In flies, asymmetric loading of small RNA duplexes into Argonaute2-containing RNA-induced silencing complex (Ago2-RISC) requires Dicer-2/R2D2 heterodimer, which acts as a protein sensor for the thermodynamic stabilities of the ends of small RNA duplexes. However, the mechanism of small RNA asymmetry sensing in mammalian RISC assembly remains obscure. Here, the authors quantitatively examined RISC assembly and target silencing activity in the presence or absence of Dicer in mammals. Their data show that, unlike the well-characterized fly Ago2-RISC assembly pathway, mammalian Dicer is dispensable for asymmetric RISC loading in vivo and in vitro.
Keywords: Argonaute, Dicer, RNA-induced silencing complex, RISC, RISC-loading complex
Abstract
In flies, asymmetric loading of small RNA duplexes into Argonaute2-containing RNA-induced silencing complex (Ago2-RISC) requires Dicer-2/R2D2 heterodimer, which acts as a protein sensor for the thermodynamic stabilities of the ends of small RNA duplexes. However, the mechanism of small RNA asymmetry sensing in mammalian RISC assembly remains obscure. Here, we quantitatively examined RISC assembly and target silencing activity in the presence or absence of Dicer in mammals. Our data show that, unlike the well-characterized fly Ago2-RISC assembly pathway, mammalian Dicer is dispensable for asymmetric RISC loading in vivo and in vitro.
INTRODUCTION
MicroRNAs (miRNAs) are small noncoding RNAs encoded in the genomes of most eukaryotic organisms. Genes coding for miRNAs are first transcribed as primary miRNAs (pri-miRNAs). Typically, pri-miRNAs are processed in the nucleus by Drosha, an RNase III enzyme, to form pre-miRNAs, and are then exported to the cytoplasm where they are processed by another RNase III enzyme, Dicer, which produces miRNA/miRNA* duplexes of ∼22 nt in length. Small interfering RNAs (siRNAs) are another class of ∼21-nt small RNAs, which are produced from long double-stranded RNAs (dsRNAs) by Dicer (for review, see Ghildiyal and Zamore 2009; Kim et al. 2009).
Dicer possesses several functional domains: an N-terminal helicase domain, a domain with unknown function (DUF283), a PAZ domain, two RNase III domains, and a dsRNA-binding domain (Supplemental Fig. S1A). In flies, there are two Dicer paralogs (Lee et al. 2004). Dicer-1 (Dcr-1) processes pre-miRNAs into miRNA/miRNA* duplexes (Lee et al. 2004; Tsutsumi et al. 2011), while Dicer-2 (Dcr-2) processes long dsRNAs to produce siRNA duplexes (Lee et al. 2004; Welker et al. 2010; Cenik et al. 2011). In contrast, mammals have a single Dicer that processes both pre-miRNAs and long dsRNAs (Provost et al. 2002; Zhang et al. 2002). Dicer proteins often have dsRNA-binding proteins as their partners: Loquacious (Loqs) for fly Dcr-1 (Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005), R2D2 for fly Dcr-2 (Liu et al. 2003), and TAR-binding protein (TRBP) or PKR activator (PACT) for mammalian Dicer (Chendrimada et al. 2005; Gregory et al. 2005; Haase et al. 2005; Maniataki and Mourelatos 2005; Lee et al. 2006). After processing by Dicer, small RNA duplexes are assembled into an effector complex known as RNA-induced silencing complex (RISC) that has an Argonaute (Ago) protein at its core (Tabara et al. 1999; Hammond et al. 2001). In flies, miRNA/miRNA* and siRNA duplexes are preferentially sorted into two Ago paralogs, Ago1 and Ago2, respectively (Okamura et al. 2004; Förstemann et al. 2007; Tomari et al. 2007). In mammals, all four Ago proteins (Ago1–Ago4) can incorporate both miRNA/miRNA* and siRNA duplexes (Liu et al. 2004; Meister et al. 2004; Yoda et al. 2010). It is known that the PIWI domain of Ago2 binds to the C-terminal region of human Dicer (Tahbaz et al. 2004; Wang et al. 2009), but no direct protein-protein interaction has been reported between Ago proteins and dsRNA-binding proteins.
RISC assembly can be divided into at least two steps (for review, see Kawamata and Tomari 2010). First, a small RNA duplex is loaded into the Ago protein forming pre-RISC with the aid of the Hsc70/Hsp90 chaperone machinery (Iki et al. 2010; Iwasaki et al. 2010; Miyoshi et al. 2010), and second, the duplex is unwound and one of the two strands (guide strand) remains retained in Ago while the other strand (passenger strand) is discarded for degradation (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005; Kawamata et al. 2009; Yoda et al. 2010). The selection of which strand serves as the guide is not random but often asymmetric (Khvorova et al. 2003; Schwarz et al. 2003). In flies and mammals, it is widely accepted that the thermodynamic asymmetry of the ends of a small RNA duplex is a key factor that determines which of the two strands of the duplex serves as the guide. In general, the strand with the less stable 5′ end tends to serve as the guide strand, while the other strand with the more stable 5′ end is more likely discarded from the Ago protein during unwinding (Khvorova et al. 2003; Schwarz et al. 2003). Importantly, the strand serving as the guide is already determined by the polarity of small RNA duplexes upon loading, before unwinding actually occurs (Tomari et al. 2004b; Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005; Kawamata et al. 2009; Yoda et al. 2010). In flies, the Dcr-2/R2D2 heterodimer, which is essential for Ago2-RISC assembly (Liu et al. 2003; Pham et al. 2004; Tomari et al. 2004a), senses the thermodynamic asymmetry of the siRNA duplex (Tomari et al. 2004b). R2D2 orients an siRNA duplex by binding to the more stable end, which positions Dcr-2 at the opposite, less stable end. The heterodimer then loads the siRNA duplex into fly Ago2 with the prearranged orientation (Tomari et al. 2004b). Thus, Dcr-2/R2D2 has two distinct functions: dicing of long dsRNAs and asymmetric loading of siRNA duplexes into Ago2.
By analogy to the functions of fly Dcr-2/R2D2 in thermodynamic asymmetry sensing and Ago2-RISC loading, it is tempting to postulate that Dcr-1 and Loqs in flies and Dicer and TRBP (or PACT) in mammals might play similar roles in RISC assembly, in addition to their essential role in dicing (Chendrimada et al. 2005; Gregory et al. 2005; Maniataki and Mourelatos 2005; MacRae et al. 2008; Miyoshi et al. 2009; Sakurai et al. 2011). Indeed, it was recently shown that, much as fly Dcr-2/R2D2, recombinant human Dicer/TRBP can bind to siRNA duplexes according to their thermodynamic asymmetry (Gredell et al. 2010; Noland et al. 2011). However, it has not been investigated if such Dicer/TRBP binding contributes to asymmetric duplex loading into human Ago proteins. From a functional point of view, immunodepletion of human Dicer from HeLa lysate to an undetectable level did not hinder target cleavage activity of siRNAs (Martinez et al. 2002), and the efficiency of target gene silencing by siRNAs was apparently uncompromised in two independent lines of Dicer-knockout mouse embryonic stem (ES) cells (Kanellopoulou et al. 2005; Murchison et al. 2005). Moreover, siRNA-initiated canonical assembly of human Ago2-RISC was recently reconstituted in vitro without Dicer and TRBP (Ye et al. 2011). These accumulating observations raise the question of whether or not Dicer/TRBP binding plays a role in asymmetric RISC loading in mammals. Here we show that, in vivo and in vitro, Dicer is dispensable for asymmetric RISC assembly in mammals.
RESULTS AND DISCUSSION
In vivo reporter assays for asymmetric RISC assembly in mammals
We used a Dicer1−/− mouse embryonic fibroblast (MEF) cell line (Yi et al. 2006; Yang et al. 2010) to assess whether Dicer is necessary for asymmetric RISC assembly of small RNA duplexes in mammalian cells (Supplemental Fig. S1A). It was recently reported that an independently established Dicer1−/− mouse ES cell line (Murchison et al. 2005) expresses a C-terminally truncated DICER1, which may potentially retain some functions of the full-length protein (Noland et al. 2011). We, therefore, performed Western blot analysis using two different antibodies against the N-terminal or the C-terminal regions of DICER1. We confirmed the lack of expression of full-length or any specific truncated forms of DICER1 in the Dicer1−/− MEF cell line we used (Supplemental Fig. S1B). Additionally, we incubated Dicer1−/− MEF lysate with radiolabeled pre-let-7 miRNA precursor and verified that the cell lysate is deficient in dicing activity (Supplemental Fig. S1C). DICER1 expression and function were effectively rescued by transfection with a DICER1-expressing construct (Supplemental Fig. S1D).
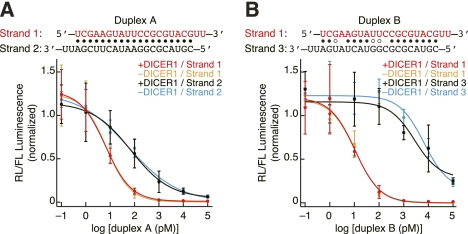
We first sought to examine the silencing efficiency of small RNA duplexes in the presence or absence of Dicer. To this end, we prepared an arbitrary, functionally asymmetric siRNA duplex (duplex A) (Fig. 1A), minimizing the complexity of mimicking natural miRNAs or targeting endogenous genes. As a matter of convenience, we refer to the two strands of the duplex as strand 1 and strand 2. Strand 1 has a less stable 5′ end than strand 2; therefore, strand 1 is more likely to function as the guide strand. Based on duplex A, we also created an artificial miRNA/miRNA*-like duplex (duplex B) (Fig. 1B), which does not require the slicer activity of Ago2 for unwinding, by introducing internal mismatches and wobble base pairs, without altering the sequence of strand 1 (Yoda et al. 2010). Duplex B is composed of strand 1 and strand 3; according to the thermodynamic asymmetry, again, strand 1 is more likely to serve as the guide strand. We then constructed reporters for each strand of these two duplexes by introducing a single target site perfectly complementary to each strand downstream from a Renilla luciferase (Rluc) reporter in the dual luciferase assay system. These constructs were cotransfected with the corresponding small RNA duplexes into Dicer1−/− MEF cells with or without DICER1 expression rescued. The incorporation of each strand into mature RISC, as a reflection of their potency to silence the respective target Rluc reporter, was assessed by dual luciferase assays.
FIGURE 1.
Mammalian Dicer is dispensable for asymmetric RISC assembly in vivo. (A,B) Silencing of Rluc reporters by increasing concentrations of duplex A (A) or duplex B (B) in Dicer1−/− MEF cells with or without DICER1 expression rescued. Mean ± s.d. (n = 3) is shown.
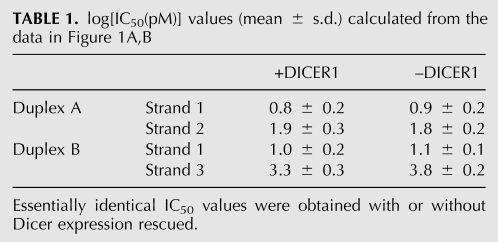
As expected, RISC was assembled asymmetrically with both duplex A and duplex B; in both cases strand 1 was more potent for target silencing. In the case of miRNA/miRNA*-like duplex B, strand 1 was two orders of magnitude more effective in silencing the reporter than strand 3 (Fig. 1B; Table 1), while in the case of siRNA duplex A, strand 1 was around one order of magnitude more potent than strand 2 (Fig. 1A; Table 1). Importantly, for both duplexes, the silencing effect of each strand was unaffected regardless of whether DICER1 expression was rescued or not (Fig. 1A,B); all the IC50 values were essentially identical in the presence or absence of DICER1 (Table 1). These findings suggest that mammalian Dicer is dispensable for duplex asymmetry sensing and RISC assembly in vivo.
TABLE 1.
log[IC50(pM)] values (mean ± s.d.) calculated from the data in Figure 1A,B
In vitro assays for asymmetric RISC assembly in mammals
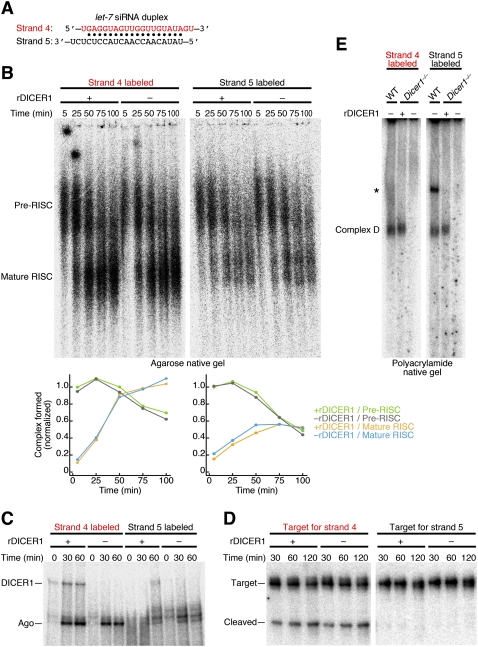
To explore any potential function of Dicer in RISC assembly, we next sought to compare the formation of pre-RISC and its conversion to mature RISC by an agarose native gel system (Tomari et al. 2004a; Kawamata et al. 2009; Yoda et al. 2010; Kawamata and Tomari 2011) in the presence or absence of Dicer. We noticed that in vitro RISC assembly activity of MEF lysate is rather weak compared to HeLa or HEK293T lysate (data not shown) and tends to fluctuate from batch to batch. Therefore, instead of comparing between Dicer1−/− MEF lysates with and without DICER1 expression rescued, we took a strategy of resupplementing Dicer1−/− MEF cell lysate with purified recombinant DICER1 (rDICER1) (Supplemental Fig. S2A). A control experiment showed that addition of rDICER1 in Dicer1−/− MEF cell lysate efficiently rescued dicing activity (Supplemental Fig. S2B). We prepared a functionally asymmetric siRNA duplex based on the let-7 sequence (let-7 siRNA duplex) (Fig. 2A), which has been intensively studied with the agarose native gel system (Kawamata et al. 2009; Yoda et al. 2010). We 5′ 32P radiolabeled either of the two strands (strand 4 and strand 5), and the duplexes were incubated in Dicer1−/− MEF lysate with or without rDICER1 resupplementation. A similar level of pre-RISC was detected when either of the two strands was radiolabeled, whereas a markedly higher level of mature RISC was detected when strand 4 was radiolabeled, precisely reflecting the asymmetry of the duplex. Importantly, the presence or absence of rDICER1 did not affect the kinetics of asymmetric loading of the duplex or subsequent unwinding (Fig. 2B). Moreover, short-wave UV crosslinking to Ago proteins (Fig. 2C) and target cleavage activity (Fig. 2D; Supplemental Fig. S2C) of each strand agreed well with the amount of mature RISC formed and were unaffected by the presence or absence of rDICER1. Therefore, mammalian Dicer is dispensable for asymmetric RISC assembly and target cleavage of small RNA duplexes in vitro.
FIGURE 2.
Mammalian Dicer is dispensable for asymmetric RISC assembly in vitro. (A) RNA duplex used in B–E. (B) Pre-RISC and mature RISC formation with strand 4 or strand 5 in the presence or absence of DICER1. Quantification is shown in the lower inset. (C) Short-wave UV crosslinking of strand 4 or strand 5 in the presence or absence of DICER1. (D) Target cleavage assay for strand 4 or strand 5 in the presence or absence of DICER1. (E) Formation of Dicer-containing complex (complex D) in the presence or absence of DICER1. Asterisk denotes a nonspecific complex (see Supplemental Fig. S2D).
It was previously reported that, in mammalian cell lysate, siRNA duplexes form a complex containing Dicer and TRBP (therein referred to as complex D or pre-RLC), which can be separated on polyacrylamide native gel (Pellino et al. 2005; Sakurai et al. 2011). Indeed, we readily detected such a complex D with radiolabeled let-7 siRNA duplex in WT MEF lysate and Dicer1−/− MEF lysate supplemented with rDICER1 (Fig. 2E; Supplemental Fig. S2D). However, unlike mature RISC, formation of complex D showed no correlation with Ago crosslinking or target cleavage activity (cf. Fig. 2B–E), confirming that Dicer plays a minimum role in RISC assembly in mammals.
Dicer is not required for asymmetric RISC assembly in mammals
In the fly Ago2-RISC assembly pathway, Dcr-2/R2D2 is absolutely required for loading of siRNA duplexes; in the absence of Dcr-2 or R2D2, no Ago2-RISC is formed from siRNA duplexes (Liu et al. 2003; Pham et al. 2004; Tomari et al. 2004a). For this reason, a complex containing Dcr-2/R2D2 is called RISC-loading complex (RLC). In contrast, whether or not Dicer is required for RISC assembly in mammals has been a subject of debate. Previous multiple lines of evidence (Martinez et al. 2002; Kanellopoulou et al. 2005; Murchison et al. 2005; Ye et al. 2011) together with our current findings indicate that Dicer is not required for RISC assembly in mammals. In this regard, we wish to refrain from calling Dicer/TRBP or Dicer/TRBP/Ago2 as RLC.
Dcr-2/R2D2 binds to siRNA duplexes in an orientation according to their thermodynamic asymmetry (Tomari et al. 2004b). Recombinant human Dicer/TRBP is also able to bind to siRNAs in an asymmetric manner, just like fly Dcr-2/R2D2 (Noland et al. 2011). However, our data suggest that such Dicer/TRBP binding plays no essential role in asymmetric RISC assembly in mammals. How, then, can the thermodynamic asymmetry of small RNA duplexes be sensed in mammals? Although we cannot exclude that there are unknown factors for this function, we favor the hypothesis that Ago proteins themselves can generally sense the thermodynamic asymmetry of small RNA duplexes. Structural studies of an archaeal Piwi protein and a bacterial Ago protein have shown that the 5′ phosphate of the guide strand is anchored in a phosphate-binding pocket at the interface between the MID and PIWI domains (Parker et al. 2005; Wang et al. 2008). In that position, pairing between the 5′ first base of the guide strand and the complementary base near the 3′ end of the passenger strand is blocked, with the ends of the two strands directed in opposite directions. This structural conformation should favor the orientation of the small RNA duplex with the end that is more easily unwound (i.e., the less stable end) toward the phosphate-binding pocket. The high homology of the MID and PIWI domains, in particular the architecture of the C-terminal carboxylate that forms the phosphate-binding pocket, suggests that the binding mode for a small RNA molecule is commonly shared by Ago proteins (Parker et al. 2005; Wang et al. 2008; Boland et al. 2011). In the case of fly Ago2-RISC assembly, Dcr-2/R2D2 binding might double-check or amplify the sensing of thermodynamic asymmetry in addition to Ago2 itself; an siRNA duplex needs to bind Dcr-2/R2D2 before entering Ago2, and yet the duplex might transiently shuttle between Dcr-2/R2D2 and Ago2, with each protein checking the asymmetry, before it is properly docked into Ago2. Supporting this view, crosslinking experiments show that the orientation of Dcr-2/R2D2 on the siRNA duplex is less asymmetric for the recombinant heterodimer than for embryo lysate (Tomari et al. 2004b). In contrast, such a double-checking system may be lacking in mammals, where Dicer/TRBP binding is uncoupled from RISC assembly. Interestingly, in dicer-mutant zebrafish embryos, injected miRNA duplexes were fully active for target silencing, indicating that RISC assembly does not require Dicer in fish as in mammals (Giraldez et al. 2006). Even in flies, simultaneous >95% depletion of Dcr-1 and Dcr-2, which abolished siRNA duplex-initiated Ago2-RISC assembly, did not affect assembly of miRNA/miRNA* duplexes into Ago1-RISC, suggesting that Dicer is dispensable for fly Ago1-RISC assembly (Kawamata et al. 2009). Therefore, although fly Ago2-RISC assembly is the best-characterized pathway to date, it is likely to represent a very unique pathway entirely dependent on Dcr-2/R2D2. Indeed, the four mammalian Ago proteins and fly Ago1 are more closely related to each other than to fly Ago2 (Kawamata et al. 2009; Yoda et al. 2010). Moreover, fly Ago1 and human Ago1–4 show a strong preference for uracil (U) at the 5′ end of the guide strand, which could, in theory, also help in sensing the thermodynamic asymmetry, whereas fly Ago2 tends to favor 5′ cytosine (C) (Czech et al. 2009; Okamura et al. 2009; Ghildiyal et al. 2010). Further study is required to clarify what makes fly Ago2 so special.
MATERIALS AND METHODS
Plasmids constructions
For the construction of the mouse DICER1 expression vector (pCAGEN-DICER1), the Dicer1 sequence was amplified from pBluescript II KS(–)-Dicer (Doi et al. 2003), using the primers Dcr-F: 5′-TTTTGGCAAAGAATTCGATGATTGAAAAGCCCTGCT-3′ and Dcr-R: 5′-TATCCTCGAGGAATTCGTGGAGCTGTGGTTCTGGTC-3′, and inserted in the EcoRI site of the pCAGEN vector using the In-Fusion Advantage PCR cloning kit (Clontech). For the SBP-tagged DICER1 construct (pCAGEN-SBP-DICER1), pCAGEN-DICER1 was amplified and linearized using the primers Dcr-F1: 5′-GCAGGCTCCGCGGCCATGGCAGGCCTGCAGCTCAT-3′ and Dcr-R1: 5′-TATCCTCGAGGAATTCTCAGCTGTTAGGAACCTGAGGC-3′. The SBP sequence was amplified from pASW (Iwasaki et al. 2010) using the primers SBP-F: 5′-TTTTGGCAAAGAATTCCCATGGACGA GAAGACCACCGGC-3′ and SBP-R: 5′-GGCCGCGGAGCCTGCTTTTT-3′, and these two fragments were fused using the In-Fusion Advantage PCR cloning kit (Clontech). Oligos perfectly complementary to each strand of duplex A and duplex B were synthesized and cloned in the XhoI and NotI sites of the psiCHECK-2 vector (Promega).
Luciferase reporter assay
Approximately 4 × 104 Dicer1−/− MEF cells (Yi et al. 2006; Yang et al. 2010) per well were seeded in 24-well plates and 24 h later were transfected with 0.8 μg of pCAGEN or pCAGEN-DICER1 using Lipofectamine 2000, according to the manufacturer's instructions. Twenty four hours after the initial transfection, cells were cotransfected with 0.25 μg of the target-bearing psiCHECK-2 constructs and with increasing concentrations of 5′ pre-phosphorylated duplex A or duplex B, using DharmaFect Duo (Dharmacon). Forty eight hours after the initial transfections, Renilla and Firefly luciferase expressions were measured using Dual Luciferase Reporter Assay System (Promega), and normalized to mock RNA transfections. Graphs were generated and IC50 values were calculated using Igor Pro software (Wavemetrics).
Antibodies
The following antibodies were used for Western blot and super shift assays: anti-DICER1 “C-terminal” (sc-30226, Santa Cruz), anti-DICER1 “N-terminal” (Kanellopoulou et al. 2005), anti-AGO2 (2897, Cell Signaling), and normal rabbit IgG (2729, Cell Signaling).
Preparation of recombinant DICER1
HEK293T cells were transfected with pCAGEN-SBP-DICER1 using FugeneHD transfection reagent, and rDICER1 was purified by streptavidin sepharose as previously described (Tsutsumi et al. 2011).
Native gel analysis, UV crosslinking, dicing, and target cleavage assays
Methods were previously described in detail and were used with minor modifications (Haley et al. 2003; Pellino et al. 2005; Yoda et al. 2010; Kawamata and Tomari 2011; Sakurai et al. 2011). In sum, in vitro RISC assembly reactions typically contained 3 μL 40× reaction mix, 10 nM target RNA, 10 nM radiolabeled RNA duplex, 4 μL cell lysate, and 1 μL rDICER1 in 10 μL reaction mixture. Samples were run on a 1.4% agarose native gel or a 4.5% polyacrylamide (29:1) native gel. For supershift assays, samples were mixed with 0.5 μL of anti-DICER1 “C-terminal” or normal rabbit IgG and incubated at 4°C for 1 h before electrophoresis. For UV crosslinking, samples were irradiated at a wavelength of 254 nm for 5 min and run on an SDS-PAGE. Dicing reactions typically contained 3 μL 40× reaction mix, 5 nM radiolabeled pre-let-7, 4 μL cell lysate, and 1 μL rDICER1 in 10 μL reaction mixture. Samples were incubated at 30°C for 30 min, digested with Proteinase K, precipitated with ethanol, and run on 10% Urea-PAGE. Target cleavage assays were done as previously described (Haley et al. 2003).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Alexander Tarakhovsky for providing the Dicer1–/– MEF cell line, Chrysi Kanellopoulou for the anti-DICER1 antisera, Kumiko Ui-Tei for pBluescript II KS(–)-Dicer, and Connie Cepko for pCAGEN. We thank Mayuko Yoda for technical assistance, Hervé Seitz, Klaus Förstemann, and members of the Tomari laboratory for their critical comments and suggestions on the manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas (“Functional machinery for noncoding RNAs”) from the Japan Ministry of Education, Culture, Sports, Science and Technology.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029785.111.
REFERENCES
- Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O 2011. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci 108: 10466–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Tanaka Hall TM, Zamore PD 2011. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol Cell 42: 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ 2009. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell 36: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, Saigo K 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr Biol 13: 41–46 [DOI] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD 2005. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3: e236 doi: 10.1371/journal.pbio.0030236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD 2007. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell 130: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD 2010. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF 2006. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Gredell JA, Dittmer MJ, Wu M, Chan C, Walton SP 2010. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry 49: 3148–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640 [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Tang G, Zamore PD 2003. In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30: 330–336 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M 2010. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y 2010. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39: 292–299 [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q 2005. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 19: 1674–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y 2010. Making RISC. Trends Biochem Sci 35: 368–376 [DOI] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y 2011. Native gel analysis for RISC assembly. Methods Mol Biol 725: 91–105 [DOI] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y 2009. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol 16: 953–960 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN 2006. The role of PACT in the RNA silencing pathway. EMBO J 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA 2008. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci 105: 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z 2005. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev 19: 2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC 2005. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Okada TN, Siomi H, Siomi MC 2009. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA 15: 1282–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi MC 2010. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol 17: 1024–1026 [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci 102: 12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland CL, Ma E, Doudna JA 2011. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell 43: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC 2009. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D 2005. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellino JL, Jaskiewicz L, Filipowicz W, Sontheimer EJ 2005. ATP modulates siRNA interactions with an endogenous human Dicer complex. RNA 11: 1719–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ 2004. A Dicer-2-dependent 80S complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94 [DOI] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21: 5864–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC 2005. Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol 3: e235 doi: 10.1371/journal.pbio.0030235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Amarzguioui M, Kim DH, Alluin J, Heale B, Song MS, Gatignol A, Behlke MA, Rossi JJ 2011. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res 39: 1510–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132 [DOI] [PubMed] [Google Scholar]
- Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC 2004. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep 5: 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD 2004a. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831–841 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD 2004b. A protein sensor for siRNA asymmetry. Science 306: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD 2007. Sorting of Drosophila small silencing RNAs. Cell 130: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y 2011. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol. 18: 1153–1158 [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ 2008. Structure of an Argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456: 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E 2009. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol 16: 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S, Bass BL 2010. Dicer's helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC 2010. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci 107: 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H 2011. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol 18: 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E 2006. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 38: 356–362 [DOI] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y 2010. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol 17: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]