The Ro 60-kDa autoantigen, a ring-shaped RNA-binding protein, traffics between the nucleus and cytoplasm in vertebrate cells. In the cytoplasm, Ro binds noncoding RNAs called Y RNAs. Y RNA binding blocks a nuclear accumulation signal, retaining Ro in the cytoplasm. Following UV irradiation, this signal becomes accessible, allowing Ro to accumulate in nuclei. The authors identified several proteins that copurify with the mouse Ro protein and report that the zipcode-binding protein ZBP1 influences the subcellular localization of both Ro and the Y3 RNA. Both Ro and Y3 RNA accumulate in nuclei when ZBP1 is depleted. Accordingly, the data indicate that ZBP1 may function as an adapter to export the Ro/Y3 RNA complex from nuclei.
Keywords: Ro autoantigen, noncoding RNA, nuclear transport
Abstract
The Ro 60-kDa autoantigen, a ring-shaped RNA-binding protein, traffics between the nucleus and cytoplasm in vertebrate cells. In some vertebrate nuclei, Ro binds misfolded noncoding RNAs and may function in quality control. In the cytoplasm, Ro binds noncoding RNAs called Y RNAs. Y RNA binding blocks a nuclear accumulation signal, retaining Ro in the cytoplasm. Following UV irradiation, this signal becomes accessible, allowing Ro to accumulate in nuclei. To investigate how other cellular components influence the function and subcellular location of Ro, we identified several proteins that copurify with the mouse Ro protein. Here, we report that the zipcode-binding protein ZBP1 influences the subcellular localization of both Ro and the Y3 RNA. Binding of ZBP1 to the Ro/Y3 complex increases after UV irradiation and requires the Y3 RNA. Despite the lack of an identifiable CRM1-dependent export signal, nuclear export of Ro is sensitive to the CRM1 inhibitor leptomycin B. In agreement with a previous report, we find that ZBP1 export is partly dependent on CRM1. Both Ro and Y3 RNA accumulate in nuclei when ZBP1 is depleted. Our data indicate that ZBP1 may function as an adapter to export the Ro/Y3 RNA complex from nuclei.
INTRODUCTION
All cells contain a diversity of ribonucleoprotein particles (RNPs), whose functions impact nearly every aspect of cell biology. Although nuclear-cytoplasmic trafficking is part of the biogenesis of some of these RNPs, most mature RNPs at steady state are localized to their sites of function in the nucleus, nucleolus, or cytoplasm. Historically, this tendency of mature RNPs to be localized to specific subcellular compartments enabled their grouping, often before functional information was available, into classes such as small nuclear RNPs (snRNPs), small nucleolar RNPs (snoRNPs), and small cytoplasmic RNPs (scRNPs) (Hannon et al. 2006). Similarly, many long noncoding RNAs are confined to the nucleus, where they function in chromatin remodeling or are structural components of nuclear subcompartments (Chen and Carmichael 2010).
One exception to this paradigm is the Ro 60-kDa protein and its associated RNAs. Ro is a ring-shaped RNA-binding protein that is important for cell survival after UV irradiation of both mammalian cells and the eubacterium Deinococcus radiodurans (Sim and Wolin 2011). In vertebrate cells, Ro traffics between nuclear and cytoplasmic compartments. In some nuclei, Ro binds misfolded noncoding RNAs and is proposed to function in noncoding RNA quality control (O'Brien and Wolin 1994; Shi et al. 1996; Labbe et al. 1999; Chen et al. 2003; Hogg and Collins 2007). In the cytoplasm, Ro is bound to ∼100-nt noncoding RNAs called Y RNAs. All vertebrates contain between two and four distinct Y RNAs (Mosig et al. 2007; Perreault et al. 2007). For example, human cells contain four Y RNAs (hY1, hY3, hY4, and hY5), while mouse cells contain only two RNAs (mY1 and mY3). Although the primary sequences of individual Y RNAs in a given species differ, all Y RNAs can fold into a secondary structure consisting of a large internal loop and a long stem formed by base-pairing the 5′ and 3′ ends. Binding of Ro to a conserved sequence within this stem stabilizes Y RNAs from degradation (Labbe et al. 1999; Chen et al. 2003; Xue et al. 2003).
Although the function of Y RNAs was mysterious for many years, recent studies indicate that one role of these RNAs is to modulate the function and subcellular location of Ro. In D. radiodurans, where the Ro ortholog is required for the heat stress-induced maturation of 23S rRNA, binding of a Y RNA to Ro inhibits this process during normal growth (Chen et al. 2007). In mouse cells, Y RNAs influence the subcellular distibution of Ro. Specifically, Y RNAs block a nuclear accumulation signal on Ro, retaining it in the cytoplasm (Sim et al. 2009). However, as Ro and a Y RNA accumulate in the nuclei after UV irradiation (Chen et al. 2003), this signal must become accessible during stress. Y RNAs may also influence the nuclear export of Ro, as Ro likely returns to the cytoplasm as a Ro/Y RNA complex (Simons et al. 1994). It is also proposed that Ro recognizes some misfolded RNAs as a Ro/Y RNA complex (Hogg and Collins 2007).
Although studies of the Ro ortholog in D. radiodurans have revealed that the bacterial Ro functions with the 3′ to 5′ exoribonucleases RNase II and RNase PH to mature 23S rRNA during heat stress (Chen et al. 2007) and with the exoribonuclease polynucleotide phosphorylase to degrade rRNA during growth in stationary phase (Wurtmann and Wolin 2010), far less is known about the proteins that function with the vertebrate Ro protein. Several proteins, including nucleolin, the splicing factors Puf60 and hnRNP I, and the interferon-inducible protein IFIT5, all interact with human Ro by binding one or more Y RNAs (Bouffard et al. 2000; Fabini et al. 2001; Fouraux et al. 2002; Hogg and Collins 2007). While these results provide evidence that the multiple Ro/Y RNA complexes found in vertebrate cells may be functionally distinct, the significance of these protein interactions for Ro function remains unknown.
To learn more about how Ro and Y RNAs are influenced by interacting proteins, we used tandem affinity purification to identify proteins that copurify with the mouse Ro protein. We report that the zipcode-binding protein ZBP1, the Y-box protein YB-1, and MOV10, a member of the DExH/D class of RNA-dependent ATPases, all associate with Ro by binding to one or more shared RNAs. We show that the interaction of ZBP1 with Ro requires mY3 RNA, and that the association of ZBP1 with the Ro/mY3 complex increases after UV irradiation. Interestingly, although Ro appears to lack a CRM1-dependent nuclear export signal (NES), nuclear export of Ro is sensitive to the CRM1 inhibitor leptomycin B. Depletion of ZBP1, which contains a CRM1-dependent NES (Nielsen et al. 2003; Oleynikov and Singer 2003), results in accumulation of both Ro and mY3 RNA in nuclei. Our data are consistent with a model in which ZBP1 functions as an adapter to export the Ro/mY3 complex from nuclei.
RESULTS
Identification of Ro-associated proteins
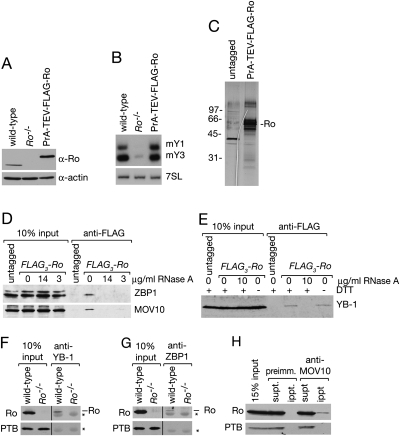
To identify proteins that copurify with mouse Ro, we constructed stable cell lines in which all Ro was fused to an epitope tag that facilitated purification. To this end, we fused Ro to a tag consisting of the two IgG-binding domains of Staphylococcus aureus Protein A, a TEV protease cleavage site, and a FLAG epitope, transfected the plasmid into Ro−/− cells and selected for stable lines. Using Western blotting with anti-Ro antibodies, the tagged protein, which was expressed under the control of the Ubiquitin C (UbC) promoter, appeared threefold higher than in wild-type cells (Fig. 1A); however, this may be an overestimate, since the Protein A tag also binds the primary and secondary IgG antibodies used for Western blotting (Forsgren and Sjoquist 1967). Northern blotting confirmed that in the presence of Protein A-TEV-FLAG-Ro, both mouse Y RNAs accumulated, indicating the fusion protein was able to stabilize Y RNAs (Fig. 1B).
FIGURE 1.
Identification of proteins that copurify with Ro. (A) Lysates from wild-type, Ro−/−, and ProteinA-TEV-FLAG-Ro cells were subjected to Western blotting with anti-Ro antibodies (top). As a loading control, the blot was reprobed for actin (bottom). (B) RNAs extracted from the cell lines were subjected to Northern blotting to detect mY1 and mY3 RNAs. As a control, the blot was reprobed to detect 7SL RNA. (C) After affinity purification, eluates from untagged and ProteinA-TEV-FLAG-Ro cells were fractionated in an SDS–polyacylamide gel and visualized by silver staining. (D,E) Lysates of untagged and FLAG3-Ro cells were incubated with the indicated concentrations of RNase A, followed by immunoprecipitation with anti-FLAG antibodies. Western blots were probed to detect ZBP1 and MOV10 (D) or YB-1 (E). Because YB-1 is important for cell survival during oxidative stress (Lu et al. 2005), we also examined whether its association with Ro varied when cells were lysed in the presence of 0.5 mM DTT (E). (F–H) Lysates of wild-type and Ro−/− fibroblasts were subjected to immunoprecipitation with rabbit anti-YB-1 (F), anti-ZBP1 (G), preimmune sera and anti-MOV10 antibodies (H). Proteins in immunoprecipitates were subjected to Western blotting to detect Ro. (*) IgG. As a negative control, the blots were reprobed to detect the polypyrimidine tract-binding protein PTB.
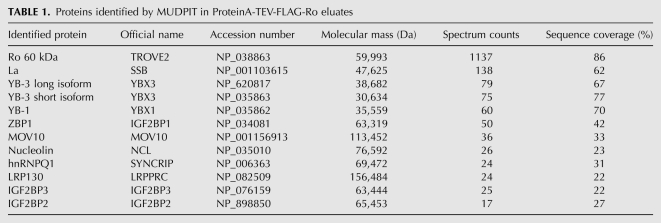
After two rounds of affinity purification from Protein A-TEV-FLAG-Ro cell lysates, silver staining revealed Ro and additional bands that were not present in a parallel purification from an untagged strain (Fig. 1C). Both samples were directly analyzed using multidimensional protein-identification technology (Yates et al. 2009). As expected, an abundant protein in the eluate from the tagged strain was the La protein, which binds newly synthesized Y RNAs in Ro RNPs (Hendrick et al. 1981; Simons et al. 1994). The other abundant proteins, based on spectral counts, consisted largely of known RNA-binding proteins, including the Y-box proteins YB-1 and YB-3, the zipcode-binding protein ZBP1 (also called IGF2BP1 and IMP1), the putative RNA helicase MOV10, nucleolin, and two proteins closely related to ZBP1, IGF2BP2 and IGF2BP3 (Table 1). Both ZBP1/IGF2BP1 (Jonson et al. 2007) and nucleolin (Fouraux et al. 2002; Chiu et al. 2006) were previously described to copurify with human Ro.
TABLE 1.
Proteins identified by MUDPIT in ProteinA-TEV-FLAG-Ro eluates
The association of Ro with ZBP1, MOV10, and YB-1 is RNA dependent
For our initial experiments, we focused on YB-1, ZBP1, and MOV10, which were among the most abundant proteins in the eluate. Similar to Ro, YB-1 accumulates in nuclei after UV irradiation (Koike et al. 1997). Consistent with a related function, Ro, YB-1, and nucleolin all copurify with human ZBP1 (Jonson et al. 2007; Weidensdorfer et al. 2009), a protein that functions in mRNA localization, translational regulation, and mRNA stability (Ross et al. 1997; Doyle et al. 1998; Nielsen et al. 1999; Hüttelmaier et al. 2005). MOV10, which associates with microRNAs as part of RISC (Meister et al. 2005), has been found with Ro, La, YB-1, and nucleolin in purifications of APOBEC3G, a cytidine deaminase involved in protecting the human genome from retroviruses and endogenous retrotransposons (Chiu et al. 2006; Kozak et al. 2006; Gallois-Montbrun et al. 2007). To validate the interactions, we performed immunoprecipitations from mouse fibroblasts, in which all Ro contained three copies of the FLAG epitope (Sim et al. 2009). Western blotting with antibodies to ZBP1, MOV10, and YB-1 confirmed that a small fraction of each protein was present in anti-FLAG immunoprecipitates from the tagged cells, but was not detected in immunoprecipitates from untagged cells (Fig. 1D,E). Treatment of the lysates with RNase A revealed that the interactions of Ro with all three proteins required RNA (Fig. 1D,E).
We also used antibodies against YB-1, ZBP1, and MOV10 to immunoprecipitate Ro RNPs from wild-type fibroblasts. Western blotting revealed that a small fraction of Ro (∼1%–2%) was present in each immunoprecipitate (Fig. 1F–H). Although the fraction of Ro associated with these proteins was low, Ro was not detected when preimmune serum was used (Fig. 1H). Moreover, the polypyrimidine tract-binding protein, hnRNPI/PTB, which associates with both Ro and ZBP1 in human cells (Fabini et al. 2001; Weidensdorfer et al. 2009), was not detected in our immunoprecipitates from mouse cells (Fig. 1F–H), thus serving as a negative control.
MOV10 and ZBP1 increase in nuclei after UV irradiation
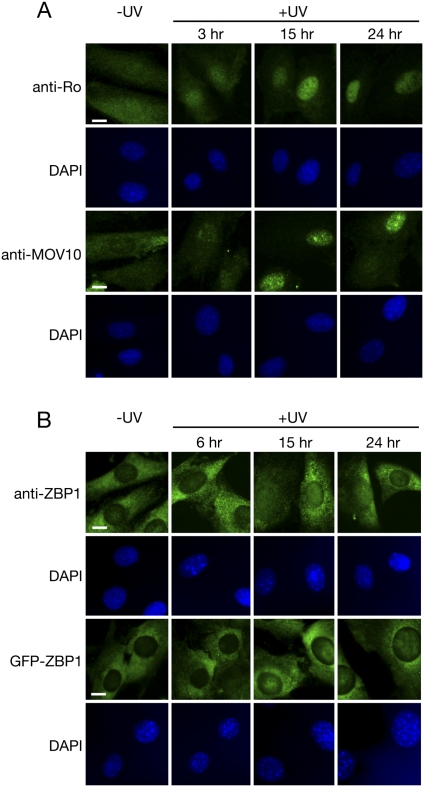
Since Ro is both nuclear and cytoplasmic in unstressed cells, but becomes strongly nuclear after UV irradiation (Chen et al. 2003), we examined the subcellular distribution of the associated proteins. The subcellular location of MOV10 is controversial, as some studies using epitope-tagged proteins in transient transfection experiments found that it is cytoplasmic and P-body associated in unstressed cells (Meister et al. 2005; Wulczyn et al. 2007), while others reported that the endogenous MOV10 is primarily nuclear (El Messaoudi-Aubert et al. 2010). Using affinity-purified anti-MOV10 antibodies, we found that the endogenous MOV10 was both nuclear and cytoplasmic in unstressed mouse fibroblasts, but accumulated strongly in nuclei after UV irradiation (Fig. 2A). In contrast to Ro, which is detected in nuclei within 3 h of irradiation (Chen et al. 2003), MOV10 was not strongly nuclear until 9 h after irradiation (Fig. 2A; data not shown). Using anti-ZBP1 antibodies for immunofluorescence, we found that, as described for human osteosarcoma cells using these antibodies (Stöhr et al. 2006), ZBP1 was almost entirely cytoplasmic in unstressed fibroblasts. However, at 15 and 24 h after UV irradiation, a small fraction of ZBP1 was detected in nuclei (Fig. 2B). Since the anti-ZBP1 antibody, which was raised against the 95% identical chicken protein (Farina et al. 2003), recognizes several bands in mouse cells (Fig. 1D), we confirmed this result by examining fibroblasts that were stably transfected with GFP-ZBP1 expressed under the control of the UbC promoter. As in the immunofluorescence experiments, GFP-ZBP1 was exclusively cytoplasmic before irradiation, but was detected in nuclei 15 and 24 h after irradiation. Notably, although ZBP1 accumulates in stress granules following arsenate or heat-shock treatment of human osteosarcoma cells (Stöhr et al. 2006), neither the endogenous ZBP1 or GFP-ZBP1 appeared to accumulate strongly in stress granules at the UV dose (10 J/m2 UVC) used in our experiments (Fig. 2B). Although YB-1 was reported to accumulate in nuclei after irradiation (Koike et al. 1997), our experiments to localize YB-1 were unsuccessful, as immunofluorescence with an antipeptide antibody (Stickeler et al. 2001) resulted in only background staining.
FIGURE 2.
Ro, MOV10, and ZBP1 all increase in nuclei after UV irradiation. (A) Wild-type fibroblasts were either unirradiated (left) or irradiated with 10 J/m2 UVC, allowed to recover for the indicated times, then fixed and subjected to immunofluorescence with anti-Ro or anti-MOV10 antibodies. Nuclei (panels below immunofluorescence) were visualized with DAPI. Bars, 10 μm. (B) Wild-type fibroblasts (top) or fibroblasts stably expressing GFP-ZBP1 were unirradiated (left) or irradiated as in A and allowed to recover for the indicated times. Cells were fixed and stained to detect ZBP1 (top) or directly mounted after fixation to detect GFP-ZBP1 (bottom). Bars, 10 μm.
Association of Ro with ZBP1 increases after UV irradiation and requires mY3 RNA
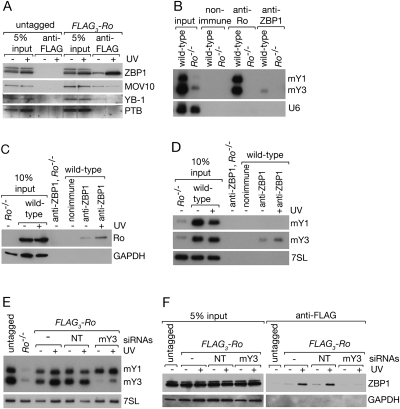
Since both ZBP1 and MOV10 were detected in nuclei ∼15 h after UV irradiation, we examined whether the interaction of either protein with Ro increased at this time. Immunoprecipitation with anti-Ro antibodies, followed by Western blotting of the immunoprecipitates, revealed that the fraction of ZBP1 associated with Ro increased approximately fourfold (Fig. 3A). The increase was specific to ZBP1, as the levels of MOV10 and YB-1 in the immunoprecipitates were unchanged (Fig. 3A). Experiments to determine whether the interaction of Ro with ZBP1 occurs in nuclei were inconclusive, because Ro leaks out of nuclei in all tested protocols for separating cultured cells into nuclear and cytoplasmic fractions (data not shown).
FIGURE 3.
The association of ZBP1 with Ro increases after UV irradiation and requires mY3 RNA. (A) Wild-type and FLAG3-Ro fibroblasts were either unirradiated or irradiated with 10 J/m2 UVC. After a 15-h recovery, cells were lysed and subjected to immunoprecipitation with anti-FLAG antibodies. Immunoblotting was performed to detect ZBP1, MOV10, and YB-1. As a negative control, the blot was reprobed for PTB. (B) Wild-type and Ro−/− lysates were subjected to immunoprecipitation with the indicated antibodies. RNAs extracted from immunoprecipitates and equivalent amounts of the lysates were subjected to Northern blotting to detect mY1 and mY3 RNAs. As a negative control, the blot was reprobed to detect U6 snRNA. (C,D) Ro−/− and wild-type fibroblasts were either unirradiated or irradiated and allowed to recover as in A. Lysates were subjected to immunoprecipitation with control nonimmune rabbit serum or with anti-ZBP1. After washing, the immunoprecipitate was divided into two aliquots. One aliquot was subjected to Western blotting to detect Ro, MOV10, and the GAPDH loading control (C). RNA extracted from the second aliquot was subjected to Northern blotting to detect mY1 and mY3 RNAs (D). As a control, the Northern blot was reprobed to detect 7SL RNA. (E,F) Either no siRNAs, nontarget control siRNAs, or siRNAs against mY3 were transfected into FLAG3-Ro fibroblasts. After 24 h, cells were irradiated with 10 J/m2 UVC and allowed to recover for 15 h. RNA extracted from half of the cells was subjected to Northern analyses to detect mY1, mY3, and 7SL RNA (E). Lysates from the remaining cells were subjected to immunoprecipitation with anti-FLAG antibodies. Lysates and proteins in immunoprecipitates were subjected to Western blotting to detect ZBP1 (F). As a control, the blot was reprobed to detect GAPDH.
Since the interaction between Ro and ZBP1 requires RNA (Fig. 1D), we examined whether either of the mouse Y RNAs was involved. Immunoprecipitations with anti-ZBP1 antibodies, followed by Northern blotting, revealed that a small fraction of mY3 RNA, but not mY1 RNA, was present in the immunoprecipitates (Fig. 3B). As expected, if ZBP1 interacts with the Ro/mY3 RNA complex, the levels of both Ro and mY3 in anti-ZBP1 immunoprecipitates increased to a similar extent after UV irradiation (Fig. 3C,D). To examine whether mY3 was important for the interaction, we used siRNAs against the large internal loop of this RNA (Sim et al. 2009) to reduce the level of mY3 RNA in FLAG3-Ro cells by >85% (Fig. 3E). Depletion of mY3 RNA eliminated the interaction of Ro with ZBP1 in both unirradiated and UV-irradiated cells (Fig. 3F). We conclude that mY3 RNA is required for the interaction of Ro with ZBP1.
Nuclear export of both Ro and ZBP1 involves CRM1
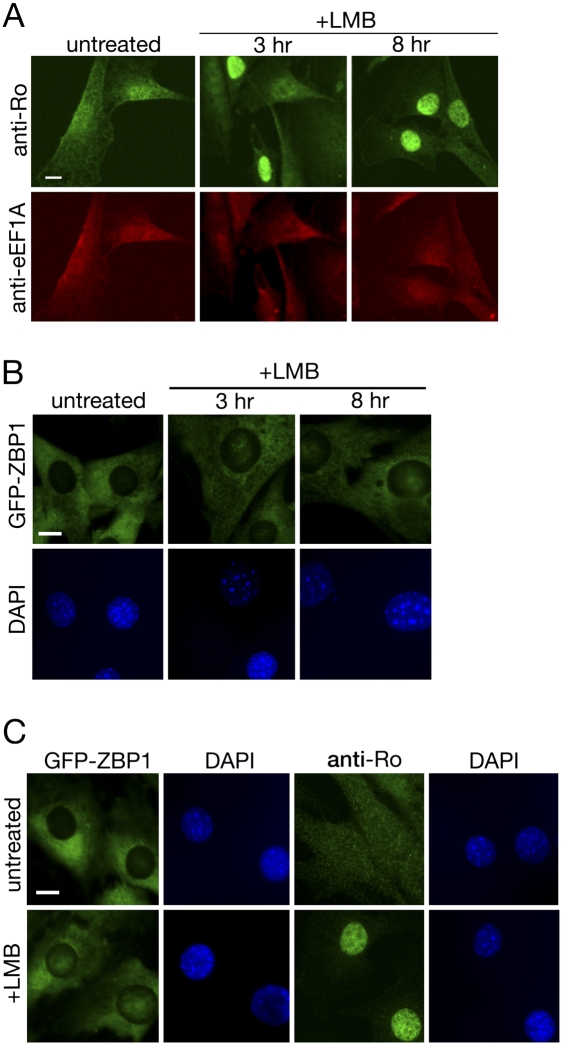
Previously, we reported that Y RNAs block a nuclear accumulation signal on the Ro surface, favoring cytoplasmic localization of Ro (Sim et al. 2009). However, since Ro may be exported from nuclei as a Ro/Y RNA complex (Simons et al. 1994), an additional role for Y RNAs in enhancing export could not be eliminated. Because ZBP1 shuttles between the nucleus and cytoplasm (Nielsen et al. 2003; Oleynikov and Singer 2003), we tested whether ZBP1 was involved in nuclear export of Ro. First, we examined whether Ro export was sensitive to leptomycin B (LMB), a specific inhibitor of the CRM1 export factor (Fornerod et al. 1997). Ro does not possess a recognizable CRM1-dependent nuclear export sequence (NES), and we have failed to detect binding of Ro to a GST-CRM1 fusion in the presence of a GTPase-deficient mutant of Ran (data not shown). However, ZBP1 has both a CRM1-dependent NES and a second NES that functions through a leptomycin B-insensitive pathway (Nielsen et al. 2003; Oleynikov and Singer 2003). Treatment of fibroblasts with LMB revealed that 60% of the cells showed nuclear accumulation of Ro within 3 h, with 80% of the cells showing nuclear accumulation by 8 h (Fig. 4A). In contrast, the subcellular location of elongation factor EF1A, which is exported by exportin-5 (Bohnsack et al. 2002), was unaffected by leptomycin B (Fig. 4A). Our result that Ro accumulates in nuclei upon LMB treatment indicates that Ro undergoes nucleocytoplasmic shuttling. Moreover, Ro is likely exported from nuclei by a pathway that involves CRM1.
FIGURE 4.
Nuclear export of Ro and ZBP1 involves CRM1. (A) Wild-type fibroblasts were either untreated or treated with 10 ng/mL of LMB. At the indicated times, cells were fixed and subjected to immunofluorescence with anti-Ro (top) and anti-eEF1A (bottom) antibodies. Bar, 10 μm. (B) Fibroblasts stably expressing GFP-ZBP1 were either untreated or treated with LMB as in A. At the indicated times, cells were fixed and mounted. Nuclei were visualized with DAPI. Bar, 10 μm. (C) Wild-type and GFP-ZBP1-expressing fibroblasts were either untreated or treated with 10 ng/mL LMB for 12 h. Wild-type cells were fixed and subjected to immunofluorescence to detect Ro, while GFP-ZBP1-expressing cells were fixed and directly mounted. Bar, 10 μm.
We also examined the localization of GFP-ZBP1 during leptomycin B treatment. In chicken fibroblasts, ZBP1 export is sensitive to leptomycin B (Oleynikov and Singer 2003); however, in mouse cells, export of a transiently transfected human ZBP1 fused to GFP was reported to be leptomycin B insensitive due to the presence of a second CRM1-independent NES (Nielsen et al. 2003). Examination of GFP-ZBP1 after leptomycin B treatment of our stable cell lines revealed that, consistent with the study in chicken cells (Oleynikov and Singer 2003), a fraction of GFP-ZBP1 became detectable in nuclei within 3 h of leptomycin B addition, and became more prominent by 12 h (Fig. 4B,C). Together with the finding that ZBP1 contains a CRM1-dependent NES (Nielsen et al. 2003; Oleynikov and Singer 2003), our results suggest that some ZBP1 is exported via a CRM1-dependent pathway. Although the fraction of ZBP1 that accumulates in nuclei during leptomycin B treatment is low, this may be because the second NES allows ZBP1 to access another export pathway when CRM1 is inactivated (Nielsen et al. 2003).
Ro and mY3 RNA accumulate in nuclei when ZBP1 is depleted
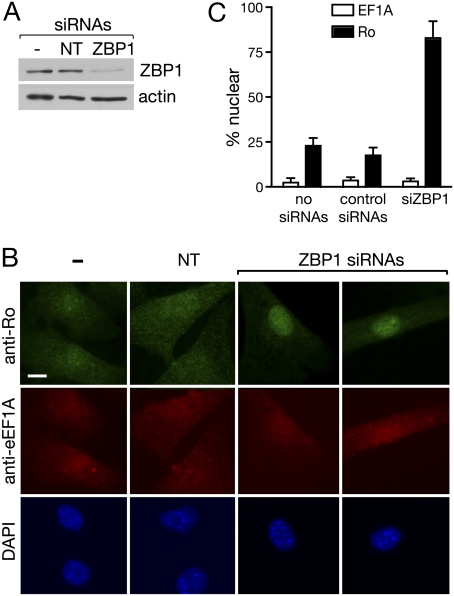
To determine whether ZBP1 was important for nuclear export of the Ro/mY3 complex, we used siRNAs to reduce ZBP1 levels in fibroblasts. By performing two rounds of transfection, we were able to reduce ZBP1 by 84% by 72 h after the first transfection (Fig. 5A). Immunofluorescence revealed that in cells depleted of ZBP1, Ro accumulated in nuclei (Fig. 5B,C). In contrast, the subcellular location of EF1A was unchanged. One explanation for the finding that siRNA-mediated depletion of ZBP1 results in a striking change in Ro subcellular distribution, yet only a small fraction of ZBP1 is detected in anti-Ro immunoprecipitates, is that the association of the Ro/Y3 complex with ZBP1 occurs transiently during export. Thus, despite the fact that only a modest fraction of Ro associates stably with ZBP1 at steady state, Ro would accumulate in nuclei over time in ZBP1-depleted cells if the ZBP interaction is required for efficient Ro nuclear export. Consistent with this hypothesis, Ro accumulates in nuclei within 3 h of LMB incubation (Fig. 4), indicating that Ro undergoes active nucleocytoplasmic shuttling.
FIGURE 5.
Ro accumulates in nuclei when ZBP1 is depleted. (A,B) siRNAs against ZBP1 or control nontarget siRNAs were transfected into wild-type fibroblasts. After 48 h, the transfection was repeated. At 72 h after the initial transfection, cells were harvested and subjected to Western blotting to detect ZBP1 (A) or immunofluorescence to detect Ro and eEF1A (B). In B, nuclei were visualized with DAPI. Bars, 10 μm. (C) Histogram showing the percentage of cells with predominantly nuclear staining of Ro and eEF1A. Data from three independent experiments were graphed.
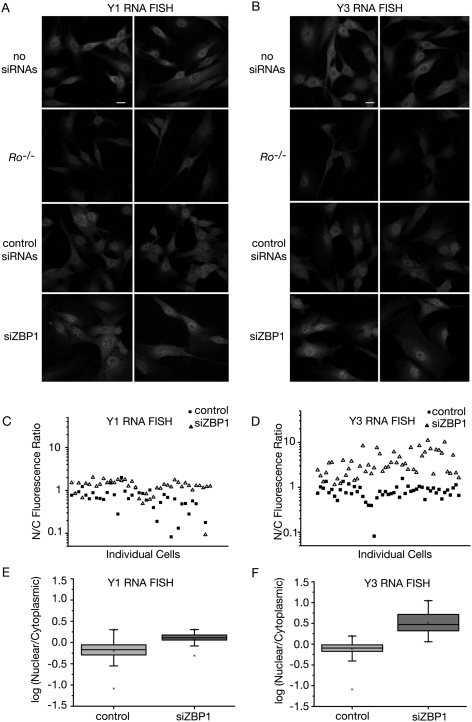
We performed fluorescence in situ hybridization (FISH) to determine whether ZBP1 depletion affected the subcellular distribution of either Y RNA. For probes, we used oligonucleotides complementary to the large internal loops, since these loops are accessible to siRNAs (Sim et al. 2009). Because the levels of both Y RNAs are greatly reduced in Ro−/− cells (Fig. 1B; Labbe et al. 1999; Chen et al. 2003), the fluorescent signals in these cells were used as a standard to determine the background fluorescence. For both mY1 and mY3, we detected stronger signals in wild-type cells than in Ro−/− cells (Fig. 6A,B), indicating that the oligonucleotides detect their specific targets. Importantly, after siRNA-mediated depletion of ZBP1, mY3 accumulated in nuclei (Fig. 6B). The change in Y3 RNA localization was less complete than observed for Ro, suggesting that this RNA may be able to access other export pathways (see Discussion). Nonetheless, quantification of the nuclear/cytoplasmic signal ratios in individual cells revealed clear differences for mY3 after ZBP1 depletion. In both untreated cells and cells receiving control siRNAs, the nuclear/cytoplasmic ratio was generally below 1, indicating that mY3 is largely cytoplasmic (Fig. 6D). After depleting ZBP1, the nuclear/cytoplasmic ratios ranged from 1.5 to 10, indicating that mY3 becomes strongly enriched in nuclei (Fig. 6D). In contrast, the mY1 nuclear/cytoplasmic ratios ranged from 0.5 to 2 after ZBP1 depletion. Thus, although some cells also showed nuclear enrichment of mY1, these changes were significantly smaller than for mY3 (Fig. 6, cf. C,E and D,F). We conclude that ZBP1 depletion results in nuclear accumulation of Ro and mY3 RNA.
FIGURE 6.
mY3 RNA accumulates in nuclei when ZBP1 is depleted. (A,B) Wild-type fibroblasts were transfected with siRNAs against ZBP1 or control siRNAs as described for Figure 5. At 72 h after the first transfection, cells were fixed and subjected to FISH for mY1 (A) and mY3 (B) RNA. As a negative control for the fluorescence signal, FISH was also performed on Ro−/− fibroblasts. Bars, 20 μm. (C,D) Scatter plots of the ratio of the nuclear and cytoplasmic signals in individual cells (logarithmic scale) after FISH to detect mY1 (C) and mY3 (D). The FISH signal was calculated by subtracting the background signal on Ro−/− fibroblasts from the signal on either control cells or cells transfected with siRNAs against ZBP1. Since there were no statistically significant differences between the fluorescence signals of untransfected cells and control siRNA-transfected cells for either mY1 or mY3 FISH, both untransfected and control siRNA-transfected cells were counted in the control group. (E,F) Box plots showing the logarithm of the nuclear/cytoplasmic signal ratios. For mY1 FISH (E), the statistical analysis was between control (n = 36) and siZBP1 (n = 40) transfected cells. The upper and lower box boundaries indicate the 25th and 75th percentiles of the data, respectively. The upper and lower bars indicate the first and 99th percentiles, respectively. The analysis for the mY3 FISH (F) was between control (n = 52) and siZBP1-treated (n = 58) cells. One-way ANOVA test between the logarithms of the nuclear/cytoplasmic ratios of the mY3 FISH signals in siZBP1-treated vs. control cells (D,F) and between the mY1 and mY3 FISH signals in siZBP1-treated cells: P < 0.001.
DISCUSSION
Although Ro binds misfolded RNAs in vertebrate cells and contributes to survival after UV irradiation, little is known about the molecular mechanisms by which Ro functions. To begin to address these questions, we used affinity purification to identify proteins that copurify with mouse Ro. This revealed several RNA-binding proteins, including two proteins shown previously to copurify with human Ro, nucleolin, and ZBP1 (Fouraux et al. 2002; Chiu et al. 2006; Jonson et al. 2007). Because the association of Ro with ZBP1 increased after UV irradiation, we focused on elucidating the functional significance of this interaction. We showed that the association of Ro with ZBP1 is mediated by RNA binding and requires mY3 RNA. Interestingly, in ZBP1-depleted cells, the normally cytoplasmic Ro and mY3 RNA accumulate in nuclei. Together with the findings that leptomycin B causes Ro to accumulate in nuclei and that ZBP1 contains a CRM1-dependent NES (Nielsen et al. 2003; Oleynikov and Singer 2003), our data suggest that ZBP1 may function as an adapter for export of the Ro/mY3 complex.
Our data, together with studies of nuclear export of human Y1 RNA in Xenopus oocytes, are consistent with a model in which export of the Ro/mY3 complex is mediated by ZBP1 via a CRM1-dependent pathway, while the less-abundant mY1 RNA is likely exported by exportin-5 (Simons et al. 1994, 1996; Rutjes et al. 2001; Gwizdek et al. 2003). Although the pathway by which Ro is exported was not examined in the oocyte studies, the finding that an hY1 RNA carrying a mutation in the Ro binding site was exported more slowly than wild-type hY1 was interpreted to mean that hY1 is exported bound to Ro (Simons et al. 1996). Our result that Ro accumulates in nuclei in the presence of leptomycin B does not rule out this possibility, since some Ro remains cytoplasmic when CRM1 is inhibited. Similarly, studies showing that injection of excess hY3 or hY4 RNAs blocks hY1 export, which were interpreted as demonstrating that these RNAs all utilize the same export pathway (Rutjes et al. 2001), could indicate that like tRNAs (Hopper et al. 2010), Y RNAs can be exported through multiple pathways. Consistent with multiple export pathways, tRNAmeti also competes with hY1 for export (Simons et al. 1996). Moreover, one explanation for the finding that nuclear accumulation of Ro in ZBP1-depleted cells (Fig. 5) is more striking than that of mY3 RNA (Fig. 6) is that mY3 can access additional export pathways.
How might ZBP1 interact with the Ro/mY3 RNA complex? Our finding that mY3 is required for complex formation, together with a recent report that hY1 and hY3 RNAs bind ZBP1 in vitro (Köhn et al. 2010), suggests that since Ro interacts with the mY3 RNA terminal stem (Wolin and Steitz 1984; Pruijn et al. 1991; Green et al. 1998; Stein et al. 2005), ZBP1 may bind sequences at the other end of mY3, such as the large internal loop. Moreover, as the interaction of ZBP1 with hY3 RNA can be competed with a zipcode-containing RNA (Köhn et al. 2010), the binding site on ZBP1 for Y3 RNA and the zipcode may overlap. Recent studies have shown that the third and fourth KH domains of ZBP1 form an antiparallel pseudodimer that interacts with a bipartite zipcode sequence from β-actin mRNA (Chao et al. 2010). Interestingly, the reported CRM1-independent NES (Nielsen et al. 2003) spans the linker between these two KH domains (Chao et al. 2010). Thus, one explanation for the striking accumulation of Ro during leptomycin B treatment is that association of the bulky Ro/Y3 complex with ZBP1 sterically blocks the CRM1-independent NES, causing Ro export to depend on the CRM1 NES. In contrast, if the binding of zipcode-containing mRNA to ZBP1 leaves the CRM1-independent NES accessible to export factors, ZBP1 export would not be strongly dependent on CRM1. We note that this model does not exclude the possibility that other CRM1-interacting proteins also contribute to Ro nuclear export.
Interestingly, although the Ro ortholog in D. radiodurans associates with the 3′–5′ exoribonuclease polynucleotide phosphorylase (Chen et al. 2007; Wurtmann and Wolin 2010), we did not identify an annotated exonuclease in our tandem affinity purification of Ro. One possibility is that the mouse Ro protein does not associate with exonucleases. Alternatively, the interaction may be of lower affinity in mammalian cells compared with bacteria. Moreover, we do not yet know how the other copurifying proteins, such as YB-1 and MOV10, affect Ro function. Future experiments will be aimed at examining the roles of these other proteins, as well as determining whether additional proteins, such as exonucleases, copurify with Ro under different conditions.
MATERIALS AND METHODS
Cell lines and cell culture
The generation of immortalized mouse wild-type, Ro−/−, and 3xFLAG-Ro fibroblast lines was described (Sim et al. 2009). To generate Protein A-TEV-FLAG-Ro cells, the IgG-binding domains of Protein A, a TEV cleavage site and a FLAG epitope were fused to mouse Ro using PCR and cloned into the Bam HI/Xho I sites of pUB6/V5-HisA (Invitrogen) to create pUB6-TFMRO. This construct places the tagged Ro under control of the human UbC promoter. After transfection into Ro−/− fibroblasts, stable lines were selected with 5 μg/mL of Blasticidin S (InvivoGen). To generate GFP-ZBP1 cells, a mouse ZBP1 cDNA (MGC IMAGE ID 30008106) was purchased from ATCC, the ORF fused to the eGFP C terminus, and cloned into pUB6/V5-HisA to generate pUB-GFP-ZBP1. Because the purchased cDNA contained a nucleotide change that replaced the conserved glutamate at position 276 with glycine, mutagenesis was used to restore the glutamate. After transfection into immortalized fibroblasts, stable lines were selected with Blastacidin S. All cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and 2 mM L-glutamine. UV irradiation was as described (Sim et al. 2009). In some experiments, 10 ng/mL of leptomycin B (Sigma-Aldrich) was added to the media and the cells fixed at the indicated times.
Purification of Ro and associated proteins
Protein A-TEV-FLAG-Ro and untagged wild-type fibroblasts (∼108 cells each) were harvested in PBS (Invitrogen) and lysed in 10 mL of buffer A (20 mM HEPES at pH 7.9, 20% glycerol, 200 mM KCl, 0.5% NP-40, 0.5 mM EDTA, 1x protease inhibitor cocktail [Roche Applied Science] and 1 mM phenylmethylsulfonyl fluoride [PMSF]) using a Dounce homogenizer. After centrifuging at 55K rpm at 4°C for 45 min in a TLA 100.2 rotor, the cleared lysate was incubated with 100 μL of IgG-Sepharose beads (GE Healthcare) at 4°C for 2 h. Beads were washed with buffer A before incubating with 100 U of TEV protease in 500 μL of TEV cleavage buffer (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.1% NP-40, 1 mM DTT) overnight at 4°C. The eluate from the cleavage reaction was incubated with anti-Flag M2-Agarose (Sigma-Aldrich) beads for 2 h, followed by washing with NET-2 (40 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.1% NP-40, 1 mM MgCl2) containing 1 mM PMSF and 1x protease inhibitor cocktail (Roche Applied Science). Ro was eluted with 0.2 mg/mL of 3xFLAG peptide in NET-2.
Mass spectrometry
After TCA precipitation, samples were resuspended in 8 M urea, 100 mM Tris (pH 8.5) and reduced with 100 mM TCEP. Iodoacetamide (55 mM final concentration) was added to alkylate cysteines. Following Lys-C (1 μg/100 μL) digestion for 4 h at 37°C, CaCl2 was added (final concentration of 1 mM), and trypsin (1 μg/100 μL) was used to digest the samples further. The peptides were pressure loaded onto a fused silica capillary desalting column containing 3 cm of 5-μm C18 resin packed into a 250-μm i.d. capillary with a 2-μm filtered union (UpChurch Scientific). The desalting column was washed with buffer containing 95% water, 5% acetonitrile, and 0.1% formic acid. After desalting, a 100 μm i.d capillary with a 5-μm pulled tip packed with 10 cm C18 material followed by 3 cm of 5-μm Partisphere strong cation exchanger (Whatman) was attached to the filter union and the entire split-column (desalting column–filter union–analytical column) was placed inline with an Agilent 1100 quaternary HPLC and analyzed using a 5-step separation described previously (Washburn et al. 2001).
As peptides eluted from the microcapillary column, they were electrosprayed directly into an LCQ-Deca mass spectrometer (ThermoFinnigan) with the application of a distal 2.4 kV spray voltage. A cycle of one full-scan mass spectrum (400–1400 m/z) followed by three data-dependent MS/MS spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcaliber datasystem.
MS/MS spectra were searched with the SEQUEST algorithm (Eng et al. 1994) against the most recent versions of the human mouse and rat databases for 2004 (Peng et al. 2003). No enzyme specificity was considered for any search. SEQUEST results were assembled and filtered using the DTASelect (version 1.9) program (Tabb et al. 2002; Cociorva et al. 2006). Filtering criteria for positive protein identifications were the identification of two unique peptides with Xcorr values of 2.0 for plus 1 spectra, 2.2 for plus 2 spectra, and 3.75 for plus 3 spectra.
Immunoprecipitations, immunoblotting, and immunofluoresence
For immunoprecipitations, cells were washed in PBS and sonicated in NET-2 containing 1 mM PMSF and 1x protease inhibitor cocktail. After centrifuging at 16,000g, lysates were incubated with anti-FLAG M2-conjugated agarose (Sigma-Aldrich) or other antibodies bound to protein A-Sepharose (GE Healthcare) as described (Wolin and Steitz 1984). For anti-FLAG immunoprecipitations, proteins were eluted with 0.2 mg/mL of 3xFLAG peptide. For other antibodies, proteins were eluted from the beads by boiling in SDS-PAGE loading buffer. RNAs in immunoprecipitates were isolated by phenol extraction and ethanol precipitation, fractionated in 5% polyacrylamide/8 M urea gels and subjected to Northern analysis. For RNase treatment, RNase A (Roche Applied Science) was added to the cleared lysates at the indicated concentrations for 20 min at room temperature (RT). Western blotting (Yoo and Wolin 1994) and immunofluorescence (Sim et al. 2009) were performed as described. Antibodies were mouse anti-FLAG M2 (Sigma-Aldrich), mouse anti-GAPDH (Sigma-Aldrich), rabbit anti-human YB1 (Stickeler et al. 2001), rabbit anti-chicken ZBP1 (Farina et al. 2003), rabbit anti-human PTB (Hall et al. 2004), mouse anti-human PTB (Invitrogen), and mouse anti-EF1A (Millipore). Anti-MOV10 antibody was generated in rabbits using the peptide CSGPRRHQNLPQEREGE. A monoclonal anti-Ro antibody that does not recognize native Ro (Xue et al. 2003) was used for Western blotting. The rabbit anti-mouse Ro antibody that recognizes native Ro (Chen et al. 2003) was used for immunofluorescence.
siRNA transfections and Northern analyses
The siRNAs used to deplete Y RNAs were mY1 sense: 5′-UGUUCUACACUUUCCCCCCUUUU-3′, mY1 antisense: 5′-AAGGGGGGAAAGUGUAGAACAUU-3′, mY3 sense: 5′-GUUACAGAUUUCUUUGUUCUU-3′, and mY3 antisense: 5′-GAACAAAGAAAUCUGUAACUU-3′. These siRNAs, as well as Smartpool siRNAs for mouse ZBP1 and the control siRNAs (siCONTROL Non-Targeting siRNA #1), were purchased from Dharmacon. Transfection was performed as described (Sim et al. 2009). For ZBP1, at 48 h after transfection, cells were subjected to a second round of transfection and incubated for an additional 24 h.
For Northern analyses, RNAs were separated in 5% polyacrylamide/8 M urea gels and transferred to Hybond-N (GE Healthcare) or ZetaProbe GT membranes (Bio-Rad). [γ32-P]ATP-labeled oligonucleotides were hybridized as described (Tarn et al. 1995). Oligonucleotide probes were mY1: 5′-AAGGGGGGAAAGTGTAGAACAGGA-3′, mY3: 5′-GAGCGGAGAAGGAACAAAGAAATCTG-3′, SRP RNA: 5′-TGCTCCGTTTCCGACCTGGGCCGGTTC-3′, and U6: 5′-ATGGAACGCTTCACGAATTTGCGAGTC-3′.
Fluorescence in situ hybridization
Wild-type, Ro−/−, and siRNA-transfected wild-type fibroblasts were plated on CC2-coated chamber slides (Nunc) and cultured overnight. FISH was performed as described (Yao et al. 2011). Briefly, cells were washed twice with PBS, fixed with 4% formaldehyde in PBS at RT, and permeabilized with 0.1% Triton in PBS. After washing with PBS + 0.05% Tween-20 (PBST), cells were washed once in 2X SSC + 0.05% Tween-20 and incubated in 2X SSC + 50% formamide at 37°C for 1 h. Oligonucleotides were labeled with the DIG-Oligonucleotide Tailing kit (Roche Applied Science), G-25 column-purified, denatured at 95°C for 5 min, and applied to the cells. Slides were sealed using a coverslip with rubber cement, and incubated in a humid chamber at 37°C overnight. Next, coverslips were removed and slides washed with 2X SSC + 50% Formamide at 37°C for 1 h, washed once in 2X SSC + 0.05% Tween-20 and three times with PBST at RT. After blocking for 1 h with 1% bovine serum albumin in PBST, the cells were incubated with anti-DIG-Rhodamine (Roche Applied Science, 1:50 dilution) at RT for 1 h. Slides were then washed three times with PBST, mounted on a coverslip using Vectashield (Vector Labs), and visualized with a Zeiss LSM510 META confocal microscope. Z-stack images were acquired and maximum intensity projection images were presented and analyzed using ImageJ. Oligonucleotide probes were mY1: 5′-GGGAAAGTGTAGAACAGGAGTTCAATCTGTAACTGACTGTGAACA-3′ and mY3: 5′-GAAGGAACAAAGAAATCTGTAACTGGTTGTGATCAATTAGTTGTAAAC-3′.
ACKNOWLEDGMENTS
We thank Bo Hu for studies of the binding of Ro to CRM1 and Thomas Cooper (Baylor College of Medicine), Robert Singer (Albert Einstein College of Medicine), and Douglas Black (University of California, Los Angeles) for gifts of antibodies. This work was supported by a postdoctoral fellowship from the Arthritis Foundation (S.S.), grants from the Ellison Medical Foundation (S.L.W.) and the National Institutes of Health (R01 GM073863 and GM048410 to S.L.W. and P41 RR011823 to J.R.Y. III), and startup funds from Yale University (J.Y.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029207.111.
REFERENCES
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Görlich D 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21: 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffard P, Barbar E, Brière F, Boire G 2000. Interaction cloning and characterization of RoBPI, a novel protein binding to human Ro ribonucleoproteins. RNA 6: 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH 2010. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev 24: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG 2010. Decoding the function of nuclear long noncoding RNAs. Curr Opin Cell Biol 22: 357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Smith JD, Shi H, Yang DD, Flavell RA, Wolin SL 2003. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr Biol 13: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Chen X, Wurtmann EJ, Van Batavia J, Zybailov B, Washburn MP, Wolin SL 2007. An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC 2006. High molecular-mass APOBEC3G complexes restrict Alu transcription. Proc Natl Acad Sci 103: 15588–15593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociorva D, Tabb D, Yates JRI 2006. Validation of tandem mass spectrometry database search results using DTASelect. Curr Protoc Bioinformatics 16: 13.14.11–13.14.14 [DOI] [PubMed] [Google Scholar]
- Doyle GA, Betz NA, Leeds PF, Fleisig AJ, Prokipcak RD, Ross J 1998. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res 26: 5036–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Messaoudi-Aubert S, Nicholls J, Maertens GN, Brookes S, Bernstein E, Peters G 2010. Role for the MOV10 RNA helicase in polycomb-mediated repression of the INK4a tumor suppressor. Nat Struct Mol Biol 17: 862–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, Mccormack AL, Yates JR 1994. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989 [DOI] [PubMed] [Google Scholar]
- Fabini G, Raijmakers R, Hayer S, Fouraux MA, Pruijn GJ, Steiner G 2001. The heterogeneous nuclear ribonucleoproteins I and K interact with a subset of Ro ribonucleoprotein-associated Y RNAs in vitro and in vivo. J Biol Chem 276: 20711–20718 [DOI] [PubMed] [Google Scholar]
- Farina KL, Hüttelmaier S, Musunuru K, Darnell R, Singer RH 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Forsgren A, Sjoquist J 1967. “Protein A” from Staphylococcus aureus. 3. Reaction with rabbit γ-globulin. J Immunol 99: 19–24 [PubMed] [Google Scholar]
- Fouraux MA, Bouvet P, Verkaart S, van Venrooij WJ, Pruijn GJ 2002. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J Mol Biol 320: 475–488 [DOI] [PubMed] [Google Scholar]
- Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol 81: 2165–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CD, Long KS, Shi H, Wolin SL 1998. Binding of the 60-kDa Ro autoantigen to Y RNAs: Evidence for recognition in the major groove of a conserved helix. RNA 4: 750–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, Ossareh-Nazari B, Brownawell AM, Doglio A, Bertrand E, Macara IG, Dargemont C 2003. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J Biol Chem 278: 5505–5508 [DOI] [PubMed] [Google Scholar]
- Hall MP, Huang S, Black DL 2004. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol Biol Cell 15: 774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Rivas FV, Murchison EP, Steitz JA 2006. The expanding universe of noncoding RNAs. Cold Spring Harb Symp Quant Biol 71: 551–564 [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA 1981. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol 1: 1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Collins K 2007. Human Y5 RNA specializes a Ro ribonucleoprotein for 5S ribosomal quality control. Genes Dev 21: 3067–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Pai DA, Engelke DR 2010. Cellular dynamics of tRNAs and their genes. FEBS Lett 584: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH 2005. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515 [DOI] [PubMed] [Google Scholar]
- Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC 2007. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics 6: 798–811 [DOI] [PubMed] [Google Scholar]
- Köhn M, Lederer M, Wächter K, Hüttelmaier S 2010. Near-infrared (NIR) dye-labeled RNAs identify binding of ZBP1 to the noncoding Y3-RNA. RNA 16: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike K, Uchiumi T, Ohga T, Toh S, Wada M, Kohno K, Kuwano M 1997. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett 417: 390–394 [DOI] [PubMed] [Google Scholar]
- Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem 281: 29105–29119 [DOI] [PubMed] [Google Scholar]
- Labbe JC, Hekimi S, Rokeach LA 1999. The levels of the RoRNP-associated Y RNA are dependent upon the presence of ROP-1, the Caenorhabditis elegans Ro60 protein. Genetics 151: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ 2005. YB-1 is important for late-stage embryonic development, optimal cellular stress responses and the prevention of premature senescence. Mol Cell Biol 25: 4625–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T 2005. Identification of novel Argonaute-associated proteins. Curr Biol 15: 2149–2155 [DOI] [PubMed] [Google Scholar]
- Mosig A, Guofeng M, Stadler BMR, Stadler PF 2007. Evolution of the vertebrate Y RNA cluster. Theory Biosci 126: 9–14 [DOI] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 19: 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Adolph SK, Rajpert-De Meyts E, Lykke-Andersen J, Koch G, Christiansen J, Nielsen FC 2003. Nuclear transit of human zipcode-binding protein IMP1. Biochem J 376: 383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien CA, Wolin SL 1994. A possible role for the 60 kd Ro autoantigen in a discard pathway for defective 5S ribosomal RNA precursors. Genes Dev 8: 2891–2903 [DOI] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH 2003. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol 13: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2: 43–50 [DOI] [PubMed] [Google Scholar]
- Perreault J, Perreault J-P, Boire G 2007. Ro-associated Y RNAs in metazoans: evolution and diversification. Mol Biol Evol 24: 1678–1689 [DOI] [PubMed] [Google Scholar]
- Pruijn GJM, Slobbe RL, van Venrooij WJ 1991. Analysis of protein-RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res 19: 5173–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjes SA, Lund E, van der Heijden A, Grimm C, van Venrooij WJ, Pruijn GJ 2001. Identification of a novel cis-acting RNA element involved in nuclear export of hY RNAs. RNA 7: 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, O'Brien CA, Van Horn DJ, Wolin SL 1996. A misfolded form of 5S rRNA is associated with the Ro and La autoantigens. RNA 2: 769–784 [PMC free article] [PubMed] [Google Scholar]
- Sim S, Wolin SL 2011. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscip Rev: RNA 2: 686–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL 2009. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell 20: 1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons FHM, Pruijn GJM, van Venrooij WJ 1994. Analysis of the intracellular localization and assembly of Ro ribonucleoprotein particles by microinjection into Xenopus laevis oocytes. J Cell Biol 125: 981–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons FHM, Rutjes SA, van Venrooij WJ, Pruijn GJM 1996. The interactions with Ro60 and La differentially affect nuclear export of hY1 RNA. RNA 2: 264–273 [PMC free article] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM 2005. Structural insights into RNA quality control: The Ro autoantigen binds misfolded RNAs via its central cavity. Cell 121: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA 2001. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J 20: 3821–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Hüttelmaier S 2006. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol 175: 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR III 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W-Y, Yario TA, Steitz JA 1995. U12 snRNA in vertebrates: Evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of intron. RNA 1: 644–656 [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR III 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19: 242–247 [DOI] [PubMed] [Google Scholar]
- Weidensdorfer D, Stöhr N, Baude A, Lederer M, Köhn M, Schierhorn A, Buchmeier S, Wahle E, Hüttelmaier S 2009. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15: 104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Steitz JA 1984. The Ro small cytoplasmic ribonucleoproteins: Identification of the antigenic protein and its binding site on the Ro RNAs. Proc Natl Acad Sci 81: 1996–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R 2007. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 21: 415–426 [DOI] [PubMed] [Google Scholar]
- Wurtmann EJ, Wolin SL 2010. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc Natl Acad Sci 107: 4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Shi H, Smith JD, Chen X, Noe DA, Cedervall T, Yang DD, Eynon E, Brash DE, Kashgarian M, et al. 2003. A lupus-like syndrome develops in mice lacking the Ro 60-kDa protein, a major lupus autoantigen. Proc Natl Acad Sci 100: 7503–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Fetter RD, Hu P, Betzig E, Tjian R 2011. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev 25: 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, Ruse CI, Nakorchevsky A 2009. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng 11: 49–79 [DOI] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL 1994. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol 14: 5412–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]