SUMMARY
Breast cancer suppressor BRCA2 is critical for maintenance of genomic integrity and resistance to agents that damage DNA or collapse replication forks, presumably through homology-directed repair of double-strand breaks (HDR). Using single-molecule DNA fiber analysis, we show here that nascent replication tracts created before fork stalling with hydroxyurea are degraded in the absence of BRCA2 but are stable in wild-type cells. BRCA2 mutational analysis reveals that a conserved C-terminal site, involved in stabilizing RAD51 filaments but not in loading RAD51 onto DNA, is essential for this fork protection but dispensable for HDR. RAD51 filament disruption in wild-type cells phenocopies BRCA2 deficiency. BRCA2 prevents chromosomal aberrations upon replication stalling, which are alleviated by inhibition of MRE11, the nuclease responsible for this novel fork instability. Thus, BRCA2 prevents rather than repairs nucleolytic lesions at stalled replication forks to maintain genomic integrity, and hence likely suppresses tumorigenesis through this novel replication-specific function.
INTRODUCTION
BRCA2 is one of the two genes frequently found mutated in hereditary breast cancers, and its mutation is also associated with ovarian and pancreatic cancer in adults, as well as brain and other tumors in children with Fanconi anemia (Gudmundsdottir and Ashworth, 2006; Moynahan and Jasin, 2010). Although it may have other cellular functions, including during cell cycle progression (Ayoub et al., 2009), the role of BRCA2 is best understood during DNA double-strand break (DSB) repair by homologous recombination, also termed homology-directed repair (HDR) (Moynahan et al., 2001), where it mediates RAD51 nucleoprotein filament formation on single-stranded (ss) DNA (Jensen et al., 2010).
Human BRCA2 has 8 conserved RAD51 interaction motifs termed BRC repeats, which are essential for HDR (Moynahan and Jasin, 2010). The importance of HDR for survival is reflected in the observation that truncations of BRCA2 which include the BRC repeats are lethal in mice during embryogenesis (Moynahan, 2002). In addition to the BRC repeats, a RAD51 interaction site has been identified in the C-terminal ~200 amino acids of BRCA2 (C-ter), which is also conserved, but which is distinct in sequence from the BRC repeats (Esashi et al., 2005). While BRCA2 truncations involving only the BRCA2 C-ter region appear developmentally normal, they confer shorter life spans, increased tumorigenesis, and hematopoietic dysfunction (McAllister et al., 2002; Navarro et al., 2006; Donoho et al., 2003).
BRCA2 plays a key role in repairing DSBs arising during replication, which are repaired by HDR (Bryant et al., 2005; Lomonosov et al., 2003; Su et al., 2008). Although DSBs in principle can be repaired via non-homologous end-joining (NHEJ), HDR is a preferred pathway during the S- and G2-phases of the cell cycle when homologous chromatids are available to template the repair process (Moynahan and Jasin, 2010). DSB repair by either pathway requires end-processing, for which the MRE11 nuclease is implicated (Mimitou and Symington, 2009). To this end, MRE11 has endonuclease activity that promotes 5’-3’ resection of DNA ends critical for HDR, as well as 3’–5’ exonuclease activity (Williams et al., 2008), which may also trim DNA ends for repair. Moreover, MRE11 is rapidly recruited to nuclear foci at stalled forks upon exposure to the replication poison hydroxyurea (HU) (Wang et al., 2000).
Given BRCA2’s central function during HDR, it has been presumed that BRCA2 is required during replication perturbation due to its function in HDR (Budzowska and Kanaar, 2009; Nagaraju and Scully, 2007). Here we investigate at the molecular level the role of BRCA2 when replication is perturbed in vivo. We find that BRCA2 has a protective function during replication fork stalling that is mechanistically distinct from repair via HDR. Specifically, BRCA2 prevents degradation of nascent strands at stalled forks by MRE11. The RAD51 binding site within the BRCA2 C-ter, which stabilizes RAD51 filaments is necessary for protection of stalled forks, but it is not required for DSB repair via HDR, providing a separation of function between these two processes. BRCA2 mutants with compromised fork protection exhibit increased spontaneous and HU-induced chromosomal aberrations that are alleviated by MRE11 inhibition. Thus, these data reveal a critical role for BRCA2 in maintaining genomic stability, and likely suppressing tumorigenesis, independent of HDR.
RESULTS
BRCA2 Protects Nascent DNA Strands at Stalled Replication Forks
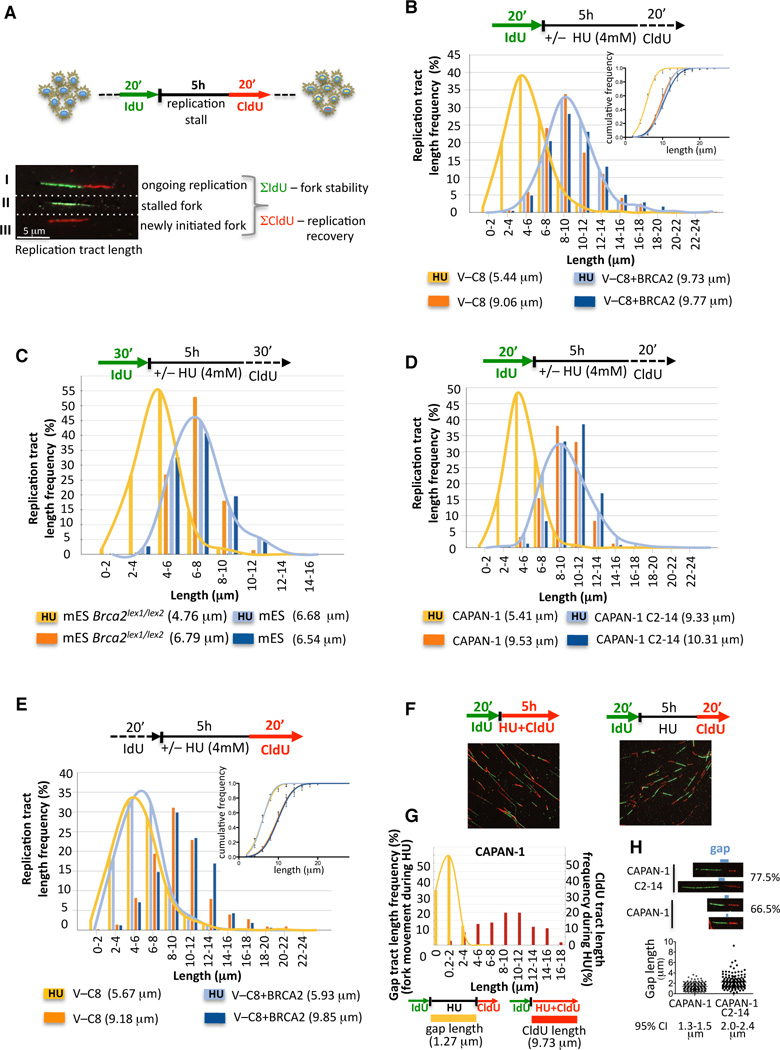
To obtain a better understanding of BRCA2’s role during DNA replication, DNA fiber analysis was utilized to monitor replication perturbation genome-wide at single-molecule resolution (Figure 1A). This procedure marks newly synthesized DNA strands just prior to (green fluorescent IdU) and after (red fluorescent CldU) exposure to HU, which transiently stalls replication by causing an imbalance in the deoxyribonucleoside triphosphate pool. Retention of the IdU label after HU treatment measures the stability of stalled forks (Σ IdU; Figure 1A).
Figure 1. BRCA2 Protects Nascent DNA Strands at Stalled Replication Forks.
(A) Schematic of single DNA fiber analysis. Green tracts, IdU; red tracts, CldU. Examples of various types of tracts are shown.
(B) IdU tract length distributions from DNA fibers from BRCA2-deficient (V-C8) and proficient (V-C8+BRCA2) hamster cells in the presence (replication stalling) or absence (unperturbed replication) of HU. Sketch above delineates experimental design. Median tract lengths are given in parentheses here and in subsequent figures. Inset, cumulative distributions.
(C) IdU tracts in Brca2lex1/lex2 and wild-type mES, with and without HU.
(D) IdU tracts in the BRCA2-deficient human CAPAN-1 and BRCA2 revertant of CAPAN-1 (C2-14), with and without HU.
(E) CldU tracts of replication after exposure to HU or media in V-C8 and V-C8+BRCA2 cells. Inset, cumulative distributions; see also Figures S1D.
(F) DNA fiber images from CAPAN-1 cells treated with HU and labeled with CldU either during HU (left panel) or after HU (right panel).
(G) Gap lengths between IdU tracts before HU and CldU tracts after HU (yellow distribution) and CldU tract lengths during HU (red distribution) in CAPAN-1 cells.
(H) Individual DNA fiber images from CAPAN-1 and C2-14 cells treated with HU and then labeled with CldU, marked to show the gap in label between the IdU and CldU labels. The % of fibers with gaps between the IdU and CldU labels is given for each cell line. Gap length frequency is shown for each cell line. See also Figure S1F.
See also Table S1 for detailed information on data sets and statistical tests, including 95% confidence interval for cumulative distributions.
In hamster cells expressing wild-type BRCA2 (V-C8+BRCA2), the median IdU tract length is maintained intact with or without HU treatment for 5 h (9.73 and 9.77 µm respectively, p=0.924, two-tailed Mann-Whitney test; Figure 1B), indicating that the integrity of stalled forks is not compromised during prolonged periods of replication stress. In contrast, nascent IdU tracts substantially shorten in cells deficient for BRCA2 (V-C8 (Wiegant et al., 2006)) when replication forks are stalled compared to unperturbed replication (5.44 and 9.06 µm, respectively, p<0.0001; Figure 1B). As nascent IdU tracts are formed prior to treatment with HU, the disappearance of IdU label occurs during HU exposure. We confirmed that IdU shortening in BRCA2-deficient cells occurs irrespective of the choice of the replication poison by replacing HU with the chemotherapeutic gemcitabine, a nucleoside analogue that inhibits DNA elongation (Figure S1A). Thus, BRCA2 functions in protecting nascent strands when replication forks are stalled.
To determine if the requirement for BRCA2 in protecting stalled replication forks is common to other cell types, we examined Brca2lex1/lex2 mouse embryonic stem (mES) cells, which express a C-terminal truncation of BRCA2 (Morimatsu et al., 1998). As with V-C8 cells, IdU tracts shorten in Brca2lex1/lex2 cells exposed to HU compared with unimpeded replication (4.76 and 6.79 µm, respectively; p=<0.0001), while the nascent tracts remain intact in wild-type mES cells (Figure 1C, Figure S1B). In addition, human CAPAN-1 cells, which express a similar BRCA2 truncation as V-C8 cells (Goggins et al., 1996), are defective in maintaining nascent tracts compared with unimpeded replication (5.41 µm and 9.53 µm, respectively; p=<0.0001; Figure 1D, Figure S1C). Reversion of the BRCA2 mutation in the CAPAN-1 cells by a nearby second-site mutation (C2-14 cells, (Sakai et al., 2008); Figure S1C) largely restores the protection of the stalled replication forks (Figure 1D). These results indicate that BRCA2 is required for the protection of stalled replication forks in multiple mammalian cell lines.
Recovery after Replication Stalling in BRCA2-Deficient Cells
Depletion of RAD51 has been reported to decrease replication restart after HU (Petermann et al., 2010) and uncouple leading and lagging-strands (Hashimoto et al., 2010). To address whether replication recovery is affected when BRCA2 is absent, CldU tract lengths formed after HU exposure were measured. Replication is substantially slowed after HU treatment, but similarly so in both control and BRCA2-deficient cells (5.93 and 5.67 µm, V-C8+BRCA2 and V-C8 after HU, respectively; Figure 1E). In these experiments, the IdU and CldU labels are often quite separated, indicating that forks stall only transiently with HU in these hamster cells. Therefore, we assessed the frequency of replication restart (IdU→CldU) when CldU was present during HU. As with replication recovery after HU, we find no significant difference between BRCA2-proficient and deficient hamster cells (Figure S1D). Further, the frequency of new replication tracts is also unaffected by BRCA2 status (Figure S1D).
Given that replication restart may be more stringently controlled in human cells than rodent cells (Petermann et al., 2010), we also assessed restart in CAPAN-1 and revertant cells. Consistent with a more efficient block in human cells, IdU and CdU tracts are more frequently joined when the CldU label is added after HU exposure than they are in hamster cells. This apparent replication restart (IdU→CldU) is not significantly impaired under this condition with or without BRCA2 (Figure S1E), although we did observe a small increase in tracts labeled only with IdU in CAPAN-1 cells (13% to 19%, p=0.279; Figure S1E).
We noticed that most restart tracts have small gaps between the IdU and CldU labels (Figure 1F, right panel), indicating limited fork progression during exposure to HU. A larger portion of tracts from CAPAN-1 C2–C14 cells have gaps (77.5%) compared with CAPAN-1 cells (66.5%), implying that the block to restart is more efficient in the absence of BRCA2 (p=0.0134, Figure 1H). To examine transient replication restart during HU, we included CldU during HU in a control experiment in CAPAN-1 cells and found that ~97% of forks continue to incorporate CldU label even in the presence of HU (Figure 1F, left panel). Surprisingly, the continuous CldU tracts formed during HU are substantially longer than the gaps between the two labels in experiments where the label is omitted during HU (compare 9.73 µm CldU tract length during HU and 1.27 µm gap length during HU without label, Figure 1G). We interpret this data to reflect asymmetric replication fork movement during HU, indicating leading and lagging strand uncoupling (Figure S1F). We reasoned that gap tract lengths can be used to assess the ability of both strands to continue replication during HU. Gap lengths are smaller in CAPAN-1 compared to CAPAN-1 C2-14 cells (p=<0.0001, Figure 1H), providing evidence that BRCA2 can suppress uncoupling of the leading and lagging strands.
Nucleolytic Degradation of Stalled Forks Is Progressive and Has Distinct Directionalities
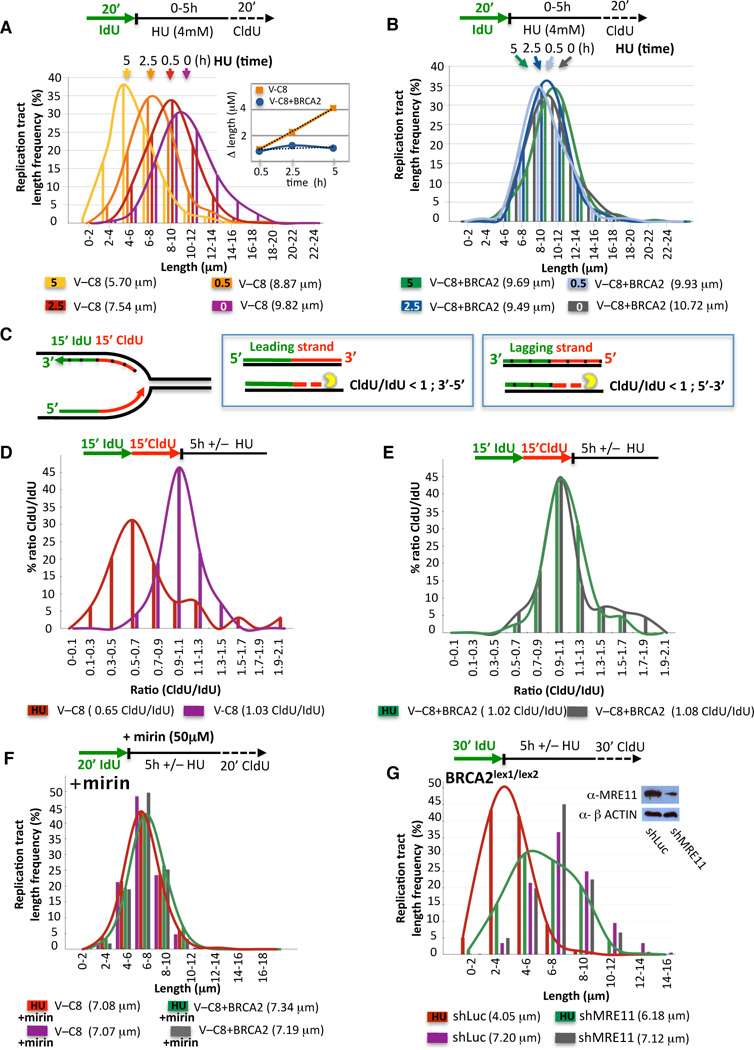
To better understand the mechanism of replication tract shortening during fork stalling, we monitored the integrity of the nascent strands by varying the exposure times to HU. Consistent with nucleolytic degradation of the stalled forks, preformed IdU tracts in V-C8 cells progressively shorten during HU (Figure 2A and inset, Figure S2A), with a rate of 0.7 µm/h, corresponding to ~1.8 kb/h. By contrast, preformed IdU tracts in cells with BRCA2 remain largely unchanged (Figure 2B, Figure S2B).
Figure 2. Inhibition of MRE11 Alleviates Nucleolytic Degradation of Stalled Forks.
(A–B) Preformed IdU tract lengths in V-C8 (A) and V-C8+BRCA2 (B) cells during different exposure times to HU. Inset, the rate of IdU tract length change is 0.7 µm/h, estimated to be ~1.8 kb/h.
(C) Sketch of design and expected outcome of nuclease directionality test. Tick marks delineate lagging strands.
(D–E) Distribution curves of the ratio of CldU/IdU tract lengths with or without HU in V-C8 (D) and V-C8+BRCA2 (E) cells.
(F) IdU tract lengths in V-C8 and V-C8+BRCA2 cells with or without HU in the presence of the MRE11 inhibitor mirin.
(E) IdU tract lengths in Brca2lex1/lex2 mES cells after shRNA treatment directed against MRE11 or control (shLuc) with or without HU. Western blot inset shows the MRE11 knockdown.
To fine map the degradation, we consecutively labeled nascent tracts with IdU and CldU for equal periods of time before HU (Figure 2C). In unperturbed V-C8 cells, IdU and CldU tracts are similar in length (CldU/IdU=1.03; Figure 2D). When challenged with HU, CldU tracts shorten while IdU tracts remain intact (CldU/IdU=0.65; Figure 2D, Figure S2C), indicating that the more recently synthesized DNA is degraded first. Thus, leading strands are degraded 3’–5’, while lagging strands are degraded 5’-3’ (Figure 2C). In contrast, the CldU/IdU ratio in VC-8+BRCA2 cells is ~1 with or without HU (Figure 2E, Figure S2C). These results imply that BRCA2 protects against degradation of stalled replication forks with opposite directionalities for the leading and lagging strands.
MRE11 Is Responsible for Nascent Strand Shortening at Stalled Forks
The slow kinetics of degradation (~1.8 kb/h) are reminiscent of another controlled degradative process, that of DNA end resection (~4 kb/h in yeast; (Fishman-Lobell et al., 1992)). We considered that MRE11, which possesses 3’–5’ exonuclease activity and also promotes 5’-3’ end resection (Mimitou and Symington, 2009; Williams et al., 2008), could promote fork degradation in the absence of BRCA2. To test this, we used mirin, a chemical inhibitor of MRE11 nuclease activity (Dupre et al., 2008). With mirin, IdU tracts are similar in length irrespective of replication stalling in both BRCA2-deficient and proficient cells (Figure 2F, Figure S2D), suggesting that the MRE11 nuclease degrades stalled forks in the absence of BRCA2. To exclude off-target effects by the inhibitor, we expressed shRNA against MRE11 in Brca2lex1/lex2 cells (Figure 2G), which like mirin, substantially protects the nascent tracts during HU (6.18 and 4.05 µm with and without MRE11 knockdown, respectively; p=<0.0001; Figure 2F). These results further implicate the MRE11 complex, specifically its nuclease activity, in fork degradation in the absence of BRCA2.
Replication Fork Protection Is Independent of Canonical NHEJ and the HDR Protein RAD54
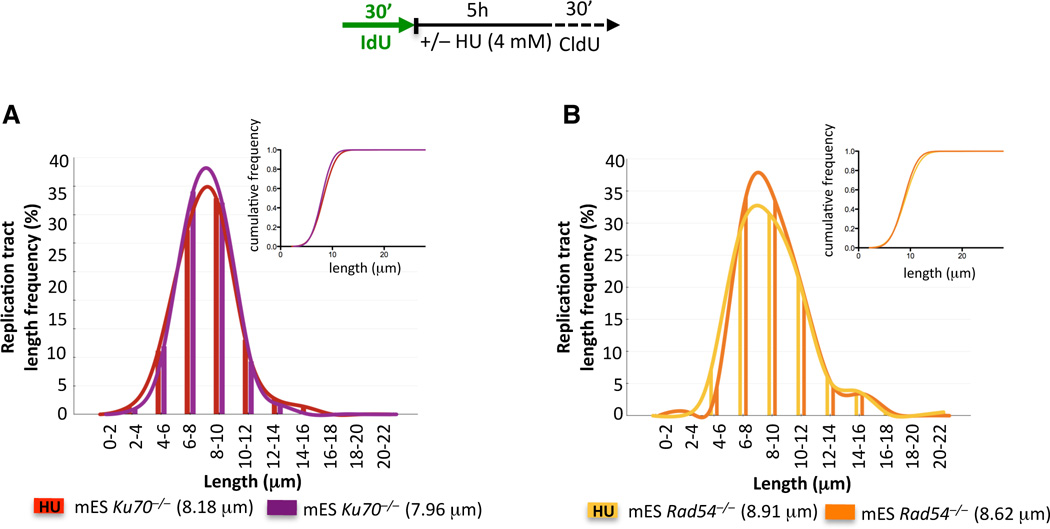
The KU heterodimer, a key component of canonical NHEJ, is important in the protection of DNA ends from nucleolytic digestion (Kass and Jasin, 2010). We tested whether loss of KU would also lead to deprotection of DNA ends at stalled replication forks. Nascent IdU tracts are maintained intact in Ku70−/− mES cells (Figure 3A), consistent with a specific role for KU at DSBs, but not at stalled forks (Pierce et al., 2001).
Figure 3. RAD54 and KU70 Deficiency Do Not Affect the Stability of Nascent Strands at Stalled Replication Forks.
IdU tracts in Rad54−/− (A) and Ku70−/− (B) mES cells with or without HU. Inset, cumulative distributions.
We next examined whether protection of stalled forks is a property of all HDR proteins. RAD54 acts during late steps of HDR (Heyer et al., 2006), downstream of BRCA2-mediated RAD51 nucleoprotein filament formation. Yet, RAD54 is not evidently involved in fork protection, as Rad54−/− mES cells exhibit similar IdU tract lengths with or without HU (Figure 3B), thus indicating that not all HDR components are required to avoid fork degradation.
Domain Requirements for BRCA2 in Replication Fork Protection
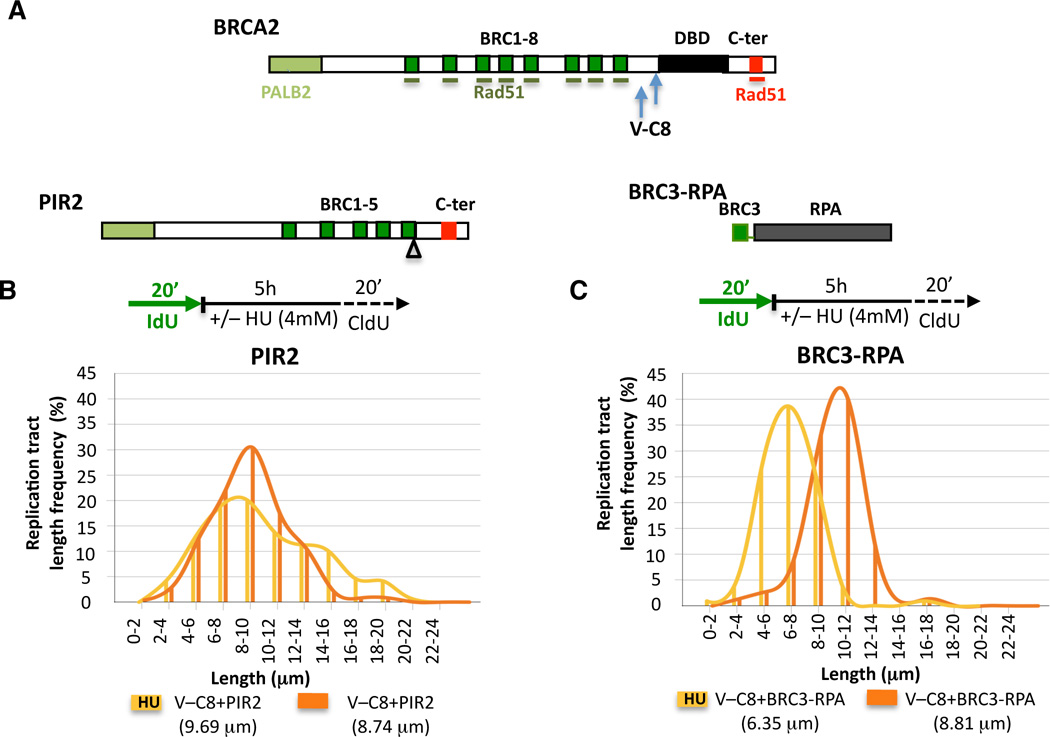
BRCA2 contains both protein and DNA interaction domains, including several BRC repeats which bind RAD51, a DNA binding domain (DBD) consisting of several DNA binding modules, and a C-terminal site (C-ter) which also binds RAD51 (Figure 4A). Given the multidomain structure of BRCA2, we sought to characterize the domain requirements for replication fork stability.
Figure 4. BRCA2 Domain Analysis Reveals Differences in Fork Stability and HDR.
(A) Graphical sketch of human BRCA2, PIR2, and BRC3-RPA. PALB2-interaction site (bright green), BRC repeats (dark green bars), DNA binding domain (DBD, black bar), C-terminal region (C-ter) with non-BRC RAD51 binding site (red bar). Arrows indicate truncations in V-C8 cells.
(B–C) IdU tracts in V-C8 cells stably expressing the PIR2 (B) or BRC3-RPA (C) peptides with or without HU.
V-C8 cells have two Brca2 alleles encoding proteins truncated for the C-ter and DBD domain (Figure 4A) (Wiegant et al., 2006) and are defective in both HDR (Saeki et al., 2006) and maintenance of fork stability (Figure 1B). As BRCA2 could directly inhibit degradation via protein-DNA interactions requiring the DBD domain, we generated a V-C8 cell line stably expressing a BRCA2 peptide missing the entire DBD domain (PIR2, Figure 4A), but which is proficient at HDR (Figure S3A; (Edwards et al., 2008)). No substantial difference in IdU tract lengths was observed with or without HU (9.69 and 8.74 µm, respectively; Figure 4B, Figure S3B). Thus, the BRCA2 DBD is not required for the protection of stalled replication forks, indicating that it is unlikely that BRCA2 directly inhibits nucleolytic degradation by binding to nascent DNA.
V-C8 cells stably expressing a peptide consisting of one BRC repeat fused to the large subunit of the ssDNA-binding protein RPA (BRC3-RPA, Figure 4A) are proficient in HDR (Saeki et al., 2006), suggesting that the main function of BRCA2 in HDR is to deliver RAD51 to ssDNA. These cells, however, exhibit shorter nascent IdU tract lengths when replication is stalled with HU (6.35 and 8.81 µm with and without HU, respectively, p<0.0001; Figure 4C, Figure S3C). Although the degradation is not as extensive as with V-C8 cells (Figure 1B), these results suggest that the delivery of RAD51 to ssDNA is not sufficient protect stalled forks.
Highly Conserved BRCA2 C-ter RAD51 Interaction Site Is Essential for Replication Fork Stability but Dispensable for HDR
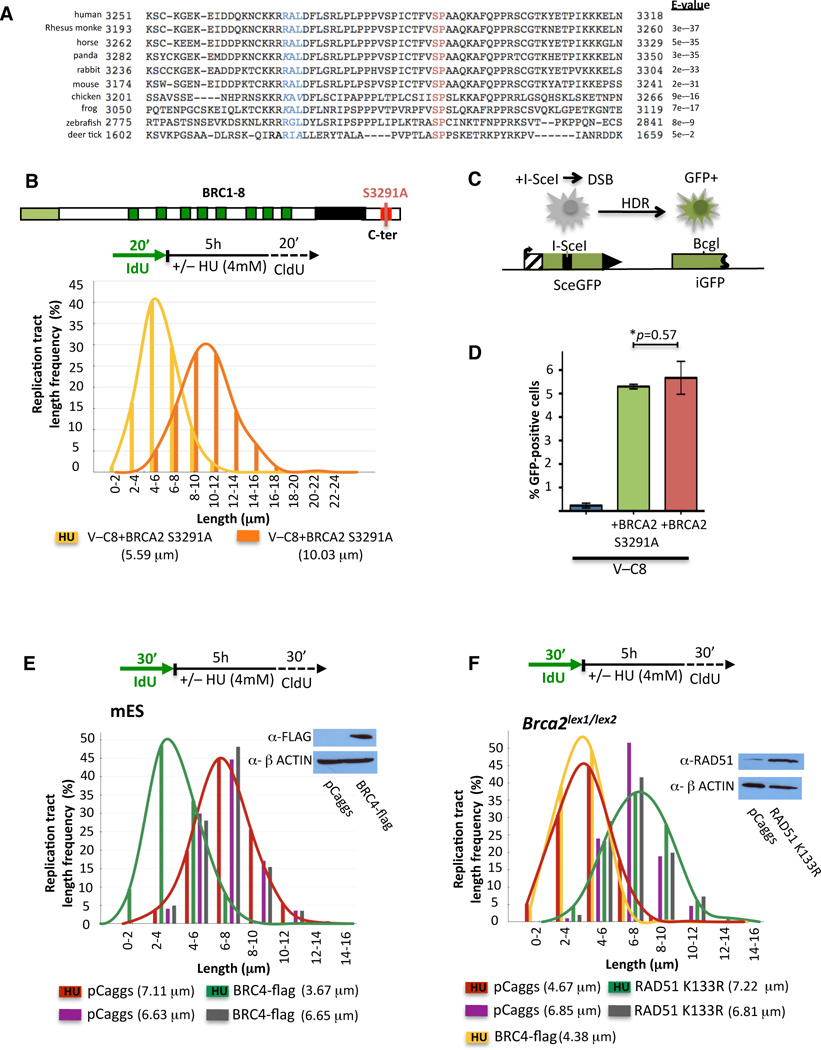
BRCA2 interacts with RAD51 through both the BRC repeats and the C-ter. While RAD51 loading through the BRC repeats does not seem to be sufficient for the protection of stalled forks, the conserved C-ter appears to provide an essential function, given our results with the Brca2lex1/lex2 cells (Figure 1C). The C-ter binds RAD51 differently than the BRC repeats in that it interacts with RAD51 oligomers and stabilizes RAD51 filaments (Davies and Pellegrini, 2007; Esashi et al., 2007). Moreover, the RAD51 binding site at the C-ter contains a cyclin dependent kinase (CDK) phosphorylation consensus sequence that is conserved throughout vertebrates (Esashi et al., 2005) as well as some invertebrates (Figure 5A, Figure S4A); phosphorylation by CDK at this site (S3291) abrogates the C-ter-RAD51 interaction (Davies and Pellegrini, 2007; Esashi et al., 2005), thereby promoting RAD51 filament disassembly which in turn promotes entry into mitosis (Ayoub et al., 2009).
Figure 5. Highly Conserved BRCA2 C-ter is Essential in Maintaining Replication Fork Stability by Stabilizing RAD51 Filaments.
(A) Blast alignments of BRCA2 C-terminal sequences; CDK phosphorylation target motif (SP, red letters); cyclin A recognition motif (RxL, blue; similar amino acids are in italics). See also Figure S4A.
(B) IdU tracts in V-C8+BRCA2 S3291A cells with or without HU.
(C) HDR assay for a DSB using the DR-GFP reporter.
(D) HDR assay in V-C8 and BRCA2 BAC-complemented cells. GFP-positive cells indicate HDR events after I-SceI expression. Error bars indicate standard deviation; p-value derived from two-tailed Student’s t-test.
(E) IdU tracts after BRC4 peptide expression in mES cells with or without HU. pCaggs, empty vector. Western blot inset shows BRC4 transient expression.
(F) IdU tracts after RAD51 K133R or BRC4 expression in Brca2lex1/lex2 mES cells with or without HU. Western blot inset shows RAD51 K133R transient expression.
To further investigate the function of the C-ter, we utilized V-C8 cells stably expressing full-length BRCA2 containing the S3291A mutation which, like phosphorylation, disrupts RAD51 binding at this site (Davies and Pellegrini, 2007; Esashi et al., 2005). As with the Brca2lex1/lex2 cells, IdU tracts shorten with HU (5.59 and 10.03 µm with and without HU, respectively, p=<0.0001; Figure 5B, Figure S4B), implicating RAD51 interaction at the C-ter in the protection of stalled replication forks, perhaps through the stabilization of RAD51 filaments.
To determine the relationship between the protection of stalled forks and HDR, we quantified HDR in V-C8+BRCA2 S3291A cells using the DR-GFP reporter, where a DSB introduced by the I-SceI endonuclease followed by HDR leads to GFP-positive cells (Figure 5C). V-C8 cells are highly defective in HDR compared with V-C8+BRCA2 cells (~25-fold; Figure 5D;(Saeki et al., 2006)). Importantly, V-C8+BRCA2 S3291A cells are as efficient for HDR as those expressing wild-type BRCA2. Thus, while the BRCA2 C-ter RAD51 interaction site is essential for the protection of stalled replication forks, it is dispensable for HDR, providing a clear separation of function mutation for the two processes.
BRCA2 Maintains Nascent Replication Tracts by Stabilizing RAD51 Filaments
To directly test the involvement of RAD51 in the protection of stalled forks, we expressed the BRC4 peptide (Saeki et al., 2006), which suppresses DNA binding of RAD51 and thus perturbs RAD51 filaments (Davies et al., 2001; Hashimoto et al., 2010). As with defective BRCA2, expression of the BRC4 peptide in wild-type mES cells leads to substantially shorter IdU tracts upon HU (3.67 and 6.65 µm with and without HU, respectively, p=<0.0001; Figure 5E, Figure S4D). These data indicate that disruption of RAD51 filaments leads to nascent strand degradation.
We next asked whether stabilization of filaments could suppress this degradation. ATP hydrolysis by RAD51 is required for efficient dissociation from DNA while association with DNA is unaffected (Benson et al., 1994; van Mameren et al., 2009). RAD51 K133R, which is devoid of ATPase activity, forms stable filaments and promotes strand exchange in vitro (Morrison et al., 1999) while suppressing turnover or completion of successful HDR in vivo (Stark et al., 2002). Over-expression of RAD51 K133R in Brca2lex1/lex2 cells exposed to HU renders IdU tracts resistant to degradation (7.22 and 6.81 µm with and without HU, respectively, p=0.077; Figure 5F, Figure S4E), indicating that stabilized RAD51 filaments rescue the protection of stalled forks and consistent with the requirement for the C-ter in this process. Expression of BRC4 has no effect in these cells (Figure 5F, Figure S4E), suggesting that BRCA2 and RAD51 are epistatic for fork protection.
Replication Fork Stalling Leads to MRE11-Dependent Genomic Instability in BRCA2-Deficient Cells
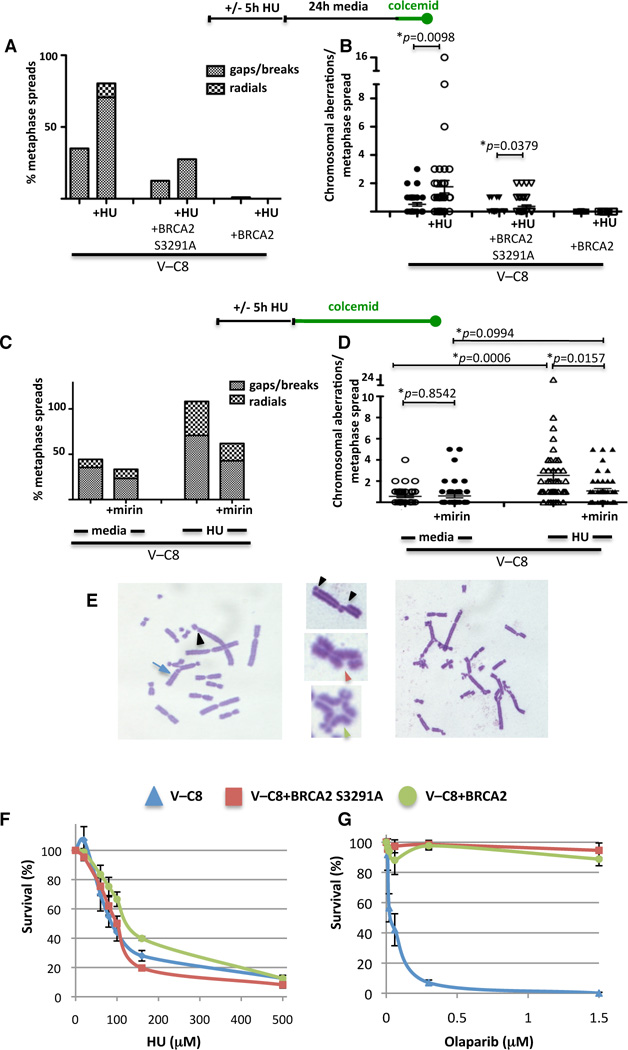
We next sought to determine the physiological consequences of replication perturbation in cells that cannot maintain the integrity of stalled replication forks. Both V-C8 and V-C8+BRCA2 S3291A cells exhibit elevated spontaneous chromosomal abnormalities compared to cells with wild-type BRCA2, revealing intrinsic genomic instability in cells expressing BRCA2 S3291A (Figure 6A). Exposure to HU substantially increases the average number of aberrations per metaphase spread in both cell lines, from 0.51 to 1.77 for V-C8 cells and from 0.12 to 0.37 for V-C8+BRCA2 S3291A cells (Figure 6B). Thus, replication stalling induces chromosomal aberrations in cells which are defective in protecting forks from degradation.
Figure 6. Replication Fork Stalling Leads to Genomic Instability in BRCA2-Deficient Cells.
(A–B) Chromosomal aberrations with or without HU treatment in the indicated V-C8 cell lines. Sketch above the graphs delineates experimental design. The % of metaphase spreads with the indicated aberrations (A) and the number of chromosome aberrations per metaphase (B) are plotted. p-value derived from two-tailed Student’s t-test.
(C–D) Chromosomal aberrations with or without HU in V-C8 cells in the presence or absence of the MRE11-inhibitor mirin. Sketch above the graph delineates experimental design. The % of metaphase spreads with the indicated aberrations (C) and the number of chromosome aberrations per metaphase (D) are plotted.
(G) Chromosomal aberrations in V-C8 cells exposed to HU include breaks (black arrowheads), gaps (red arrowhead), triradials, quadradials (green arrowhead) and other translocation (blue arrow).
(F–G) Survival of indicated V-C8 cell lines upon continuous exposure to HU (F) and the Parp-inhibitor olaparib (G).
Treatment of cells with HU has been reported to lead to DSB formation after prolonged exposure times, while treatment with HU for a few hours, as performed here, does not cause such lesions (Hanada et al., 2007; Petermann et al., 2010). Given that the degradation of forks is not correlated to HDR (Figure 5D), we sought to further distinguish genomic instability arising from DSBs from genomic instability arising from degraded forks by treating cells with colcemid immediately after HU exposure. As with delayed colcemid treatment, breaks/gaps increase in V-C8 cells with HU exposure, and triradial/quadriradial chromosomes increase even further (Figure 6C and 6E), suggesting that unprotected replication forks expose potential sites for aberrant interchromosomal interactions. Overall, the average number of chromosomal aberrations per metaphase in V-C8 cells increases from 0.56 to 2.5 with HU (Figure 6D). Similarly, aberrations increase with HU in V-C8+BRCA2 S3291A cells, while the number of aberrations in cells expressing wild-type BRCA2 is substantially lower (Figures S5A–C). Thus, cells in which stalled replication forks degrade exhibit greater genomic instability.
As the MRE11-inhibitor mirin alleviates degradation of stalled forks in BRCA2-deficient cells (Figure 2F), we asked whether inhibition of MRE11 could also alleviate HU-induced chromosome aberrations. Both breaks/gaps and radial chromosomes were reduced in V-C8 cells treated with HU and mirin compared to HU alone (Figure 6C). Overall, the average number of chromosomal aberrations per metaphase decreased more than 2-fold when MRE11 nuclease activity was inhibited during fork stalling (1.07 and 2.54 with and without mirin, respectively, p=0.015; Figure 6D). As inhibiting the degradation of stalled replication forks in BRCA2-deficient cells is associated with reduced numbers of chromosome aberrations, these data support a relationship between degradation of stalled replication forks by MRE11 and genomic instability.
Replication Stalling In BRCA2-Deficient Cells Does Not Reduce Cellular Survival
To investigate long-term effects of stalled replication, cell survival was examined following exposure to agents that have differential effects on replication. After continuous exposure to HU, V-C8 cells and V-C8+BRCA2 S3291A show only modest sensitivity to HU relative to cells expressing wild-type BRCA2 (Figure 6F), suggesting that fork degradation has little effect on cell survival. We also pulsed cells under the conditions used in the DNA fiber experiments, and again saw no significant difference in survival (data not shown). Thus, although chromosomal aberrations increase, cell survival is not compromised, indicating that BRCA2 deficiency will be associated with increased mutagenesis when replication is perturbed.
V-C8 cells are exquisitely sensitive to poly(ADP-ribose) polymerase (PARP) inhibitors like olaparib (Figure 6G; (Farmer et al., 2005)). By contrast, cells expressing either wild-type BRCA2 or the S3291A mutant are similarly resistant to this drug (Figure 6G). Given that PARP inhibition leads to ssDNA breaks which are converted to DSBs during S phase, efficient repair of these DSBs is consistent with the BRCA2 S3291A mutant being proficient at DSB repair by HDR. Supporting this, similar results were obtained with other agents that require HDR for their repair, i.e., 6-thioguanine and mitomycin C (Figures S5D and S5E).
DISCUSSION
BRCA2 Protects Stalled Replication Forks by Blocking Degradation
As HDR is important during perturbed DNA replication, it has been presumed that the key function of BRCA2 in this regard is to promote repair of collapsed replication forks via HDR (Figure S6A). We show here that BRCA2 prevents rather than repairs nucleolytic lesions at stalled forks, revealing a conceptual and mechanistic difference with consequences for tumorigenesis, as discussed below. This novel role for BRCA2 in protecting newly replicated strands at stalled replication forks from degradation was unforeseen. Importantly, using a separation of function BRCA2 mutant, we find that fork protection is distinct from HDR, yet critical in maintaining genomic stability when DNA replication is stressed with HU.
Requirement for Evolutionary Conserved BRCA2 C-ter in Stabilizing RAD51 Filaments during Replication Stalling
BRCA2 is crucial for RAD51 filament formation on ssDNA (Jensen et al., 2010; Moynahan and Jasin, 2010). To this end, BRCA2 binds monomeric RAD51 via its BRC motifs (Davies and Pellegrini, 2007), directing RAD51 onto ssDNA while preventing RAD51 from nucleating filaments on dsDNA (Jensen et al., 2010). We show here that fork protection specifically requires a RAD51 interaction site at the BRCA2 C-terminus (C-ter), which differs from BRC motifs in that it stabilizes RAD51 filaments by binding to the interface of two adjacent RAD51 molecules (Davies and Pellegrini, 2007; Esashi et al., 2007). Our results extend the recent observation of a role for RAD51 in protecting nascent strands from degradation (Hashimoto et al., 2010) and provide critical mechanistic insight. Specifically, we show that RAD51 filaments stabilized with an ATPase–defective RAD51 reverse the degradation of stalled forks in a BRCA2 mutant. Conversely, conditions that destabilize RAD51 filaments, such as BRC4 expression or BRCA2 C-ter mutation, render cells susceptible to fork degradation. RAD51 filament dissociation is triggered by ATP hydrolysis in conjunction with the release of tension stored within the extended DNA strand of the nucleoprotein filament (van Mameren et al., 2009). As replication fork structures exert force on the DNA (Postow et al., 2001), they may require greater efforts at filament stabilization than frank DSBs. As the C-ter site is highly conserved throughout vertebrates and even invertebrates, it may reflect an evolutionarily conserved mechanism for the protection of stalled forks.
MRE11 Nuclease and Stalled Replication Fork Degradation
Our results imply that fork degradation occurs 3’–5’ on leading strands and 5’-3’ on lagging strands promoted by MRE11. The intrinsic nucleolytic activities of MRE11 (Williams et al., 2008) may be sufficient to promote fork degradation; alternatively, MRE11 could act in conjunction with other nucleases. For example, as part of the MRN complex MRE11 could initiate degradation of nascent lagging strands, while other 5’-3’ nucleases (EXO1, CtIP) could be involved in extensive degradation, akin to DNA end resection (Mimitou and Symington, 2009). Additionally, MRE11 could interact with other nucleases for leading strand degradation, for example, WRN, which possesses 3’–5’ exonuclease activity that is stimulated by MRE11 (Cheng et al., 2004) and which is recruited by MRN to replication foci upon HU treatment (Franchitto and Pichierri, 2004).
Although Exo1 degrades stalled replication forks that reverse into “chicken foot” structures in checkpoint defective yeast cells (Cotta-Ramusino et al., 2005), checkpoint activation has been reported to be intact in BRCA2-deficient cells (Lomonosov et al., 2003). Given that another consequence of MRE11 inhibition is disruption of ATM signaling (Dupre et al., 2008), it is possible that EXO1 promotes resection of lagging strands in these checkpoint-compromised cells. Such strand selective degradation would create ssDNA stretches not immediately suitable for aberrant NHEJ, which could explain the observed decrease in HU-induced radial structures when MRE11 is inhibited. Whether directly or indirectly, MRE11 promotes deleterious strand processing at stalled forks in BRCA2-deficient cells.
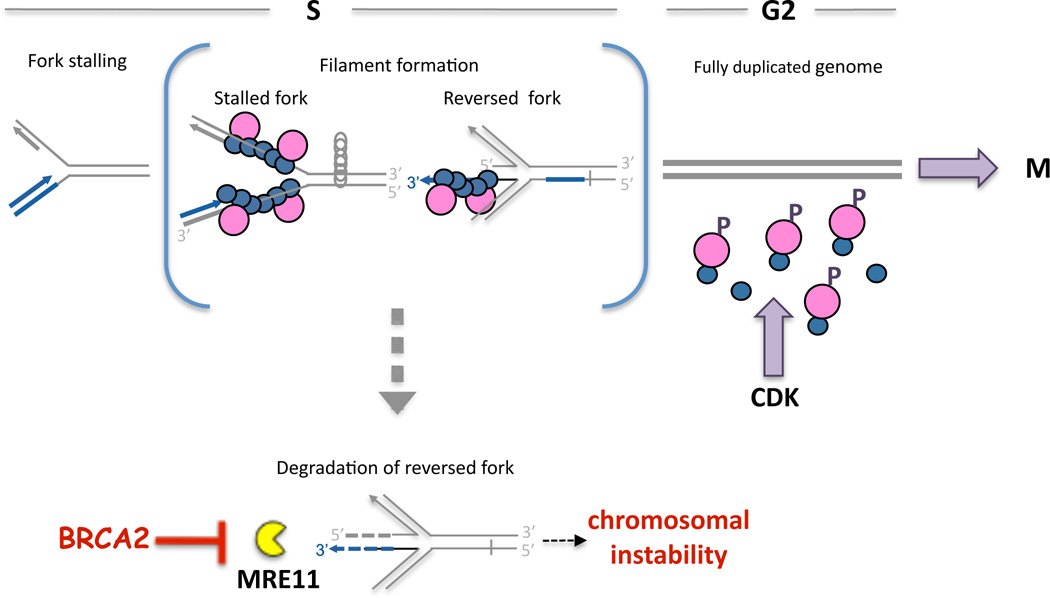
BRCA2-Mediated Fork Protection
Given our data, we propose a model in which BRCA2 prevents nucleolytic attack of stalled replication fork intermediates that have reversed to form a “chicken foot” structure (Figure 7, reversed fork). Fork reversal regresses the stalling site back into the duplex region where, in the case of a template lesion, it is accessible for nucleotide or base excision repair. If the leading strand has progressed farther than the lagging strand, fork reversal will result in a 3’ ssDNA overhang, which can be extended by additional end-resection to support RAD51 filament assembly. Alternatively, blocked leading strands with continued lagging strand synthesis produces 5’ protruding ends upon fork reversal that are extendable by MRE11-promoted processing (Figure S6B). As BRCA2 binds to 3’ and 5’ tailed DNA substrates, promotes RAD51 filaments on both 3’ and 5’ ssDNA overhangs (Mazloum and Holloman, 2009), and blocks RAD51 nucleation on dsDNA (Jensen et al., 2010), BRCA2, by stabilizing RAD51 filaments, can probably limit both 3’–5’ degradation of the reversed leading strand (Figure 7, broken blue arrow) as well as excessive 5’-3’ processing of the lagging strand (Figure 7, broken grey bar).
Figure 7. Model for HDR-independent Role of BRCA2 During DNA Replication.
A replication fork can stall for various reasons, including insufficient pools of nucleotides as from HU. BRCA2 (pink circle) stabilizes RAD51 filaments on stalled replication forks (blue circles), thereby preventing fork reversal promoted by positive supercoils ahead of the fork (grey circles). Alternatively, stabilized filaments directly protect a reversed fork. Once the replication stall is removed, genome duplication can proceed until completed. BRCA2 is then no longer required for fork protection, such that CDK phosphorylates the BRCA2 C-ter, allowing residual RAD51 filaments to dissociate to promote progression into M-phase. Importantly, in the absence of BRCA2, nascent strands of the stalled fork are unprotected and degraded by MRE11 (yellow pacman), leading to chromosomal instability.
Continued unwinding at a stalled fork can result in positive supercoiling of the parental strands upfront of the fork, providing sufficient energy for fork reversal in the absence of the replisome (Postow et al., 2001). However, in yeast extensive fork reversal is observed as a consequence of defective checkpoint signaling, such that reversed forks are considered pathological structures which are rare in wild-type backgrounds (Foiani et al., 2000; Sogo et al., 2002). Besides or in addition to protection of reversed forks, therefore, BRCA2 could stabilize the replisome and the stalled fork structure itself to prevent formation of reversed forks, which could then be degraded (Figure 7). For template lesions, RAD51-covered forks could then recruit translesion synthesis polymerases, similar to what is thought to occur in Escherichia coli when error-free repair is not possible (Schlacher and Goodman, 2007).
The protective role of BRCA2 is independent of HDR, but we propose that it could relate to the licensing of cell-cycle progression (Figure 7). In the presence of stalled replication forks, CHK1-mediated inhibition of CDK prevents phosphorylation of BRCA2 C-ter. Eventually when dNTP pools are restored or lesions are removed, replication can restart, through fork regression or other means. Once the genome is fully duplicated and stable RAD51 filaments are no longer required to protect the nascent DNA strands, CDK phosphorylates the BRCA2 C-ter, thus promoting RAD51 disassembly, which has been shown to promote entry into mitosis (Ayoub et al., 2009).
Recovery after Replication Stalling in BRCA2-Deficient Cells
We did not observe any obvious defect in replication recovery or initiation of new replication tracts with disruption of either BRCA2 (Figure 1E and Figure S1D) or RAD51 in rodent cells (Figure S4F), seemingly contradicting a previous report implicating RAD51 in replication restart in a human cell line (Petermann et al., 2010). Formally, this could be due to differences in cell systems, as replication in rodent cells does not appear to be as stringently inhibited by HU as in human cells, and indeed in BRCA2-deficient human cells we did observe a small, although insignificant, decrease in forks that were dual-labeled after exposure to HU (Figure S1E).
Upon further examination, however, we provide evidence that fork uncoupling increases with HU in BRCA2–deficient cells, extending recent findings implicating RAD51 in this process (Hashimoto et al., 2010). The reported decrease in apparent replication restart in RAD51-depleted human cells (Petermann et al., 2010), therefore, could also reflect increased uncoupling of leading and lagging strands when monitoring progression of one strand only. Importantly, inhibition of MRE11 does not affect fork uncoupling (Hashimoto et al., 2010), while we observe fewer aberrations by inhibiting MRE11. Therefore, fork uncoupling, which is a frequent event in wild-type cells (Sogo et al., 2002), may have minor consequences on chromosomal stability compared to nascent strand degradation described here.
Unprotected Replication Forks are a Source of Chromosomal Instability
We observe that while replication stress does not substantially compromise survival of BRCA2-deficient cells, it does lead to gross chromosome aberrations, including breaks and radial chromosome structures. The lesions created by degradation are structurally distinct from bona fide DSBs such as created by inhibition of PARP, which causes severe cell death in HDR deficient cells. As replication stress occurs with oncogene activation (Negrini et al., 2010), our results imply that BRCA2-deficient cells are likely particularly vulnerable to oncogene activation for the generation of chromosome aberrations leading to tumorigenesis. Whether cells die or undergo mutagenesis has important consequences with regard to tumorigenesis. For example, intestinal BRCA2 deficiency does not promote tumor formation due to elimination of cells by apoptosis (Hay and Clarke, 2005). By contrast, survival of cells with acquired mutations, such as those arising during replication stalling as shown here, encourages initiation and progression of tumorgenesis. In support of fork degradation-dependent, HDR-independent mechanisms of tumorigenesis, mutations affecting the BRCA2 C-ter CDK phosphorylation site are found in individuals affected with breast cancer (Esashi et al., 2005).
We propose that the HDR-independent role of BRCA2 in preventing the degradation of stalled replication forks and the resultant chromosome rearrangements has important implications for therapy. BRCA2 mutants that are defective in maintaining fork stability but remain proficient for HDR will be insensitive to chemotherapeutics such as PARP inhibitors, which specifically exploit the defect in repair of DSBs. Moreover, chemotherapeutics that elicit transient replication stress may not only be ineffective in killing cells but also induce mutagenesis and genomic instability. Collectively, these results and concepts change our understanding of BRCA2 and MRE11 during replication and repair.
EXPERIMENTAL PROCEDURES
Cell lines, chemicals and drugs
Hamster cells, including V-C8 (Wiegant et al., 2006), V-C8+BRC3-RPA (Saeki et al., 2006), V-C8+PIR2, and V-C8 cells complemented with human BACs (V-C8+BRCA2 and V-C8+BRCA S3291A, see Supplemental Information), were grown in DMEM with 10% FBS. CAPAN-1 and CAPAN-1 C2-14 (Sakai et al., 2008) were grown in RPMI 1640 with 20% FBS and 1 mM sodium pyruvate. Brca2lex1/lex2 (Morimatsu et al., 1998), Ku70−/− (Gu et al., 1997), and Rad54−/− (Essers et al., 1997) mES cells were cultured in standard ES media. Brca2lex1/lex2 cells express a BRCA2 peptide deleted for the C-ter ~200 amino acids encoded by exon 27. Except for olaparib (Selleck Chemicals), all chemicals were from Sigma-Aldrich.
DNA fiber assay
Cells were labeled with IdU (50 µM), followed by exposure to hydroxyurea (4 mM), gemcitabine (1 µM), or untreated media, and chased with CldU (50 µM). DNA fibers were essentially spread as described (Jackson and Pombo, 1998) before standard detection of IdU and CldU tracts (primarys: α-IdU, α-BrdU from BD Biosciences; α-CldU, α-BrdU from Novus Biologicals and secondarys: Alexa Fluors 488 and 555, respectively, from Invitrogen). Fibers were imaged (Olympus BX60 microscope) and analyzed using ImageJ software. Statistics were calculated using Prism software (see Table S1). The rate for nascent tract replication was estimated using the conversion of 2.59 kb/µm (Jackson and Pombo, 1998).
Cell Transfections, Survival Assays, and Metaphase Spreads
For HDR assays, 5 × 106 V-C8 cells or derivatives were transfected with 50µg pCBASce, the I-SceI expression vector, by electroporation at 280V, 1000µF (Saeki et al., 2006) and 48 h later, GFP-positive cells were quantified using a FACScan and CellQuest software (Becton Dickinson). For other assays, 5 × 106 or 10 × 106 mES cells were transfected with expression vectors for BRC4 (BRCA2 aa 1511–1578) (Saeki et al., 2006), RAD51K133R (Stark et al., 2002) and MRE11 shRNA (5’ACAGGAGAAGAGATCAACT in pSuper) by electroporation at 250V, 950 µF. Expression was assessed 36 h after transfection with antibodies against Flag (Sigma), RAD51 (Santa Cruz), or MRE11 (Abcam).
For survival assays, 3000 cells were seeded in a 24-well plate the day before continuous treatment with the indicated drugs. The number of viable cells was determined when confluency reached ~80% for the untreated cells using Cell Titer 96 AQeous One Solution Cell Proliferation Assay (Promega). For metaphase spreads, 2 × 105 cells were seeded the day before HU (4 mM) and treated with colcemid (0.1 µg/ml, Gibco), as indicated. For metaphase spreads, cells were swollen with 0.075 M KCL (12 min, 37°C), fixed with methanol/acetic acid (3:1), dropped onto a microscope slide, stained with 5% Giemsa, and mounted with Cytoseal 60 (Fisher Scientific) prior to imaging with an Olympus BX60 microscope.
HIGHLIGHTS.
BRCA2 protects stalled replication forks from nucleolytic degradation
RAD51 filament stabilization by BRCA2 C-ter separates fork protection from HDR
Replication stalling of forks unprotected by BRCA2 causes genomic instability
MRE11 inhibition prevents fork degradation and reduces resultant genomic instability
Supplementary Material
ACKNOWLEDGEMENTS
We thank Francesca Cole, Jeannine LaRocque, Koji Nakanishi, Yu Zhang and other members of the Jasin lab and for reagents and discussions, Marie-Emilie Terret (MSKCC) and Margaret Leversha (MSKCC) for technical assistance, Julia Sidorova (Seattle) for critical discussions along with Duncan Wright (MSKCC) and Reginald Hill (UCLA), and Toshiyasu Taniguchi (Seattle), Ouathek Ouerfelli (MSKCC), and Berry Sleckman (St. Louis) for reagents. K.S. is the Berger Foundation Fellow of the Damon Runyon Cancer Research Foundation (DRG 1957-07). This work was supported by NIH grants R01CA121110 (H.W.) and R01GM54668 and P01CA94060 (M.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes extended Experimental Procedures, six figures, and one table and can be found with this article online.
REFERENCES
- Ayoub N, Rajendra E, Su X, Jeyasekharan AD, Mahen R, Venkitaraman AR. The carboxyl terminus of Brca2 links the disassembly of Rad51 complexes to mitotic entry. Curr Biol. 2009;19:1075–1085. doi: 10.1016/j.cub.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Budzowska M, Kanaar R. Mechanisms of Dealing with DNA Damage-Induced Replication Problems. Cell Biochemistry and Biophysics. 2009;53:17–31. doi: 10.1007/s12013-008-9039-y. [DOI] [PubMed] [Google Scholar]
- Cheng WH, von Kobbe C, Opresko PL, Arthur LM, Komatsu K, Seidman MM, Carney JP, Bohr VA. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J Biol Chem. 2004;279:21169–21176. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho G, Brenneman MA, Cui TX, Donoviel D, Vogel H, Goodwin EH, Chen DJ, Hasty P. Deletion of Brca2 exon 27 causes hypersensitivity to DNA crosslinks, chromosomal instability, and reduced life span in mice. Genes Chromosomes Cancer. 2003;36:317–331. doi: 10.1002/gcc.10148. [DOI] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J, Rudin N, Haber JE. Two Alternative Pathways of Double-Strand Break Repair That Are Kinetically Separable and Independently Modulated. Molecular and Cellular Biology. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M, Pellicioli A, Lopes M, Lucca C, Ferrari M, Liberi G, Muzi Falconi M, Plevani P. DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat Res. 2000;451:187–196. doi: 10.1016/s0027-5107(00)00049-x. [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pichierri P. Werner syndrome protein and the MRE11 complex are involved in a common pathway of replication fork recovery. Cell Cycle. 2004;3:1331–1339. doi: 10.4161/cc.3.10.1185. [DOI] [PubMed] [Google Scholar]
- Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- Gu Y, Jin S, Gao Y, Weaver D, Alt F. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay T, Clarke AR. DNA damage hypersensitivity in cells lacking BRCA2: a review of in vitro and in vivo data. Biochem Soc Trans. 2005;33:715–717. doi: 10.1042/BST0330715. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon Clusters are Stable Units of Chromosome Structure: Evidence That Nuclear Organization Contributes to the Efficient Activation and Propagation of S Phase in Human Cells. The Journal of Cell Biology. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonosov M, Anand S, Sangrithi M, Davies R, Ventikaraman AR. Stabilization of stalled DNA replicaiton forks by the BRCA2 breast cancer susceptibility protein. Genes & Development. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazloum N, Holloman WK. Brh2 promotes a template-switching reaction enabling recombinational bypass of lesions during DNA synthesis. Mol Cell. 2009;36:620–630. doi: 10.1016/j.molcel.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister KA, Bennett LM, Houle CD, Ward T, Malphurs J, Collins NK, Cachafeiro C, Haseman J, Goulding EH, Bunch D, et al. Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res. 2002;62:990–994. [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu M, Donoho G, Hasty P. Cells deleted for Brca2 COOH terminus exhibit hypersensitivity to gamma-radiation and premature senescence. Cancer Res. 1998;58:3441–3447. [PubMed] [Google Scholar]
- Morrison C, Shinohara A, Sonoda E, Yamaguchi-Iwai Y, Takata M, Weichselbaum RR, Takeda S. The essential functions of human Rad51 are independent of ATP hydrolysis. Mol Cell Biol. 1999;19:6891–6897. doi: 10.1128/mcb.19.10.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene. 2002;21:8994–9007. doi: 10.1038/sj.onc.1206177. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is Required For Homology-Directed Repair of Chromosomal Breaks. Molecular Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- Nagaraju G, Scully R. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair (Amst) 2007;6:1018–1031. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro S, Meza NW, Quintana-Bustamante O, Casado JA, Jacome A, McAllister K, Puerto S, Surralles J, Segovia JC, Bueren JA. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol Ther. 2006;14:525–535. doi: 10.1016/j.ymthe.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc Natl Acad Sci U S A. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H, Siaud N, Christ N, Wiegant WW, van Buul PP, Han M, Zdzienicka MZ, Stark JM, Jasin M. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci U S A. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Goodman MF. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat Rev Mol Cell Biol. 2007;8:587–594. doi: 10.1038/nrm2198. [DOI] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Stark JM, Hu P, Pierce AJ, Moynahan ME, Ellis N, Jasin M. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277:20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- Su X, Bernal JA, Venkitaraman AR. Cell-cycle coordination between DNA replication and recombination revealed by a vertebrate N-end rule degron-Rad51. Nat Struct Mol Biol. 2008;15:1049–1058. doi: 10.1038/nsmb.1490. [DOI] [PubMed] [Google Scholar]
- van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJ, Wuite GJ. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes & Development. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Wiegant WW, Overmeer RM, Godthelp BC, van Buul PP, Zdzienicka MZ. Chinese hamster cell mutant, V-C8, a model for analysis of Brca2 function. Mutational Research. 2006;600:79–88. doi: 10.1016/j.mrfmmm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.