Ski2 is a cytoplasmic RNA helicase that functions together with the exosome in the turnover and quality control of mRNAs. Ski2 is conserved in eukaryotes and is related to the helicase Mtr4, a cofactor of the nuclear exosome involved in the processing and quality control of a variety of structured RNAs. The authors have determined the 2.4 Å resolution crystal structure of the 113 kDa helicase region of Saccharomyces cerevisiae Ski2. The structure shows that Ski2 has an overall architecture similar to that of Mtr4, with a core DExH region and an extended insertion domain. Biochemically, the Ski2 insertion has broad substrate specificity, binding both single-stranded and double-stranded RNAs. Therefore, it seems possible that the Ski2 and Mtr4 insertion domains have evolved with different properties tailored to the type of transcripts that are the substrates of the cytoplasmic and nuclear exosome.
Keywords: RNA degradation, exosome, helicase, structure
Abstract
Ski2 is a cytoplasmic RNA helicase that functions together with the exosome in the turnover and quality control of mRNAs. Ski2 is conserved in eukaryotes and is related to the helicase Mtr4, a cofactor of the nuclear exosome involved in the processing and quality control of a variety of structured RNAs. We have determined the 2.4 Å resolution crystal structure of the 113 kDa helicase region of Saccharomyces cerevisiae Ski2. The structure shows that Ski2 has an overall architecture similar to that of Mtr4, with a core DExH region and an extended insertion domain. The insertion is not required for the formation of the Ski2–Ski3–Ski8 complex, but is instead an RNA-binding domain. While this is reminiscent of the Mtr4 insertion, there are specific structural and biochemical differences between the two helicases. The insertion of yeast Mtr4 consists of a β-barrel domain that is flexibly attached to a helical stalk, contains a KOW signature motif, and binds in vitro-transcribed tRNAiMet, but not single-stranded RNA. The β-barrel domain of yeast Ski2 does not contain a KOW motif and is tightly packed against the helical stalk, forming a single structural unit maintained by a zinc-binding site. Biochemically, the Ski2 insertion has broad substrate specificity, binding both single-stranded and double-stranded RNAs. We speculate that the Ski2 and Mtr4 insertion domains have evolved with different properties tailored to the type of transcripts that are the substrates of the cytoplasmic and nuclear exosome.
INTRODUCTION
The exosome is a conserved and essential macromolecular complex that degrades RNA substrates processively from the 3′ end (Mitchell et al. 1997). In the eukaryotic nucleus, the exosome is involved in the maturation of ribosomal RNAs, small nuclear RNAs, and small nucleolar RNAs (Allmang et al. 1999; van Hoof et al. 2000a; Houalla et al. 2006). It functions in the turnover of pre-mRNAs and cryptic unstable transcripts (Bousquet-Antonelli et al. 2000; Hilleren et al. 2001; Wyers et al. 2005). It is also required in quality-control mechanisms that target aberrant nuclear RNAs such as hypomodified tRNAiMet (Kadaba et al. 2004; Vanacova et al. 2005). In the cytoplasm, the exosome is involved in bulk mRNA turnover (Anderson and Parker 1998; van Hoof et al. 2000b) and also participates in surveillance pathways for the degradation of aberrant mRNAs that contain a premature stop codon (Lejeune et al. 2003; Mitchell and Tollervey 2003; Takahashi et al. 2003; Gatfield and Izaurralde 2004) or lack one altogether (van Hoof et al. 2002).
The 10-subunit core of the eukaryotic exosome is identical in the nuclear and cytoplasmic compartments (for review, see Lorentzen et al. 2008a; Lykke-Andersen et al. 2009). Nine subunits form a barrel-like structure (Exo-9) with a prominent central channel (Liu et al. 2006). The structure of Exo-9 is similar to that of the archaeal exosome and bacterial PNPase, but lacks the catalytic activity that is characteristic of these prokaryotic complexes (Büttner et al. 2005; Lorentzen et al. 2005, 2007). The nuclease activity of the core exosome complex is conferred by the tenth subunit, Rrp44 (Liu et al. 2006; Dziembowski et al. 2007; Lebreton et al. 2008; Schaeffer et al. 2009; Schneider et al. 2009). Both Rrp44 and the catalytically inactive Exo-9 subunits are essential in yeast. The Exo-9 subcomplex modulates the activity of Rrp44 (Liu et al. 2006; Dziembowski et al. 2007; Lorentzen et al. 2008b) and binds RNA substrates, guiding them through the central channel to reach the exoribonuclease active site (Bonneau et al. 2009). The Exo-9 structure is also thought to recruit peripheral factors, such as the nuclear ribonuclease Rrp6 (Liu et al. 2006; Cristodero et al. 2008) and the cytoplasmic protein Ski7 in yeast (Araki et al. 2001; Dziembowski et al. 2007).
These peripheral factors associate with the core exosome to form an outer shell that is compartment specific (for review, see Lebreton and Seraphin 2008). In the nucleus, the exosome functions together with the helicase Mtr4, which associates with a poly(A) polymerase (Trf4/Trf5) and a zinc finger protein (Air1/Air2) to form the TRAMP complex (LaCava et al. 2005). In the cytoplasm, the exosome functions together with the Ski complex (Anderson and Parker 1998). The Ski (Superkiller) proteins were originally identified from recessive mutations that exacerbated the “killer” phenotype, that is, the ability of yeast strains containing a dsRNA virus to produce a toxin that kills other strains (Toh-E and Wickner 1979; Ridley et al. 1984; Johnson and Kolodner 1995). These studies showed that the Superkiller mutations resided in a helicase (Ski2), a tetratricopeptide-repeat (TPR) protein (Ski3), and a WD40 protein (Ski8) in addition to the cytoplasmic 5′–3′ exoribonuclease (Xrn1). The Ski2, Ski3, and Ski8 proteins were later found to associate in a complex in vivo (Brown et al. 2000). The Ski complex has been implicated in many 3′–5′ cytoplasmic degradation pathways mediated by the exosome, including normal RNA turnover (Anderson and Parker 1998; van Hoof et al. 2000b; Araki et al. 2001), nonsense-mediated decay (Mitchell and Tollervey 2003), nonstop decay (van Hoof et al. 2002), and RNA interference (Orban and Izaurralde 2005). The Ski and exosome complexes interact not only genetically, but also physically via the yeast Ski7 protein (Araki et al. 2001).
The presence of a helicase in both the Ski and TRAMP complexes is intriguing. These exosome-associated helicases are thought to contribute to substrate recognition, to unwind secondary structure elements in the nucleic acids, or to remove bound proteins, and eventually to present favorable single-stranded RNA substrates to the exosome (Lebreton and Seraphin 2008; Houseley and Tollervey 2009). The parallel between the nuclear and cytoplasmic regulators of the exosome is further compounded by the fact that Ski2 and Mtr4 share significant sequence similarity (∼35% sequence identity in the predicted helicase region). Previous structural work has shown that Mtr4 has a helicase core similar to that found in other members of the DExH family (Jackson et al. 2010; Weir et al. 2010), including the archaeal DNA helicase Hel308 (Büttner et al. 2007) and the splicing helicase Prp43 (He et al. 2010; Walbott et al. 2010). In addition, Mtr4 features a 200 aa insertion that contains a helical stalk and a β-barrel domain. The latter is structurally and functionally similar to KOW domains, which were shown to bind structured RNAs in ribosomal proteins (Kyrpides et al. 1996; Selmer et al. 2006; Zhang et al. 2009). Consistently, the KOW domain of yeast Mtr4 is required for 5.8S rRNA processing in vivo (Jackson et al. 2010) and binds transcribed tRNAiMet in vitro (Weir et al. 2010). Thus, the specific structural features of Mtr4 are in line with its biological functions in ribosomal RNA processing and quality control (de la Cruz et al. 1998; van Hoof et al. 2000a). Sequence alignments and secondary structure predictions suggest that Ski2 has a helicase and an insertion domain similar to those in Mtr4. However, the potential RNA substrates that Ski2 encounters in the cytoplasm are different from those that are recognized by Mtr4 in the nucleus. This raises the question as to whether Ski2 has specific features as compared with Mtr4. To address this question, we have analyzed the structural and biochemical properties of Ski2 from Saccharomyces cerevisiae.
RESULTS AND DISCUSSION
Structure determination of the 113 kDa helicase region of yeast Ski2
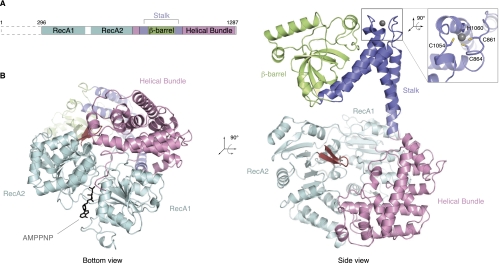
To initiate the biochemical and structural characterization of Ski2, we expressed and purified the full-length (f.l.) S. cerevisiae ortholog (residues 1–1287) from insect cells. Ski2 is predicted to have a low-complexity N-terminal region that has previously been shown to interact with Ski3 by yeast two-hybrid and coimmunoprecipitation analyses (Wang et al. 2005). Limited proteolysis of f.l. Ski2 in combination with N-terminal sequencing and mass spectrometry indicated the presence of a protease-resistant fragment of ∼113 kDa that lacked the N-terminal 295 residues and included the predicted helicase region (data not shown). We therefore engineered a construct of yeast Ski2 encompassing residues 296–1287 (designated Ski2-ΔN) (Fig. 1A). Yeast Ski2-ΔN was obtained from expression in insect cells and crystallized in the presence of adenosine 5′-(β,γ-imido)triphosphate (AMPPNP), a nonhydrolyzable ATP analog.
FIGURE 1.
Ski2 consists of a DExH helicase core and a protruding insertion domain. (A) Schematic representation of the domain organization of Ski2. The N-terminal low-complexity region not included in the crystal structure is indicated by a dashed box (residues 1–295 in S. cerevisiae), and the helicase region is shown with the domains in different colors as in the crystal structure below. (B) The S. cerevisiae Ski2-ΔN crystal structure (lacking the N-terminal 295 residues). This composite model was generated from the Ski2-ΔN and the Ski2-insert crystal structures (see text). The two views are related by a 90° rotation as indicated. In the DExH core, the RecA domains are colored in cyan and the helical bundle domain in light pink. The unwinding β-hairpin is highlighted in red, and AMPPNP (black) is depicted in stick representation. The stalk helices and the β-barrel in the insertion domain are shown in blue and green, respectively. A zinc ion present in the stalk region is shown as a gray sphere. A close-up shows the CCCH-type coordination of the zinc ion.
The crystal structure of Ski2-ΔN was solved to 2.4 Å resolution by the multiwavelength anomalous diffraction (MAD) method using a gold derivative (see Table 1 for data collection and refinement statistics). The DExH helicase core of Ski2-ΔN could be built and refined almost in its entirety (with the exception of a disordered region between residues 542 and 606) and included electron density for an AMPPNP molecule. An additional domain could be identified that was inserted between residues 830 and 1086, consisting of a helical part and a globular region. However, the electron density for the globular domain, in particular, was weak and did not allow us to trace the polypeptide chain with an unambiguous amino acid register. The structure of Ski2-ΔN bound to AMPPNP was refined to 2.4 Å resolution, with Rwork of 23.8%, Rfree of 27.5%, and good stereochemistry (Table 1). We proceeded by engineering and crystallizing the region corresponding to the inserted domain (residues 835–1085, designated Ski2-insert). The structure of Ski2-insert was determined de novo by the single anomalous diffraction (SAD) method using a crystal derivatized with gold. Crystals of Ski2-insert contained five independent molecules in the asymmetric unit. After density modification procedures, the electron density of Ski2-insert was continuous and allowed unambiguous model building. The structure of Ski2-insert was refined at 3.25 Å resolution, with Rwork of 23.6% and Rfree of 25.7% (Table 1). The model of Ski2-insert was then placed into the electron density of Ski2-ΔN with relatively minor rigid-body adjustments to give a composite model as is discussed below.
TABLE 1.
Crystallographic data collection, phasing, and refinement statistics for the Ski2-ΔN and Ski2-insert structures
Ski2 contains a conserved DExH core
Ski2-ΔN contains two distinct structural features: a compact DExH helicase core and an elongated insertion (Fig. 1B). The helicase core of Ski2 is similar to that previously described for other DExH proteins, including the DNA helicase Hel308 (Büttner et al. 2007), the splicing helicase Prp43 (He et al. 2010; Walbott et al. 2010), and the exosome helicase Mtr4 (Jackson et al. 2010; Weir et al. 2010). Briefly, the core consists of a circular arrangement of two RecA domains (RecA1 and RecA2) and a helical bundle (Fig. 1B, left). The two RecA domains contain the characteristic helicase signature motifs that mediate substrate binding and ATP hydrolysis (for review, see Pyle 2008). Also conserved is the unwinding β-hairpin in the RecA2 domain (residues 741–752, in red in Fig. 1B). This β-hairpin is characteristic of DExH-box helicases and has been shown in the Hel308 structure to wedge between the two strands of a DNA duplex that is being unwound at the 5′ end as it approaches the DExH core (Supplemental Fig. 1A; Büttner et al. 2007).
In the AMPPNP-bound structure of Ski2-ΔN, the two RecA domains face each other in a closed conformation (Fig. 1B, left). The conformation is similar to that of other DExH proteins determined in the presence of nucleic acids. For instance, both RecA domains of Ski2 superpose to those of the Mtr4–ADP–RNA structure (Weir et al. 2010), with a root mean square deviation (RMSD) of 1.3 Å over 373 Cα atoms. The overall orientation of the two RecA domains in Ski2 and other DExH proteins is restrained by the interaction with the helical bundle. AMPPNP binds between the two RecA domains with the adenosine base sandwiched by Phe328 of RecA1 and Arg767 of RecA2 (Supplemental Fig. 1B). The phosphates of AMPPNP are arranged around motif I in a canonical conformation, but electron density for the γ-phosphate is weak. This may be caused by the absence of magnesium, which would be required to properly coordinate the phosphate groups, but was not present in the crystallization condition of Ski2-ΔN.
An insertion domain protrudes from the Ski2 DExH core
The DExH core of Ski2 has an insertion of about 250 residues that occurs in the middle of the helical bundle domain (Fig. 1A). The insertion forms an elongated structure that protrudes from the core, extending by ∼50 Å from the unwinding β-hairpin that marks the position of the 5′ end of RNA bound to the DExH core (Fig. 1B, right; Supplemental Fig. 1A). The insertion folds into two pairs of antiparallel helices (α1–α4 and α2–α3), which connect to a distal β-barrel domain (Fig. 2A). The insertion is a rather flexible feature of the structure. First, the crystallographic temperature factors increase significantly as compared with the core (60 Å2 for the DExH core, 120 Å2 for helices α1–α4, 145 Å2 for helices α2–α3, and 130 Å2 for the barrel). Second, we observe domain movements when comparing the insertion domain in the two structures that we determined: Superposition of the helices α1–α4 in the Ski2-ΔN and Ski2-insert structures results in a rotation of ∼15° in the position of helices α2–α3 and the β-barrel (Supplemental Fig. 1C). Despite the flexibility, the electron density is well defined in the structure of Ski2-insert (Supplemental Fig. 1D).
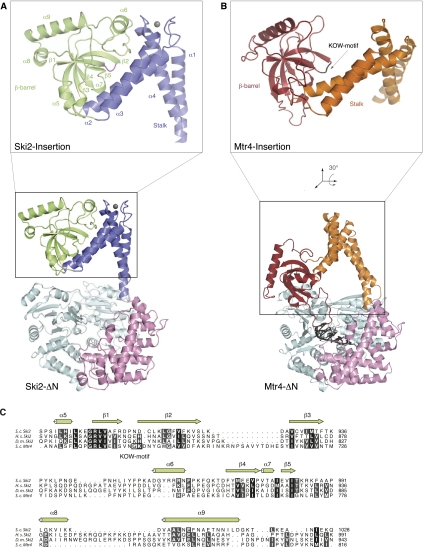
FIGURE 2.
The exosome helicases Ski2 and Mtr4 have a similar architecture. (A) The structure of yeast Ski2-ΔN as shown in Figure 1B, with the same color scheme as in Figure 1. The close-up view shows the insertion domain with the secondary structure elements labeled. The β1–β2 loop is highlighted in magenta. (B) The structure of yeast Mtr4-ΔN (pdb code 2xgj) is shown in the same orientation as Ski2-ΔN in A, after optimal superposition of the DExH core. The domains in the DExH core are colored as in Ski2-ΔN, the stalk is shown in orange, and the β-barrel in red. For the close-up view, the insertion domain has been reoriented as indicated so that the β-barrel is in the same orientation as the Ski2 β-barrel in A. The β1–β2 loop in the β-barrel containing the KOW signature motif is depicted in black. (C) A structure-based sequence alignment of the β-barrel domains of S. cerevisiae (S.c.) Ski2 and Mtr4. The secondary structure elements of Ski2 are indicated. The alignment includes Ski2 sequences from Homo sapiens (H.s.) and Drosophila melanogaster (D.m.). Conservation is indicated in shades of gray and the KOW motif of Mtr4 is indicated by a black box. Sequence conservation between Ski2 and Mtr4 is low in the insertion region and mostly restricted to structural residues that define the β-barrel. A comprehensive sequence alignment is shown in Supplemental Figure 2.
In contrast to the DExH core, the insertion is poorly conserved even among yeast species (Supplemental Fig. 2). Yet, it is a characteristic feature of exosome helicases: in Mtr4 an insertion occurs at an equivalent position in the sequence and with a comparable topology of secondary structure elements (Fig. 2A,B). The structure of the helical stalk of Ski2 is similar to that of Mtr4 in that it forms an L-like shape with the α1–α4 and α2–α3 pairs of helices roughly perpendicular to each other. In the case of yeast Ski2, the two pairs of helices are connected by a zinc-binding CCCH-type motif (Fig. 1B, right). Although no zinc was present in the purification or crystallization buffers, additional electron density is present at the center of a coordination sphere formed by Cys861, Cys864, Cys1054, and His1060. The density is consistent with a zinc ion, the presence of which was confirmed by X-ray fluorescence (data not shown). This zinc-binding site has a structural role in forming the hinge region that connects the stalk and the β−barrel domain. The four residues that coordinate the zinc ion are conserved in Ski2 orthologs in fungi, but not, for example, in metazoans (Supplemental Fig. 2), where other intramolecular interactions might serve a similar structural role.
The core structure of the β-barrel domain of Ski2 is also similar to that of Mtr4. The corresponding β-strands superpose with an RMSD of 2.6 Å, while the connecting loops vary in sequence, length, and conformation. The evolutionarily conserved residues are mostly restricted to positions on the β-strands pointing into the hydrophobic core (Fig. 2C). A key difference between the Ski2 and Mtr4 β-barrels is located in the loop connecting strands β1 and β2. The β1–β2 loop of yeast Mtr4 contains a KOW-domain signature motif (Gly686 and Lys687), a surface feature that is characteristic of domains involved in binding structured RNAs in ribosomal proteins such as bacterial L24 and eukaryotic L26 (Kyrpides et al. 1996; Selmer et al. 2006; Zhang et al. 2009). The KOW motif is not present in the corresponding β1–β2 loop of Ski2. In addition, this loop is not accessible to solvent (Fig. 2A). The barrel and the stalk of Ski2 interact via an extensive intramolecular interface (Fig. 2A; Supplemental Fig. 1D), suggesting that they form a single structural unit. These structural differences are also reflected in the biochemical properties of the two domains: while the β-barrel of Mtr4 is stable when expressed and purified in isolation (Weir et al. 2010), we could only obtain expression of a soluble β-barrel-containing fragment of Ski2 when including the α2–α3 helices and the zinc-binding site (data not shown).
The Ski2 insertion contributes to RNA binding
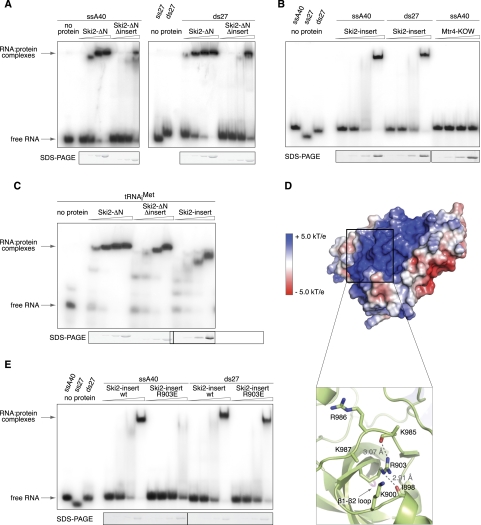
The overall structural similarity between the two exosome helicases suggests that the insertion of Ski2 functions analogously to that of Mtr4 in terms of contributing a second RNA-binding site in addition to the one expected in the DExH core. To experimentally test whether the Ski2 insertion participates in RNA binding, we used electrophoretic mobility shift assays (EMSA) and compared the RNA-binding properties of Ski2 in the presence and absence of the insertion domain (Ski2-ΔN and Ski2-ΔN-Δinsert) (Fig. 3A). The Ski2-ΔN-Δinsert mutant was engineered by replacing residues 835–1085 with a linker sequence (Gly–Ser–Arg–Gly) and behaved like Ski2-ΔN in biochemical purifications. A single-stranded poly(A) 40-mer RNA (ssA40) bound strongly to Ski2-ΔN, but markedly less well to Ski2-ΔN-Δinsert (Fig. 3A, left). A similar pattern was detected when using a double-stranded 27-mer (ds27) as substrate (Fig. 3A, right). We conclude that the insertion domain increases the affinity of Ski2 for RNA. Consistently, the insertion alone (Ski2-insert, residues 835–1085) showed robust binding to single-stranded and double-stranded RNAs (ss40 and ds27) (Fig. 3B). The Ski2 insertion was also able to bind unmodified tRNAiMet in vitro (Fig. 3C), pointing to the broad substrate-binding properties of this domain. This contrasts to the specialized KOW domain of Mtr4, which binds unmodified tRNAiMet (Weir et al. 2010), but does not show significant binding to single-stranded RNA (Fig. 3B).
FIGURE 3.
The insertion domain of Ski2 binds single- and double-stranded RNA substrates. (A) RNA binding to fragments of Ski2 that contain the DExH core with and without the insertion domain (Ski2-ΔN and Ski2-ΔN-Δinsert, respectively). Electrophoretic mobility shift assays (EMSA) were carried out with a single-stranded poly(A) 40-mer (left) or double-stranded 27-mer (right) that were labeled at the 5′ end with [32P]phosphate. Identical concentrations (0.25 μM, 0.75 μM, 2.25 μM, 6.75 μM) were used for both Ski2-ΔN and Ski2-ΔN-Δinsert and all other proteins in this figure. (Bottom) A Coomassie-stained SDS-PAGE of the protein sample used in the corresponding lanes of the gel-shift assay. Deletion of the insertion domain significantly decreases the affinity for single-stranded and double-stranded RNA. (B) EMSAs as described above with single-stranded (ssA40) and double-stranded RNA (ds27) using the Ski2 insertion domain (residues 835–1085). As compared with the Mtr4 KOW domain (residues 667–818), which fails to bind single-stranded RNA at this condition, the Ski2 insertion binds single- and double-stranded RNA with comparable affinity. (C) EMSAs show that the Ski2 insertion binds in vitro-transcribed tRNAiMet with an apparently comparable affinity to that for single- or double-stranded RNA. (D) An electrostatic surface potential analysis reveals a prominent positively charged patch on the surface of the Ski2 β-barrel. The domain is shown in a similar orientation to that used in Figure 2A, but without the α1 and α4 helices (which have been omitted for clarity). Positive electrostatic potential is shown in blue, negative potential in red. The close-up view at bottom shows how the conserved Arg903 organizes the positively charged surface by coordinating two loops with several basic residues (indicated). The side-chains of Lys985 and Lys987 are disordered. Hydrogen bonds are indicated by dashed lines together with distances. (E) EMSAs suggest that mutation of Arg903 to glutamic acid impairs binding of the Ski2-insert to single-stranded RNA (A40) but does not affect interaction with double-stranded RNA (ds27).
As discussed above, the KOW sequence motif is not present in the equivalent β1–β2 loop in the Ski2-insertion. We thus asked which surface features of the Ski2 insert are involved in the interaction with RNA. Calculation of an electrostatic surface potential revealed a prominent positively charged patch on the opposite side of the β1–β2 loop, stretching from the hinge region to the tip of the domain (Fig. 3D, top). This surface patch is partly organized by hydrogen bonds between the conserved Arg903 and the carbonyl groups of two adjacent loops, which are characterized by several positively charged residues (Fig. 3D, bottom). The central arginine residue is also present in yeast Mtr4 (Arg678), while the specific features of the loops diverge between Ski2 and Mtr4 (Fig. 2C). To perturb the positively charged surface patch of the Ski2 insertion, we mutated Arg903 to a glutamic acid. We purified the Ski2-insert R903E mutant with a similar protocol as the wild-type Ski2-insert and compared their binding properties using EMSA assays (Fig. 3E). We found that binding to single-stranded RNA was impaired in the Ski2-insert R903E mutant, while binding to double-stranded RNA was not affected. These results indicate that the charged surface patch on the Ski2 β-barrel domain is the major binding site for single-stranded RNA. They also suggest the presence of a different or more complex binding site for the recognition of double-stranded RNAs. We conclude that the insertion domain of Ski2 binds both single-stranded and structured RNA substrates. It increases the RNA-binding capabilities of the helicase by providing a second interaction site in addition to that of the DExH core.
The Ski2 insertion is not required to form the Ski complex
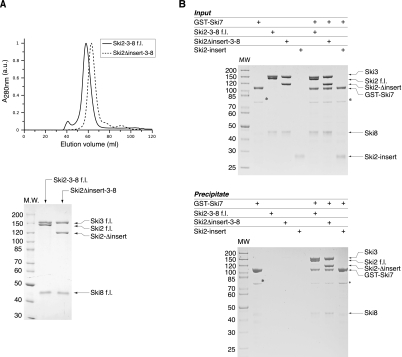
We next tested whether the insertion of Ski2 is required to mediate protein–protein interactions with the other subunits of the Ski complex. The core of the Ski complex is formed by the interaction of Ski2 with a large TPR protein (Ski3) and a small WD40 protein (Ski8). Yeast two-hybrid and coimmunoprecipitation data suggest that the N-terminal region of Ski2 mediates binding to Ski3, which in turn interacts with Ski8 (Wang et al. 2005). In line with these data, coexpression of f.l. Ski2, Ski3, and Ski8 in insect cells resulted in a stable ternary complex that eluted as a single peak in size-exclusion chromatography (Fig. 4A). Removal of the insertion in Ski2 did not affect complex formation: Ski2-Δinsert comigrated with Ski3 and Ski8 in size-exclusion chromatography (Fig. 4A). We next tested whether the insertion of Ski2 is required to bind Ski7, an outer-layer protein of the Ski complex that mediates the interaction with the exosome (Araki et al. 2001; Wang et al. 2005). In in-vitro pull-down experiments with purified proteins, f.l. GST–Ski7 efficiently precipitated both the Ski2–Ski3–Ski8 and the Ski2Δinsert–Ski3–Ski8 complexes, but not Ski2-insert alone (Fig. 4B). We conclude that the Ski2 insertion domain is not required for the formation of the Ski complex.
FIGURE 4.
The Ski2 insertion domain is not required for the protein–protein interactions of the Ski complex. (A) Analytical gel-filtration of f.l. Ski2–Ski3–Ski8 and Ski2Δinsert–Ski3–Ski8 complexes. Both complexes were copurified and then injected on an analytical size-exclusion chromatography column (top). Peak fractions were analyzed by SDS-PAGE (bottom). All three components comigrated in the case of both complexes, indicating that deletion of the insertion domain of Ski2 does not impair Ski complex formation. (B) Pull-down experiments with GST-tagged f.l. Ski7 (residues 1–747) and untagged f.l. Ski2–Ski3–Ski8 and Ski2Δinsert–Ski3–Ski8 complexes. Input samples (top) and samples precipitated on glutathione–Sepharose beads (bottom) were analyzed by SDS-PAGE. The proteins corresponding to the bands are indicated on the right side of both panels. Both complexes were efficiently precipitated by GST–Ski7, while the insertion alone (Ski2-insert) did not bind to the bait. (*) A contamination in the GST–Ski7 sample.
CONCLUSIONS
The two main regulators of exosome activity, the cytosolic Ski and the nuclear TRAMP complexes, contain related RNA helicases. The Ski2 and Mtr4 helicases have a similar architecture with two distinct structural modules: a helicase core and an insertion region. The helicase core is typical of the DExH family of helicases, and is expected to mediate ATP-dependent RNA unwinding and remodeling activities. The insertion contains a long stalk and a β-barrel domain. Removal of the insertion in Ski2 and Mtr4 does not compromise the formation of the respective complexes with Ski3 and Ski8 and with Trf4–Air2 (Weir et al. 2010), but significantly reduces the RNA-binding properties of the two helicases. The insertion domains of both Ski2 and Mtr4 bind RNA in vitro. Their position above the entry site of the RNA-binding channel in the helicase core suggests that they contribute to recruiting RNA substrates to the unwinding site.
Despite these overarching similarities, the β-barrel domains of Mtr4 and Ski2 have specific structural and functional differences. In yeast Mtr4, the β-barrel domain has a KOW signature motif, is involved in 5.8S rRNA maturation in vivo (Jackson et al. 2010), binds unmodified tRNAiMet (Weir et al. 2010), and discriminates against single-stranded RNA in vitro. In Ski2, the β-barrel domain does not exhibit a preference for binding structured RNAs, but is rather promiscuous in its RNA-binding properties. The β-barrel of Ski2 does not contain a KOW motif in the corresponding loop (β1–β2). Mutagenesis studies suggest that binding of the Ski2 β-barrel to single-stranded RNA is mediated by the opposite surface, which features several positively charged residues. We speculate that the β-barrel domains of Ski2 and Mtr4 have specialized to reflect the type of transcripts that are presented to the exosome by the two helicases. This specialization contributes to the differential processing of substrates by the cytoplasmic and nuclear exosome.
MATERIALS AND METHODS
Protein purification
Ski2 full length (1–1287) and Ski2-ΔN (296–1287) were expressed in Hi5 insect cells from recombinant baculoviruses. Coding sequences including an N-terminal 6xHis-tag, followed by a 3C protease site, were subcloned into a pFastBac1 vector (Invitrogen). Generation of bacmids and viruses as well as protein expression was done according to standard procedures. Cells were lysed osmomechanically after resuspending the pellet in a buffer containing 10 mM Tris-Cl (pH 7.4), 10 mM sodium chloride, 2 mM magnesium chloride, and 1 mM β-mercaptoethanol. The cleared lysate was supplemented with 250 mM sodium chloride and 20 mM imidazole, and loaded on a Ni-NTA-sepharose column. The protein was eluted by an imidazole gradient to 300 mM, and the eluate was further purified on a heparin–Sepharose column. The His-tag was cleaved using 3C protease. The cleaved protein was collected as flow-through from a second Ni-NTA column and subjected to a final size-exclusion step in 10 mM HEPES (pH 7.4), 200 mM sodium chloride, and 1 mM β-mercaptoethanol. Full-length Ski2–Ski3–Ski8 and Ski2Δinsert–Ski3–Ski8 complexes were obtained by coexpression of corresponding Ski2 constructs with full-length Ski3 and Ski8 in insect cells. Purification of complexes was essentially carried out as described above.
Wild-type Ski2-insert (residues 835–1085) was cloned in a modified pBR322 vector containing an N-teminal 6xHis tag, followed by a 3C protease site. Proteins were expressed in E. coli BL21 (DE3) Gold pLysS at 18°C after induction with 0.1 mM IPTG. Cells were resuspended in a buffer containing 20 mM Tris-Cl (pH 7.5), 500 mM sodium chloride, 20 mM imidazole, and 1 mM β-mercaptoethanol, and lysed by sonication. The soluble fraction was loaded on a Ni-NTA column and His-tagged protein was eluted with buffer supplemented with 300 mM imidazole and dialyzed in a low-salt buffer for cleavage of the His-tag with 3C protease. The cleaved protein was further purified with a Q–Sepharose column and a final size-exclusion column in 10 mM HEPES (pH 7.5), 200 mM sodium chloride, and 1 mM β-mercaptoethanol. The Ski2-insert mutant R903E was purified in the same manner.
Full-length Ski7 (residues 1–747) was expressed in E. coli BL21 (DE3) Gold pLysS at 18°C from a construct containing an N-terminal GST-tag. Cells were resuspended in a buffer containing 20 mM Tris-Cl (pH 7.4), 500 mM sodium chloride, 2 mM magnesium chloride, and 1 mM β-mercaptoethanol. After sonication, the soluble lysate was bound to GSH–Sepharose beads (GE Healthcare), washed, and eluted with loading buffer supplemented with 20 mM glutathione. The protein was dialyzed in loading buffer containing 5 mM EDTA and subjected to size-exclusion chromatography in 20 mM Tris-Cl (pH 7.4), 200 mM sodium chloride, 2 mM magnesium chloride, and 1 mM β-mercaptoethanol.
Crystallization
Ski2-ΔN was concentrated to 15 mg/mL, and AMPPNP was added to a final concentration of 0.5 mM. Initial hits were typically obtained in 15% (w/v) PEG 3350 and 0.1 M HEPES (pH 7.0) by vapor diffusion experiments in sitting drops at 4°C. Iterative microseeding in 3% (w/v) PEG 3350, 5% (v/v) ethylene glycol, and 0.1 M HEPES (pH 7.0) at a protein concentration of 20 mg/mL improved crystal size and diffraction quality. Gold-derivatized Ski2-ΔN crystals were obtained by soaking native crystals for 6 h in mother liquor containing 10 mM gold cyanide. Prior to flash freezing in liquid nitrogen, crystals were briefly soaked in mother liquor that was stepwise supplemented with ethylene glycol to a final concentration of 25% (v/v).
Crystals of Ski2-insert were grown by vapor-diffusion at 4°C in two different conditions, resulting in the same crystal form. The protein was concentrated to 40 mg/mL and mixed 1:1 (v/v) with 3.5 M sodium formate and 0.1 M MES (pH 6.5) (condition 1, native data set) or with 16% (w/v) PEG 3350, and 0.14 M sodium iodide (condition 2, derivative data set) (Table 1). Gold-derivatization was carried out as described above. Prior to flash freezing in liquid nitrogen, the crystals were briefly soaked in mother liquor containing 25% (v/v) glycerol.
Structure solution
Data were collected at 100 K at Swiss Light Source and processed using XDS (Kabsch 2010) and SCALA (Evans 2006). The Ski2-ΔN crystals contain one molecule per asymmetric unit. Phases were obtained with a three-wavelength multiple-anomalous dispersion experiment on gold-derivatized crystals. The PHENIX suite (Adams et al. 2010) was used for substructure solution, phasing, and density modification. An initial model was built automatically with BUCCANEER (Cowtan 2006) and extended manually in COOT (Emsley et al. 2010). Structure refinement against native data was carried out with phenix.refine (Adams et al. 2010).
For the structure of Ski2-insert, phases were determined from a single wavelength anomalous dispersion experiment and essentially calculated as described above. Five gold-derivatization sites were found, corresponding to five molecules in the asymmetric unit. Density modification was carried out using fivefold noncrystallographic symmetry averaging. The backbone was built manually in the experimental electron density. The initial model was extended by subsequent rounds of manual building and refinement against the native data. The structures of Ski2-ΔN and Ski2-insert were combined to a final model as outlined in the results section. Maximum likelihood coordinate error estimates of the final refined models were 0.76 Å for Ski2-ΔN and 0.94 Å for Ski2-insert.
Electrophoretic mobility shift assays (EMSA)
Single-stranded RNA oligos were purchased (biomers.net) and tRNAiMet was in vitro transcribed and purified as described (Weir et al. 2010). RNA substrates were 5′-labeled with [32P]phosphate by polynucleotide kinase treatment. Double-stranded 27-mer RNA oligos were generated by adding a 1.2-fold molar excess of unlabeled complementary strand to the labeled RNA oligo (5′-CCCCACCACCAUCACUUAAAAAAAAAA-3′), followed by incubation at 95°C for 5 min and annealing by slow cooling. For a typical EMSA reaction, 0.5 pmol of substrate was mixed with the indicated amounts of protein and 1 U of RiboLock RNase inhibitor (Fermentas). 10x buffer was added to final concentrations of 20 mM HEPES (pH 7.5), 50 mM potassium acetate, 5 mM magnesium acetate, 0.1% (v/v) NP-40, and 2 mM dithiothreitol. Reactions were carried out in a volume of 10 μL with a final salt concentration of 75 mM sodium chloride and 50 mM potassium chloride in all samples. Samples were incubated at 4°C for 30 min and separated on a native 6% (w/v) polyacrylamide gel. Gels were analyzed by phosphorimaging.
Pull-down assays
GST-tagged prey protein (4 μg) was mixed with equal molar amounts of bait protein. Buffer was added to a volume of 200 μL and final concentrations of 10 mM HEPES (pH 7.5), 75 mM sodium chloride, 2 mM magnesium acetate, 0.1% (v/v) NP-40, 1 mM dithiothreitol, and 12.5% (v/v) glycerol (buffer A). A total of 40 μL of a 50% (v/v) suspension of GSH–Sepharose beads (GE Healthcare) were added, and the reaction was incubated for 30 min at 4°C. Beads were washed three times with buffer A before eluting the precipitated protein. Samples were analyzed on a 4%–12% (w/v) Bis-Tris polyacrylamide gradient gel (Invitrogen).
DATA DEPOSITION
The coordinates and the structure factors have been deposited in the Protein Data Bank with accession codes 4a4z (Ski2-ΔN) and 4a4k for (Ski2-insert).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Jérôme Basquin, Karina Valer-Saldaña, and Sabine Pleyer at the MPI-Martinsried crystallization facility, and the staff of the PX beamlines at the Swiss Light Source (Villigen) for assistance during data collection. We also thank Peter Brick and members of our laboratory for suggestions and critical reading of the manuscript. This study was supported by the Boehringer Ingelheim Fonds to F.H.; by the Max Planck Gesellschaft, the Sonderforschungsbereich SFB646, the Gottfried Wilhelm Leibniz Program of the Deutsche Forschungsgemeinschaft (DFG), and the Center for Integrated Protein Science Munich (CIPSM) to E.C.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029553.111.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Parker RP 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J 20: 4684–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E 2009. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139: 547–559 [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Brown JT, Bai X, Johnson AW 2000. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6: 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner K, Wenig K, Hopfner KP 2005. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 20: 461–471 [DOI] [PubMed] [Google Scholar]
- Büttner K, Nehring S, Hopfner KP 2007. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol 14: 647–652 [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62: 1002–1011 [DOI] [PubMed] [Google Scholar]
- Cristodero M, Bottcher B, Diepholz M, Scheffzek K, Clayton C 2008. The Leishmania tarentolae exosome: purification and structural analysis by electron microscopy. Mol Biochem Parasitol 159: 24–29 [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Séraphin B 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578 [DOI] [PubMed] [Google Scholar]
- He Y, Andersen GR, Nielsen KH 2010. Structural basis for the function of DEAH helicases. EMBO Rep 11: 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Houalla R, Devaux F, Fatica A, Kufel J, Barrass D, Torchet C, Tollervey D 2006. Microarray detection of novel nuclear RNA substrates for the exosome. Yeast 23: 439–454 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ 2010. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J 29: 2205–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AW, Kolodner RD 1995. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol 15: 2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR, Ouzounis CA 1996. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci 21: 425–426 [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Seraphin B 2008. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim Biophys Acta 1779: 558–565 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456: 993–996 [DOI] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687 [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E 2005. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol 12: 575–581 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E 2007. RNA channelling by the archaeal exosome. EMBO Rep 8: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Conti E 2008a. Structural organization of the RNA-degrading exosome. Curr Opin Struct Biol 18: 709–713 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E 2008b. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell 29: 717–728 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Brodersen DE, Jensen TH 2009. Origins and activities of the eukaryotic exosome. J Cell Sci 122: 1487–1494 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol Cell 11: 1405–1413 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Orban TI, Izaurralde E 2005. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11: 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM 2008. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys 37: 317–336 [DOI] [PubMed] [Google Scholar]
- Ridley SP, Sommer SS, Wickner RB 1984. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol Cell Biol 4: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A 2009. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 16: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Leung E, Brown J, Tollervey D 2009. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res 37: 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV IV , Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Araki Y, Sakuno T, Katada T 2003. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J 22: 3951–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-E A, Wickner RB 1979. A mutant killer plasmid whose replication depends on a chromosomal “superkiller” mutation. Genetics 91: 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R 2000a. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Staples RR, Baker RE, Parker R 2000b. Function of the Ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol Cell Biol 20: 8230–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264 [DOI] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189 doi: 10.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, Henry Y, Leulliot N 2010. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J 29: 2194–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lewis MS, Johnson AW 2005. Domain interactions within the Ski2/3/8 complex and between the Ski complex and Ski7p. RNA 11: 1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JR, Bonneau F, Hentschel J, Conti E 2010. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci 107: 12139–12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, Cate JH 2009. Structures of the ribosome in intermediate states of ratcheting. Science 325: 1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]