Abstract
Background
Congenital deletions affecting 3q11q23 have rarely been reported and only five cases have been molecularly characterised. Genotype—phenotype correlation has been hampered by the variable sizes and breakpoints of the deletions. In this study, 14 novel patients with deletions in 3q11q23 were investigated and compared with 13 previously reported patients.
Methods
Clinical data were collected from 14 novel patients that had been investigated by high resolution microarray techniques. Molecular investigation and updated clinical information of one cytogenetically previously reported patient were also included.
Results
The molecular investigation identified deletions in the region 3q12.3q21.3 with different boundaries and variable sizes. The smallest studied deletion was 580 kb, located in 3q13.31. Genotype—phenotype comparison in 24 patients sharing this shortest region of overlapping deletion revealed several common major characteristics including significant developmental delay, muscular hypotonia, a high arched palate, and recognisable facial features including a short philtrum and protruding lips. Abnormal genitalia were found in the majority of males, several having micropenis. Finally, a postnatal growth pattern above the mean was apparent. The 580 kb deleted region includes five RefSeq genes and two of them are strong candidate genes for the developmental delay: DRD3 and ZBTB20.
Conclusion
A newly recognised 3q13.31 microdeletion syndrome is delineated which is of diagnostic and prognostic value. Furthermore, two genes are suggested to be responsible for the main phenotype.
Keywords: Developmental delay, microdeletion, overgrowth syndrome, genotype-phenotype, 3q
Introduction
Deletions affecting the proximal long arm of chromosome 3 are rarely reported in the literature. Hitherto, 14 patients have been described with deletions of various sizes and different breakpoints within the 3q11q23 region. The deletions were investigated in nine of the patients by standard karyotyping1–8 and only five cases have been investigated by molecular methods.9–14 The 14 patients had a range of different phenotypes including cranial and facial dysmorphisms, developmental retardation, and genital and peripheral musculoskeletal abnormalities. However, determining a proper genotype—phenotype correlation has been hampered by the few cases with molecularly defined deletions as well as by the limited number of patients described.
The advent of high resolution microarray techniques has greatly facilitated the investigation of chromosomal disorders, enabling the identification of disease-causative genes for known syndromes—for example, CHARGE syndrome (OMIM 214800) and 9q subtelomeric deletion syndrome (OMIM 610253) as reviewed in Vissers et al.15 In addition, a number of novel microdeletion and microduplications syndromes have been delineated, starting with the first described 17q21.31 microdeletion syndrome in 2006 (reviewed in Vissers et al 15). Moving from a cytogenetic approach to an ever more sensitive molecular karyotyping has reversed the strategy behind the identification of novel syndromes—that is, patients having similar/overlapping genetic rearrangements are identified before the clinical characteristics of a syndrome are defined. Furthermore, the collection of clinical and genetic information in databases such as DECIPHER,16 ISCA,17 and ECARUCA18 has been crucial for the comparison between patients with rare aberrations.
Using a reverse genetics approach and a joint collaborative effort through DECIPHER, we describe 14 novel patients carrying microscopic or submicroscopic deletions in the region 3q12.3q21.3. In addition, a molecular investigation is presented of a previously reported 3q-deletion patient.5 This study also presents a review of the 13 previously reported patients. A newly recognised 3q13.31 microdeletion syndrome is identified, characterised by developmental delay, postnatal growth above the mean, characteristic facial features, and abnormal male genitalia. The phenotype is associated with a 0.6 Mb critical region harbouring two strong candidate genes for the developmental delay, the DRD3 and ZBTB20 genes.
Patients and methods
Patients
In the present study, 15 patients were included with deletions in the proximal long arm of chromosome 3. One patient, case 2, was previously described clinically and cytogenetically by Ogilvie et al 1998,5 while the remaining 14 patients are novel. Clinical information was systematically collected from clinicians, using supplementary table 1.
The WHO Child Growth Standards were used to standardise birth height, weight, and occipitofrontal circumference (OFC) for all novel patients and for the previously reported patients where growth parameters were given.19 WHO standards are available up to 19 years of age for height and up to 10 years of age for weight. To assess OFC after birth the German head circumference references were used, which extend up to 18 years of age.20
The clinical investigations and genetic analyses were performed according to the guidelines in the Declaration of Helsinki and were approved by the ethics committee of Uppsala University. Informed consent was obtained from all family members and specific permission to publish photographs was obtained.
Methods
Molecular investigation of the 15 patients was conducted using different array platforms (table 1 and supplementary material) according to the manufacturer's instructions. The identified deletions were confirmed using karyotyping or fluorescence in situ hybridisation (FISH) (supplementary material) and parental testing was performed when parental DNA was available (13/15 cases). Deletions in cases 1 and 2 were microscopically visible and had initially been investigated using GTG banding.5 The positions of the deletions were mapped to the human NCBI/hg18 assembly of the UCSC genome browser (http://genome.ucsc.edu/).
Table 1.
Molecular characterisation of 3q12q21 deletions in present and previously published patients
| Case | Chromosomal band | Start (hg18) | End (hg18) | Size (Mb) | No of RefSeq genes | Inheritance | Method |
| Case1 | q12.3–q13.31 | 103332789 | 118628997 | 15.30 | 64 | De novo | Affymetrix GeneChip 250K Nsp/G-banding |
| Case2 | q12.3–q13.33 | 103481815 | 122521004 | 19.04 | 89 | De novo | Affymetrix GeneChip 250K Nsp/G-banding |
| Case3 | q13.11–q13.31 | 105180963 | 118326520 | 13.15 | 63 | De novo | 1 Mb clone array/FISH |
| Case4 | q13.11–q21.3 | 105782523 | 128177975 | 22.40 | 144 | De novo | Agilent array CGH 400K/Q-PCR |
| Case5 | q13.11–q13.33 | 105911706 | 120983616 | 15.07 | 78 | De novo | Agilent array CGH 44K/Q-PCR |
| Case6 | q13.13–q13.33 | 110116098 | 122633570 | 12.52 | 73 | De novo | Agilent array CGH 44K/FISH RP11-233L3 |
| Case7 | q13.13–q13.31 (inv(3)(q13.1;q26.3)) | 111722434 | 117006477 | 5.28 | 39 | De novo | Agilent array CGH 44K/FISH RP11-105H23 |
| Case8 | q13.2–q13.32 | 112976429 | 120183473 | 7.2 | 38 | Not tested | Affymetrix GeneChip 250K Nsp |
| Case9 | q13.2–q13.31 | 113680819 | 116466363 | 2.79 | 24 | De novo | Agilent array CGH 44K/FISH RP11-572C15 |
| Case10 | q13.2–q13.31 | 113764648 | 116429950 | 2.67 | 22 | De novo | BlueGnome CytochipV2/FISH RP11-572C15 |
| Case11 | q13.2–q13.31 | 113764648 | 116429950 | 2.67 | 22 | De novo | BlueGnome CytochipV2/FISH RP11-572C15 and RP11-58D2 |
| Case12 | q13.2–q13.31 | 114490215 | 116264578 | 1.77 | 15 | De novo | Affymetrix genome-wide human SNP Array6.0 |
| Case13 | q13.31–q13.31 | 115335356 | 115916848 | 0.58 | 5 | De novo | Affymetrix GeneChip 250K Nsp |
| Case14 | q13.31–q13.31 | 116922662 | 118098190 | 1.18 | 3 | Absent in mother | Agilent Array CGH 44K/FISH RP11-91D11 |
| Case15 | q13.32–q21.2 | 119261437 | 126585699 | 7.32 | 63 | De novo | SpectralChip CC4-V0.3/FISH RP11-169N13 |
| Kosaki10* | q12.2–q13.2 | 101480701 | 114803431 | 13.32 | 70 | De novo | Spectral Genomics human BAC array 2500/G-banding |
| Malan11* | q13.11–q13.33 | 104531502 | 122804242 | 18.27 | 91 | De novo | Agilent 244K/FISH |
| Simovich14* | q13.11–q13.12 | 105652857 | 108151059 | 2.50 | 2 | De novo | Illumina HumanHap550 Beadchip/FISH RP11-91B3 |
| Shimojima13* | q13.2–q13.31 | 114321633 | 116406833 | 1.9 | 16 | De novo | Agilent array CGH 105A/FISH |
| Hou9/Sato12* | q11.2–q13.31 t(3;12)(q13.2;p11.2) | 97002372 | 116490074 | 19.49 | 107 | De novo | BAC array CGH/FISH |
| Lawson-Yuen4* | q13.1–q13.3 | 104400000 | 123400000 | De novo | Chromosome analysis | ||
| Okada6* | q12–q21 | 99800000 | 131500000 | De novo | G-banding, Q | ||
| Fujita1* | q12–q23 | 99800000 | 144400000 | De novo | G-banding | ||
| Genuard2* | q13.12–q21.3 | 107800000 | 131500000 | De novo | G-banding | ||
| Jenkins3* | q11–q21 | 91700000 | 131500000 | De novo | G-banding | ||
| Ogilvie5* | q12–q21 | 99800000 | 131500000 | De novo | G-banding | ||
| McMorrow7* | q12–q21 | 99800000 | 131500000 | De novo | G-banding, Q and C | ||
| Arai8* | q12–q22 | 99800000 | 131500000 | De novo |
Previously published cases with the reference indicated; Start- and endpoints in italic indicates maximum estimated start and end.
BAC, bacterial artificial chromosome; CGH, comparative genomic hybridisation; FISH, fluorescence in situ hybridisation; SNP, single nucleotide polymorphism.
Results and discussions
Molecular details
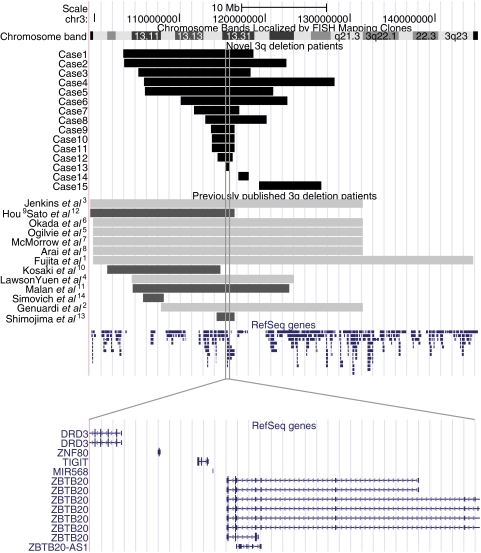
We present the clinical and molecular features of 15 novel patients harbouring deletions of the proximal long arm of chromosome 3. One of the patients was reported cytogenetically in the late 1990s5 (case 2). The present study also provides a review of 13 previously reported patients.1–14 The deletions are mapped within 3q12.3q21.3 and they range in size from the smallest of 580 kb (case 13) to the largest of 22.4 Mb (case 4) (figure 1 and table 1). Most of the deletions have different breakpoints, although the breakpoints in cases 9, 10, 11, and 12 are in close proximity (figure 1); the breakpoints of these patients are approximately located at 113.5–116.5 Mb. In all but one case, the deletion was the sole identified aberration. In case 7, an inversion was identified (inv(3)(q13.1q26.3)) and the deletion was located at the 3q13.1 inversion breakpoint. The deletions showed a de novo occurrence in 13 cases. Parental DNA was not available for testing in case 8, whose 7.2 Mb deletion is likely to have arisen de novo because of the size and gene content of this deleted region. Carrier testing in the parents of case 14 was only possible in the mother's DNA, which revealed a normal chromosome 3.
Figure 1.
A physical map of the chromosomal region 3q11.2 to 3q23, illustrating the deletions. The deletions identified in novel patients are shown in black, previously reported deletions that have been molecularly characterised are shown in dark grey, and previously reported deletions that have been cytogenetically characterised are shown in light grey. RefSeq genes are indicated in blue. The grey solid box illustrates the shortest region of overlapping deleted region, and a zoomed view shows the five RefSeq genes within this region (bottom panel).
The shortest region of overlapping deletion (SRO) is delineated by case 13, with estimated breakpoints at genomic positions 115.33—115.39 Mb (figure 1). This 580 kb segment includes five RefSeq genes: DRD3, ZNF80, TIGIT, MIR568, and ZBTB20. The SRO is shared by 13 of the novel patients, and by 11 of the previously reported patients—that is, 24 cases in total. The SRO is within the previously reported smallest deletion which was 1.9 Mb in size.13 However, the 2.5 Mb deletion identified in the case presented by Simovich et al is located at 3q13.11q12 and hence does not overlap with the region defined in the present study.14 Four cases do not have an overlapping deletion with the SRO, namely case 14, case 15 (both from this study), Kosaki et al,10 and Simovich et al.14
Clinical data
In total, clinical data from 28 patients, both novel and previously published cases, were collected and are summarised in supplementary table 1. Photographs of some of the novel patients (cases 1, 2, 4, 5, 6, 7, 9, 12 and 15) are shown in figure 2. The clinical findings in the 24 patients sharing the SRO (115.33—115.39 Mb) are summarised separately, and the frequency of these features was calculated (supplementary table 1, frequency column). These features include normal pregnancy and delivery at term with a few exceptions.1 4 5 11 14 Developmental delay is the most prevalent feature, present in 19/21 cases. Two cases did not suffer from developmental delay (case 7) or had not been examined at the time of the report due to the patient's young age (case 4). However, case 7 displayed attention deficit disorder. There are eight patients presenting with autism or attention deficits and one with epilepsy, including case 7. In 15 of 17 cases speech was delayed, and in three of these 15 patients the speech was minimal/no meaningful words were used/communication by hands by the age of 4.5, 8, and 18 years. Muscular hypotonia was found in 12/15 patients. Interestingly, muscular hypotonia was suggested by Shimojima et al to be the only common finding along with developmental delay in patients with 3q13 deletions13. The brain and central nervous system were also affected: five patients had agenesis of the corpus callosum, three patients had ventriculomegaly, and one had alobar holoprosencephaly. In total, seven patients displayed skull malformations: two with dolichocephaly, two with plagiocephaly, and three with brachycephaly. Furthermore, 10/13 patients presented with broad and prominent forehead.
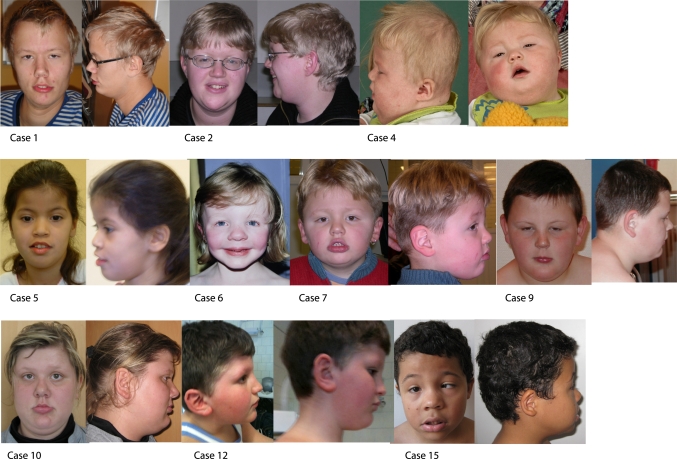
Figure 2.
Photographs of cases 1, 2, 4, 5, 6, 7, 9, 10, 12, and 15. Physical characteristics of note are short philtrum, protruding lips with full lower lips and tented upper lips, hypertelorism, and antimongoloid slanted eyes present in several cases.
Distinct recognisable facial features, including short philtrum, protruding lips with full lower lips and tented upper lips, antimongoloid slanted eyes, and hypertelorism, were apparent in several cases (figure 2). In total, the facial features in the 24 patients were short philtrum in 6/6, epicanthal folds in 8/14, hypertelorism in 7/17, antimongoloid slant in 7/13, a high arched palate in 7/10, and ptosis in 4/11. Ocular malformation included strabismus in 6/14 and myopia in 4/8. The ears were large in 5/15 and were low set in 4/15.
There was a high prevalence of abnormal external male genitalia, affecting 11/15 males. The abnormalities included micropenis (4/15), microorchidism (2/15), cryptorchidism (7/15), and shawl scrotum (2/15). All female patients had normal genitalia.
Skeletal malformations were a frequent finding, present in as many as 16/24 patients. The skeletal malformations included scoliosis, lordosis, thoracic kyphosis, joint contractures, and peripheral malformations affecting the hands and feet. Of note were the proximally set thumbs present in three novel cases (cases 2, 4, and 9).
Growth parameters were assessed in the 24 patients sharing the SRO and 4/11 (cases 4, 5, 9, and 10) had a birth OFC >85th centile (7/11 had a birth OFC >50th centile). OFC was also available at a later age and 9/20 had an OFC >85th centile (11/20 had an OFC >50th centile). Case 6 is noteworthy, having a birth OFC between 15–50th centile and an OFC between 85–97th centile at the age of 4 years and 10 months. The weight and length parameters were also reviewed and these were normal at birth, 5/16 had a weight >50th centile, 5/12 had a length >50th centile, and none of the patients displayed a weight or length >85th centile. At the time of report, 10/19 had a weight >50th centile and 9/19 had a weight >85th centile. Regarding height at the time of report, 13/21 were >50th centile and 10/21 were >85th centile. Hence, a postnatal growth pattern above the mean was observed among these patients. A larger region, encompassing 18.2 Mb in q13.11q13.33, has previously been identified in a screening of patients with syndromic overgrowth, and the present report delineates the overgrowth candidate region to 3q13.31. In DECIPHER, most of the listed microdeletion/microduplication syndromes are associated with short stature, while there is one that is characterised by tall stature—the 15q26 overgrowth syndrome. Regarding OFC, there is one listed microdeletion/microduplication syndrome in DECIPHER with macrocephaly, the 1q21.1 microduplication syndrome, in comparison with microcephaly that is present in 13 of the DECIPHER listed syndromes. Known overgrowth syndromes are Sotos syndrome, Beckwith—Wiedeman syndrome, Simpson—Golabi—Behmel syndrome, Klinefelter syndrome, homocystinuria and Marfan syndrome.21 The molecular knowledge about overgrowth syndromes is thus fairly limited and, in this context, the present report provides novel clues to finding genes involved in growth.
Candidate genes
The proximal long arm of chromosome 3 is a gene dense region (figure 1) with 145 genes within the estimated boundaries (chr3:103.32–128.18 Mb) of the 15 novel patients. Hence, a number of genes could potentially contribute to the phenotypic features of these patients. Regarding the five RefSeq genes present in the SRO, two (DRD3 and ZBTB20) are particularly interesting with respect to developmental delay, the neuropsychiatric features, and the structural brain, central nervous system, and skull malformations. DRD3 encodes D3 subtype of the dopamine receptors, which is localised to the limbic areas of the brain, and is involved in locomotion, cognition, emotion, and affection as well as neuroendocrine secretion.22 Targeted mutation of DRD3 is associated with hyperactivity in mice, and recent association studies in patients with neuropsychiatric disorders have explored the contribution of DRD3 variants to their phenotype.23–25 The ZBTB20 gene belongs to the BTB/POZ zinc finger family and is expressed in the developing hippocampal neurons.26 Downregulation of ZBTB20 disturbs the normal maturation of a certain type of neurons in the hippocampus, and changes in the cortical cytoarchitecture—for example, lack of the posterior part of the corpus callosum—were observed in transgenic mice models.27 28 One additional interesting aspect of ZBTB20, with respect to the observed postnatal overgrowth in the patients, is the fact that it regulates genes involved in growth and metabolism.29
In addition to the SRO and the genes therein, the present study provides clues about other 3q genomic regions harbouring important genes with respect to normal development. First, the deletion in case 14, telomeric of the SRO, contains LSAMP and GAP43, two strong candidate genes for developmental delay. LSAMP encodes the limbic system associated membrane protein, and studies in both human and mice models have demonstrated the involvement of LSAMP in neuropsychiatric features and behaviour.30 31 GAP43 is involved in neurite outgrowth, neurotransmission, and synaptic plasticity among other functions and GAP43 was also recently identified as a candidate gene for autism and autistic-like manifestations in human and mice.32–34 In addition, Gap43 +/− mice display decreased corpus callosum and hippocampal commissure volume.34 Secondly, the present study supports the previous suggestion that 3q11 could harbour a locus for agenesis of the corpus callosum (ACC).2 35 Five previously published 3q deletion patients exist who displayed ACC.2 4 5 7 35 Here we present one novel patient (case 6) with ACC, having a deletion that can help with further refining of the ACC critical region. As discussed above, strong candidate genes involved in ACC are ZBTB20 and GAP43.
Further support underlining the importance of DRD3, ZBTB20, LSAMP, and GAP43 in contributing to the phenotype in patients with 3q13 deletions is their haploinsufficiency score, as defined by the study by Huang et al.36 There are 49 genes of the total of 145 genes in 3q12-q21 that have a haploinsufficiency score of <50%, and these four candidate genes are among those 49 (supplementary table 2).
Conclusion
The present study describes a newly recognised 3q13.31 microdeletion syndrome based on 24 novel and previously reported patients and suggests candidate genes responsible for the developmental delay. In addition, the age of the patients in this report, ranging from infant to 20 years, provides prognostic information for patients with this microdeletion syndrome.
Supplementary Material
Acknowledgments
We thank the patients and their parents for participating in this study.
Footnotes
Funding: This study was supported by grants from the Sävstaholm Foundation, the Borgström Foundation, foundations at the Medical Faculty of Uppsala University, the Swedish Research Council, the Brazilian research foundations CNPq and FAPESP, and the Italian Ministry of Health.
Competing interests: None.
Patient consent: Obtained.
Ethics approval: This study was approved by the ethics committee of Uppsala University.
Contributors: All co-authors were responsible for the clinical and molecular investigations of their patients. AMM was responsible for the coordination and data collection from the other co-authors, the study design, genotype—phenotype correlation, and writing the manuscript. GA and MLB participated in the study design, genotype—phenotype correlation, and edited the manuscript. GA supervised the clinical data interpretation. All co-authors read and critically revised the manuscript and approved the final version.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Fujita H, Meng J, Kawamura M, Tozuka N, Ishii F, Tanaka N. Boy with a chromosome del (3)(q12q23) and blepharophimosis syndrome. Am J Med Genet 1992;44:434–6 [DOI] [PubMed] [Google Scholar]
- 2.Genuardi M, Calvieri F, Tozzi C, Coslovi R, Neri G. A new case of interstitial deletion of chromosome 3q, del(3q)(q13.12q21.3), with agenesis of the corpus callosum. Clin Dysmorphol 1994;3:292–6 [PubMed] [Google Scholar]
- 3.Jenkins MB, Stang HJ, Davis E, Boyd L. Deletion of the proximal long arm of chromosome 3 in an infant with features of Turner syndrome. Ann Genet 1985;28:42–4 [PubMed] [Google Scholar]
- 4.Lawson-Yuen A, Berend SA, Soul JS, Irons M. Patient with novel interstitial deletion of chromosome 3q13.1q13.3 and agenesis of the corpus callosum. Clin Dysmorphol 2006;15:217–20 [DOI] [PubMed] [Google Scholar]
- 5.Mackie Ogilvie C, Rooney SC, Hodgson SV, Berry AC. Deletion of chromosome 3q proximal region gives rise to a variable phenotype. Clin Genet 1998;53:220–2 [DOI] [PubMed] [Google Scholar]
- 6.Okada N, Hasegawa T, Osawa M, Fukuyama Y. A case of de novo interstitial deletion 3q. J Med Genet 1987;24:305–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMorrow LE, Reid CS, Coleman J, Medeiros A, D'Andrea M, Santucci T, McCormack MK. A new interstitial deletion of the long arm of chromosome 3. Am J Hum Genet 1986;39:124 [Google Scholar]
- 8.Arai K, Matukiyo H, Takazawa H. A case report of partial deletion of the long arm of the no 3 chromosome. Med Genet Res 1982;4:1–4 [Google Scholar]
- 9.Hou JW. Congenital arhinia with de novo reciprocal translocation, t(3;12)(q13.2;p11.2). Am J Med Genet A 2004;130A:200–3 [DOI] [PubMed] [Google Scholar]
- 10.Kosaki R, Fukuhara Y, Kosuga M, Okuyama T, Kawashima N, Honna T, Ueoka K, Kosaki K. OEIS complex with del(3)(q12.2q13.2). Am J Med Genet A 2005;135:224–6 [DOI] [PubMed] [Google Scholar]
- 11.Malan V, Chevallier S, Soler G, Coubes C, Lacombe D, Pasquier L, Soulier J, Morichon-Delvallez N, Turleau C, Munnich A, Romana S, Vekemans M, Cormier-Daire V, Colleaux L. Array-based comparative genomic hybridization identifies a high frequency of copy number variations in patients with syndromic overgrowth. Eur J Hum Genet 2010;18:227–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato D, Shimokawa O, Harada N, Olsen OE, Hou JW, Muhlbauer W, Blinkenberg E, Okamoto N, Kinoshita A, Matsumoto N, Kondo S, Kishino T, Miwa N, Ariga T, Niikawa N, Yoshiura K. Congenital arhinia: molecular-genetic analysis of five patients. Am J Med Genet A 2007;143:546–52 [DOI] [PubMed] [Google Scholar]
- 13.Shimojima K, Saito K, Yamamoto T. A de novo 1.9-Mb interstitial deletion of 3q13.2q13.31 in a girl with dysmorphic features, muscle hypotonia, and developmental delay. Am J Med Genet A 2009;149A:1818–22 [DOI] [PubMed] [Google Scholar]
- 14.Simovich MJ, Bland SD, Peiffer DA, Gunderson KL, Cheung SW, Yatsenko SA, Shinawi M. Delineation of the proximal 3q microdeletion syndrome. Am J Med Genet A 2008;146A:1729–35 [DOI] [PubMed] [Google Scholar]
- 15.Vissers LE, de Vries BB, Veltman JA. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J Med Genet 2010;47:289–97 [DOI] [PubMed] [Google Scholar]
- 16.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet 2009;84:524–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST, Richard G, Compton JG, Fuller AE, Gliem TJ, Huang S, Collinson MN, Beal SJ, Ackley T, Pickering DL, Golden DM, Aston E, Whitby H, Shetty S, Rossi MR, Rudd MK, South ST, Brothman AR, Sanger WG, Iyer RK, Crolla JA, Thorland EC, Aradhya S, Ledbetter DH, Martin CL. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med 2011;13:777–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feenstra I, Fang J, Koolen DA, Siezen A, Evans C, Winter RM, Lees MM, Riegel M, de Vries BB, Van Ravenswaaij CM, Schinzel A. European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA); an online database for rare chromosome abnormalities. Eur J Med Genet 2006;49:279–91 [DOI] [PubMed] [Google Scholar]
- 19.WHO Multicentre Growth Reference Study Group WHO Child Growth Standards based on length/height, weight and age. Acta Paediatr Suppl 2006;450:76–85 [DOI] [PubMed] [Google Scholar]
- 20.Schienkiewitz A, Schaffrath Rosario A, Dortschy R, Ellert U, Neuhauser H. German head circumference references for infants, children and adolescents in comparison with currently used national and international references. Acta Paediatr 2011;100:e28–33 [DOI] [PubMed] [Google Scholar]
- 21.Sabin MA, Werther GA, Kiess W. Genetics of obesity and overgrowth syndromes. Best Pract Res Clin Endocrinol Metab 2011;25:207–20 [DOI] [PubMed] [Google Scholar]
- 22.Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine receptors: from structure to function. Physiol Rev 1998;78:189–225 [DOI] [PubMed] [Google Scholar]
- 23.Accili D, Fishburn CS, Drago J, Steiner H, Lachowicz JE, Park BH, Gauda EB, Lee EJ, Cool MH, Sibley DR, Gerfen CR, Westphal H, Fuchs S. A targeted mutation of the D3 dopamine receptor gene is associated with hyperactivity in mice. Proc Natl Acad Sci U S A 1996;93:1945–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoenicka J, Aragues M, Ponce G, Rodriguez-Jimenez R, Jimenez-Arriero MA, Palomo T. From dopaminergic genes to psychiatric disorders. Neurotox Res 2007;11:61–72 [DOI] [PubMed] [Google Scholar]
- 25.Beninger RJ, Banasikowski TJ. Dopaminergic mechanism of reward-related incentive learning: focus on the dopamine D(3) receptor. Neurotox Res 2008;14:57–70 [DOI] [PubMed] [Google Scholar]
- 26.Mitchelmore C, Kjaerulff KM, Pedersen HC, Nielsen JV, Rasmussen TE, Fisker MF, Finsen B, Pedersen KM, Jensen NA. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J Biol Chem 2002;277:7598–609 [DOI] [PubMed] [Google Scholar]
- 27.Nielsen JV, Blom JB, Noraberg J, Jensen NA. Zbtb20-induced CA1 pyramidal neuron development and area enlargement in the cerebral midline cortex of mice. Cereb Cortex 2010;20:1904–14 [DOI] [PubMed] [Google Scholar]
- 28.Nielsen JV, Nielsen FH, Ismail R, Noraberg J, Jensen NA. Hippocampus-like corticoneurogenesis induced by two isoforms of the BTB-zinc finger gene Zbtb20 in mice. Development 2007;134:1133–40 [DOI] [PubMed] [Google Scholar]
- 29.Sutherland AP, Zhang H, Zhang Y, Michaud M, Xie Z, Patti ME, Grusby MJ, Zhang WJ. Zinc finger protein Zbtb20 is essential for postnatal survival and glucose homeostasis. Mol Cell Biol 2009;29:2804–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Catania EH, Pimenta A, Levitt P. Genetic deletion of Lsamp causes exaggerated behavioral activation in novel environments. Behav Brain Res 2008;188:380–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koido K, Koks S, Must A, Reimets A, Maron E, Shlik J, Vasar V, Vasar E. Association analysis of limbic system-associated membrane protein gene polymorphisms in mood and anxiety disorders. European Neuropsychopharmacology 2006;16(Suppl 1):S9 [Google Scholar]
- 32.Allen-Brady K, Miller J, Matsunami N, Stevens J, Block H, Farley M, Krasny L, Pingree C, Lainhart J, Leppert M, McMahon WM, Coon H. A high-density SNP genome-wide linkage scan in a large autism extended pedigree. Mol Psychiatry 2009;14:590–600 [DOI] [PubMed] [Google Scholar]
- 33.Denny JB. Molecular mechanisms, biological actions, and neuropharmacology of the growth-associated protein GAP-43. Curr Neuropharmacol 2006;4:293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaccaria KJ, Lagace DC, Eisch AJ, McCasland JS. Resistance to change and vulnerability to stress: autistic-like features of GAP43-deficient mice. Genes Brain Behav 2010;9:985–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Driscoll MC, Black GC, Clayton-Smith J, Sherr EH, Dobyns WB. Identification of genomic loci contributing to agenesis of the corpus callosum. Am J Med Genet A 2010;152A:2145–59 [DOI] [PubMed] [Google Scholar]
- 36.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet 2010;6:e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.