triplex formation between pso-TFO and TR-GFP (pso-TFO:TR-GFP), intercalation of the psoralen into duplex DNA, and exposure to 365 nm ultraviolet light (UVA), a site-specific ICL forms in TR-GFP (Fig. 16.1d). Although typically not as high as with DSB-induced HR, GFP+ cells are obtained (i.e., several percent for DR-GFP compared with a few percent or less for TR-GFP), indicating ICL-induced HR. HR is dependent on known HR factors such as BRCA1 and BRCA2 (10). Such an approach has previously been used with a supF gene reporter (16). Several modifications of this approach are possible. For example, the TR-GFP reporter has been modified to contain an origin of replication (OriP) from Epstein–Barr virus (EBV) for replication in human cells expressing the EBV nuclear antigen, EBNA1 (10, 17). This modification allows the examination of HR coupled to DNA replication. Further, the TFO “tail” can be removed, by using a pso-TFO with a disulfide bond between the psoralen moiety and the TFO (pso-SS-TFO) (10, 15).
In this chapter, we provide a protocol to quantify ICL-induced HR in which the site-specific ICL is formed in cells after transfection of the pso-TFO:TR-GFP triplex and exposure to UVA (Fig. 16.1e). A procedure to verify in vivo ICL formation is also provided.
2. Materials
2.1. Cell Culture
Human osteosarcoma cell line U2OS (ATCC HTB-96) which grows in D-MEM high glucose (GIBCO 31053-036) and 15% fetal calf serum, supplemented with 1× penicillin–streptomycin–L-glutamine (GIBCO 10378-016).
U2OS-CEP cells, which are U2OS cells stably transfected with the pCEP4 plasmid (Invitrogen V044-50), selected with 0.4 mg/ml hygromycin B (Roche 10843555001) supplemented in the medium.
Phosphate buffered saline (PBS).
37°C, 5% CO2 incubator.
2.2. Plasmids
Prepare with standard protocols. Unless plasmids are prepared to high purity with cesium chloride ultracentrifugation, avoid repeated freeze thawing:
2.3. Transfection
Electroporation system (Bio-Rad Gene Pulser II).
0.4-cm-Gap cuvettes (Bio-Rad).
Opti-MEM media (Invitrogen).
10-cm Tissue culture plates.
2.4. ICL Formation
TFO-containing oligonucleotides are from Eurogentec. Sequences are presented in Fig. 16.2a. Nucleotide modifications to enhance the stability of the triplex are locked nucleic acids (LNA, 2′O-4′C methylene bridge) (lower case) and 5′-methylated cytosines (italics) (21) (see Note 3). The 10× stock solution is 100 μM in H2O for each:
As a negative control for ICL formation, the TFO without psoralen.
For ICL formation, the pso-TFO, which is the TFO conjugated with psoralen at the 5′-nucleotide through a (CH2)6 linkage to the phosphate.
As a negative control for triplex formation, pso-mTFO, which cannot form a triplex with the target sequence in the TR-GFP plasmids.
10× TFO buffer: 500 mM HEPES (pH 7.2), 500 mM NaCl, 100 mM MgCl2, 5 mM spermine.
UVA irradiator: A UVA lamp with a sensor to accurately measure the dose (J/cm2). Alternatively, we are using a UV Stratalinker 2400 (Stratagene) with UVA bulbs (365 nm).
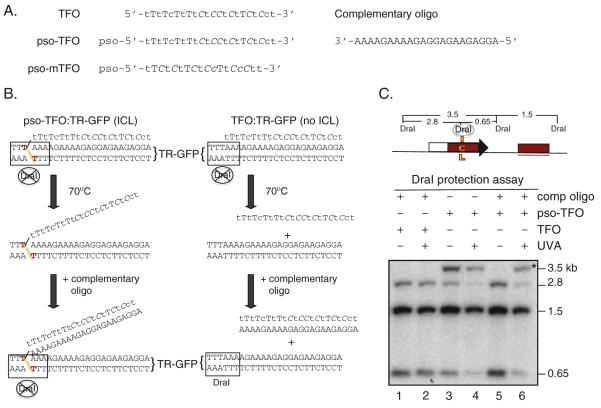
Fig. 16.2.
DraI protection assay. a Oligonucleotide sequences of TFOs and the oligonucleotide complementary to the TFO and pso-TFO used in this study. Several nucleotides within the TFO are modified to enhance the stability of the triplex. Lower case nucleotides, locked nucleic acids (LNA); cytosines in italics, methylated at the 5′ position. Psoralen (pso) is attached to the 5′ nucleotide through a (CH2)6 linkage to the phosphate. b DraI protection assay to distinguish ICL formation from triplex formation. After UVA, the triplex is heated to dissociate the TFO from the duplex; with cooling, the free TFO anneals to the complementary oligonucleotide preventing it from reannealing to the duplex, permitting DraI cleavage (right panel). By contrast for cross-linked TR-GFP, pso-TFO does not dissociate from TR-GFP because of the covalent linkage of psoralen with TR-GFP (left panel). DraI is unable to cleave in this case. However, heating and cooling steps with the complementary oligonucleotide traps pso-TFO that is not cross-linked and prevents non-covalent triplex formation during analysis (not shown). c DraI protection assay results. After extraction of DNA from the transfected cells and digestion by DraI, samples were analyzed by Southern blotting. The GFP probe is underlined. The fragment protected by the TFO and/or the ICL is 3.5 kb; if unprotected, the DraI-cleaved fragments are 2.8 and 0.65 kb. For the unconjugated TFO samples, no 3.5-kb fragment was detected without or with UVA irradiation when the complementary oligonucleotide was added prior to digestion with DraI(lanes 1 and 2, respectively) (Fig. 16.2b, right panel). In the absence of the complementary oligonucleotide, both unirradiated and UVA-irradiated pso-TFO samples showed the 3.5-kb DraI-resistant fragment (lanes 3 and 4, respectively). In contrast, in presence of the complementary oligonucleotide (lanes 5 and 6), only the pso-TFO treated sample which was UVA irradiated showed the 3.5-kb fragment corresponding to ICL formation (asterisk) (Fig. 16.2b, left panel).
2.5. DraI Protection Assay
Complementary oligonucleotide (Fig. 16.2a), 10× stock solution at 500 μM.
DraI restriction enzyme (40 U/μl; Roche).
HindIII restriction enzyme (New England Biolabs).
Materials for DNA extraction from mammalian cells. We use the QIAamp DNA Blood Mini Kit (Qiagen).
Standard materials for agarose gel electrophoresis and Southern blotting (12).
2.6. Flow Cytometry
We use a FACScan (BD), although any flow cytometry analyzer will suffice. Cells are gated by forward and side scatter, and fluorescence is analyzed on the FL1 and FL2 channels (12). GFP+ cells are determined from the FL1 shift from the majority of negative population. Consult a flow cytometry facility if you are uncertain about using a flow cytometer or do not have one accessible for your own use.
3. Methods
The assays involve transient transfection of the HR reporter plasmids into mammalian cells followed by flow cytometry 48 h later to quantify GFP+ cells. Although the DR-GFP assay was originally developed with the reporter integrated into the chromosome (11), the non-integrated reporter assays can speed the analysis tremendously. However, transfections must give reproducible efficiencies, which can be evaluated using a second marker that fluoresces in a different channel from GFP.
3.1. DR/TR-GFP Recombination Assays
We typically perform DR-GFP and TR-GFP assays in parallel to compare DSB and ICL-induced HR.
3.1.1. DR-GFP Assay
The day before transfection, plate U2OS cells at ~50% confluence such that on the day of transfection, they are still subconfluent. Using confluent cells reduces transfection efficiency and HR levels.
For transfection, trypsinize cells, pellet, and rinse once. Each transfection uses 5×106 cells/800 μl in Opti-MEM.
Add cells to cuvette.
Add 20 μg pDR-GFP-hprt and 20 μg pCBASce or pCAGGS to cuvette, mix cells and DNA well, but gently, and electroporate immediately at 950 μF/250 V.
Plate cells in 10-cm plates and incubate for 48 h.
Perform flow cytometry analysis. We typically get up to 10% GFP+ cells under these conditions.
3.1.2. TR-GFP Assay
The overall scheme is presented in Fig. 16.1e (see Note 4):
For triplex formation, mix 20 μg pTR-GFP-hprt with 4 μl TFO (TFO:TR-GFP), pso-TFO (pso-TFO:TR-GFP), or pso-mTFO (pso-mTFO/TR-GFP), each at a final concentration of 10 μM, and 4 μl of 10× TFO buffer, for a final volume of 40 μl.
Incubate at room temperature for 30 min to allow the triplex to form. The efficiency of triplex formation is checked using the DraI protection assay as described in Section 3.2.
Add 40 μl triplex mix to cells in cuvette and electroporate, as described in steps 1–4 of Section 3.1.1.
Plate cells in 10-cm plates and incubate for 1 h at 37°C.
Aspirate media completely, as residual phenol red in the media may absorb UVA, and rinse cells once with PBS. Be careful not to dislodge newly attached cells.
Add 1 ml PBS.
Place cells in Stratalinker and irradiate at 0.15 J/cm2 UVA. Avoid drying the cells. For unirradiated control samples, skip this step.
Add medium and incubate for 48 h at 37°C.
Perform flow cytometry analysis. We typically get up to a few percent GFP+ cells under these conditions.
3.2. DraI Protection Assay
The ICL is formed within a DraI restriction site such that the efficiency of ICL formation can be tested by resistance to DraI cleavage (Fig. 16.2b). As TFO binding also protects from DraI cleavage, the unconjugated TFO is removed by heating and then trapped by the addition of a complementary oligonucleotide (22):
After UVA irradiation (step 7 of Section 3.1.2), extract DNA from cells (QIAamp DNA Blood Mini kit). Measure the DNA concentration.
Prepare duplicates of 1 μg of each DNA preparation in 20 μl of 1X DraI buffer. In one set, have a 50 μM final concentration of complementary oligonucleotide.
Incubate at 70°C for 10 min to dissociate the TFO from the plasmid DNA. Slowly cool to room temperature. At this step, the complementary oligonucleotide binds the dissociated TFO, preventing it from reannealing to the plasmid. The excess complementary oligonucleotide captures all of the dissociated TFO.
Add 1 μl DraI to each sample and incubate for 1 h at 37°C.
Heat inactivate at 65°C for 20 min.
Run samples on a 0.8% agarose gel and perform Southern blotting.
Probe with the 800-bp HindIII fragment from pDR-GFP-hprt.
With complete ICL formation, the 2.8- and 0.65-kb DraI fragments are converted to a 3.45-kb fragment. The example shows substantial but not complete ICL formation (Fig. 16.2c).
3.3. TR-OriP-GFP Assay
As ICL repair may be coupled with DNA replication (23), the TR-GFP assay was modified so that the reporter could replicate in human cells, by adding OriP to the plasmid, forming TR-OriP-GFP (Fig. 16.1b), and expressing EBNA1 in U2OS cells (10):
The assays are identical to those described in Section 3.1.2, except that the plasmid (TR-OriP-GFP) and cells (U2OS-CEP) are different. U2OS cells can also be used as a negative control, since the TR-OriP-GFP plasmid will not replicate in those cells.
Acknowledgments
This work was supported by the Byrne Fund and National Institutes for Health grants P01CA94060 (M.J.) and R01GM54668 (M.J.).
4. Notes
This plasmid is based on pDR-GFP but additionally contains Hprt genomic sequences which can be used for gene targeting the reporter in mouse cells (18).
These plasmids contain an EBV origin of replication cloned between the GFP repeats of pDR-GFP and its derivative pTR-GFP.
These nucleotide modifications decrease the dissociation rate constant for triplex formation and confer an entropic gain (21).
The ICL can also be formed in vitro prior to transfection of pso-TFO:TR-GFP triplexes, although we usually obtain lower HR levels than with ICL formation in cells (10).
References
- 1.Guainazzi A, Schärer OD. Using synthetic DNA interstrand crosslinks to elucidate repair pathways and identify new therapeutic targets for cancer chemotherapy. Cell Mol Life Sci. 2010;67:3683–3697. doi: 10.1007/s00018-010-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinz JM. Role of homologous recombination in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:582–603. doi: 10.1002/em.20577. [DOI] [PubMed] [Google Scholar]
- 3.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 4.Moynahan ME, Cui TY, Jasin M. Homology-directed DNA repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- 5.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 6.Kraakman-van der Zwet M, et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol Cell Biol. 2002;22:669–679. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 8.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakanishi K, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, Brunet E, Jasin M. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2029. doi:10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce AJ, Jasin M. Measuring recombination proficiency in mouse embryonic stem cells. Methods Mol Biol. 2005;291:373–384. doi: 10.1385/1-59259-840-4:373. [DOI] [PubMed] [Google Scholar]
- 13.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slabicki M, et al. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol. 2010;8:e1000408. doi: 10.1371/journal.pbio.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin JY, Glazer PM. Repair of DNA lesions associated with triplex-forming oligonucleotides. Mol Carcinog. 2009;48:389–399. doi: 10.1002/mc.20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raha M, Wang G, Seidman MM, Glazer PM. Mutagenesis by third-strand-directed psoralen adducts in repair-deficient human cells: high frequency and altered spectrum in a xeroderma pigmentosum variant. Proc Natl Acad Sci USA. 1996;93:2941–2946. doi: 10.1073/pnas.93.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 21.Brunet E, et al. Exploring cellular activity of locked nucleic acid-modified triplex-forming oligonucleotides and defining its molecular basis. J Biol Chem. 2005;280:20076–20085. doi: 10.1074/jbc.M500021200. [DOI] [PubMed] [Google Scholar]
- 22.Brunet E, Corgnali M, Cannata F, Perrouault L, Giovannangeli C. Targeting chromosomal sites with locked nucleic acid-modified triplex-forming oligonucleotides: study of efficiency dependence on DNA nuclear environment. Nucleic Acids Res. 2006;34:4546–4553. doi: 10.1093/nar/gkl630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raschle M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]