Bacterial cold shock proteins (CSPs) regulate the cellular response to temperature downshift. Their general principle of function involves RNA chaperoning and transcriptional antitermination. Here the authors present two crystal structures of CSP B from Bacillus subtilis (Bs-CspB) in complex with either a hexanucleotide (5′-UUUUUU-3′) or heptanucleotide (5′-GUCUUUA-3′) single-stranded RNA (ssRNA). Hydrogen bonds and stacking interactions between RNA bases and aromatic sidechains characterize individual binding subsites. Additional binding subsites which are not occupied by the ligand in the crystal structure were revealed by NMR spectroscopy in solution on Bs-CspB·RNA complexes.
Keywords: cold shock protein, cold shock domain, cold shock response, OB-fold, RNA binding proteins, single-stranded RNA
Abstract
Bacterial cold shock proteins (CSPs) regulate the cellular response to temperature downshift. Their general principle of function involves RNA chaperoning and transcriptional antitermination. Here we present two crystal structures of cold shock protein B from Bacillus subtilis (Bs-CspB) in complex with either a hexanucleotide (5′-UUUUUU-3′) or heptanucleotide (5′-GUCUUUA-3′) single-stranded RNA (ssRNA). Hydrogen bonds and stacking interactions between RNA bases and aromatic sidechains characterize individual binding subsites. Additional binding subsites which are not occupied by the ligand in the crystal structure were revealed by NMR spectroscopy in solution on Bs-CspB·RNA complexes. Binding studies demonstrate that Bs-CspB associates with ssDNA as well as ssRNA with moderate sequence specificity. Varying affinities of oligonucleotides are reflected mainly in changes of the dissociation rates. The generally lower binding affinity of ssRNA compared to its ssDNA analog is attributed solely to the substitution of thymine by uracil bases in RNA.
INTRODUCTION
Bacillus subtilis, a Gram-positive mesophilic bacterium, dwells in the upper layers of soil, where it may have to respond to stress induced by an abrupt reduction of ambient temperature. The cold shock response thus elicited is characterized by a transient inhibition of many genes and increased expression of cold-induced genes. After acclimation the expression of these genes decreases, and bulk protein synthesis is restored (Graumann et al. 1996). The cold-induced proteins include a family of proteins named major cold shock proteins (CSPs). They are ubiquitous proteins in the domain of Eubacteria and Archaea (Graumann and Marahiel 1998; Giaquinto et al. 2007). CSPs appear to serve various cellular functions in the context of stress response, but they are not antifreeze proteins.
The best characterized CSPs in terms of cellular function are those from Escherichia coli. The genes cspA, cspB, cspG, and cspI have been reported to be transiently induced upon cold shock whereas other csp genes are not (Phadtare 2004). From the observation that double and triple deletions of csp genes do not result in cold sensitivity and the growth defect at 15°C of a quadruple-csp deletion strain can be complemented by overproduction of almost any of the CSP homologs, it can be concluded that CSPs are able to substitute for each other during cold acclimation (Xia et al. 2001). Among the first molecular functions attributed to Ec-CspA, a major CSP of E. coli, was the transcriptional stimulation of cold shock-sensitive genes, such as hns, encoding a DNA-binding protein, and gyrA, encoding a subunit of DNA gyrase (La Teana et al. 1991; Jones et al. 1992). Transcriptional stimulation by Ec-CspA was linked to the protein's affinity to single-stranded DNA (Brandi et al. 1994).
The influence of CSPs on transcription was described in a different study (Bae et al. 2000). Ec-CspA, Ec-CspC, and Ec-CspE reduced the efficiency of transcription termination in vitro. This could be confirmed by in vivo experiments because overexpression of cspC and cspE in E. coli induced transcription of genes downstream from multiple transcription terminators. Therefore, Ec-CspC and Ec-CspE were characterized as transcription antiterminators. Finally, the inability of Ec-CspE mutants to destabilize nucleic acid secondary structures resulted in the inability to antiterminate transcription. In this sense, Ec-CspE is essential for a transcription antitermination function of the protein (Phadtare et al. 2002a,b; Phadtare 2004).
In vitro, Ec-CspA has been shown to prevent the formation of secondary structures in RNA at low temperatures (Jiang et al. 1997) and was therefore designated as mRNA chaperone. This activity may be essential for efficient translation initiation within E. coli at reduced growth temperature (Weber and Marahiel 2002) but has been challenged for CSPs in B. subtilis (Hofweber et al. 2005).
Recently, it has been shown that E. coli cspA mRNA undergoes a structural rearrangement upon temperature downshift from 37°C to cold shock temperatures, leading to a more efficiently translated structure (Giuliodori et al. 2010). The unusually long 5′-untranslated region (5′ UTR) of Ec-CspA mRNA contains a “cold box” sequence, for which homologous sequences exist in the mRNA of several other CSPs in E. coli (Jiang et al. 1996). This sequence forms a stable stem–loop structure and stabilizes the mRNA at low temperature (Xia et al. 2002). The cold box may be the target for CSP autoregulation by binding of CspA to its own mRNA (Jiang et al. 1996; Bae et al. 1997; Mitta et al. 1997; Fang et al. 1998; Graumann and Marahiel 1998; Horn et al. 2007) although this concept was questioned in a recent study (Giuliodori et al. 2010).
Two similar cold box sequences, nonhomologous to those in E. coli, are present in the 5′ UTR of cspB and cspC mRNA of B. subtilis (Graumann et al. 1997). The high uridine content of the cold box sequences suggests that they may be bound by Bs-CspB which was found to bind thymidine-rich DNA oligonucleotide ligands with high affinity (Lopez et al. 2001; Max et al. 2006). The preferential binding of Bs-CspB to pyrimidine-rich single strands could be confirmed by a DNA microarray approach identifying the heptameric consensus sequence 5′-GTCTTTG/T-3′ (Morgan et al. 2007).
Several CSP structures were determined by X-ray crystallography (Schindelin et al. 1993, 1994; Mueller et al. 2000; Ren et al. 2008; Morgan et al. 2009) and NMR spectroscopy (Schnuchel et al. 1993; Kremer et al. 2001) including Ec-CspA, Nm-Csp from Neisseria meningitidis, Bc-Csp from Bacillus caldolyticus, Bs-CspB from B. subtilis, Tm-Csp from Thermotoga maritima, and St-CspE from Salmonella typhimurium. B. subtilis contains three CSP homologs (CspB, CspC, CspD) (Graumann et al. 1997). CSPs are small proteins comprising 65–70 amino acids and bind to single-stranded DNA and RNA with micromolar to subnanomolar dissociation constants (KD values). The unique structural feature of all CSPs is the cold shock domain (CSD), a five-stranded antiparallel β-barrel. Crystal structures of Bs-CspB and Bc-Csp (Max et al. 2006, 2007) bound to the oligodeoxyribonucleotide dT6, and solution NMR experiments (Zeeb et al. 2006) characterizing the binding of dT7 to Bs-CspB, identified a common DNA binding surface around the ribonucleoprotein (RNP) motifs I (K13-V20) and II (V26-F30) of the CSPs.
Here, we present crystal structures of Bs-CspB in complex with the oligoribonucleotides rU6 (5′-UUUUUU-3′) and rC7 (5′-GUCUUUA-3′). In both structures, one CSP is associated with one RNA molecule. This is in contrast to the previously determined crystal structures of CSPs in complex with single-stranded DNA, Bs-CspB·dT6 or Bc-Csp·dT6 where the DNA strands bridged between adjacent Bs-CspB molecules in the crystal lattice or bound to a domain-swapped Bc-Csp dimer. Nevertheless, the Bs-CspB·RNA complex structures described here demonstrate nucleotide binding to the same protein subsites contacting DNA bases in the Bs-CspB·dT6 and the Bc-Csp·dT6 structures. RNA binding is dominated by stacking interactions between bases and aromatic protein sidechains. In addition, NMR titration experiments provide a picture of the binding interface of Bs-CspB in solution. For a thermodynamic characterization, binding of oligonucleotides to Bs-CspB was assayed by isothermal titration calorimetry as well as fluorescence titrations and stopped-flow fluorescence spectroscopy, providing a clear picture of the interaction of the CSP to its natural RNA substrate.
RESULTS
Crystal structure determination of Bs-CspB·rU6 and Bs-CspB·rC7
Bs-CspB was cocrystallized with either hexauridine (rU6) or rC7 in space group P212121 (Table 1), and diffraction data were collected up to 1.68 Å and 1.38 Å, respectively. Initial phases for both data sets could be obtained by molecular replacement using the crystal structure of free Bs-CspB (1CSP) (Schindelin et al. 1993). The asymmetric units of the two crystals contain two protein molecules and two RNA strands in case of Bs-CspB·rU6 and two protein molecules and one RNA strand in case of Bs-CspB·rC7. The structures were refined to final Rwork/Rfree values of 18.0/23.5% and 15.4/19.4%, respectively. The resulting electron densities of the complexes are well defined for the protein molecules, but the terminal nucleotides of the RNA strands exhibit high B factors.
TABLE 1.
Bs-CspB·rU6 and Bs-CspB·rC7: Data collection and refinement statistics
Overall structure of Bs-CspB·rU6 and Bs-CspB·rC7
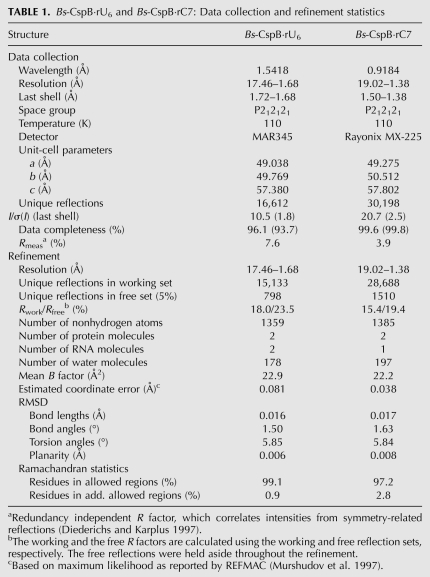
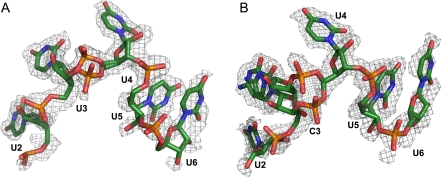
The structure of free Bs-CspB (1CSP) and the structures of Bs-CspB bound to either rU6 or rC7 superimpose very well with an r.m.s.d. of 0.34 Å and 0.29 Å for all protein backbone atoms, respectively. In the Bs-CspB·rU6 complex five of the six ribonucleotides are represented by electron density revealing the binding mode of an RNA molecule to the CSP (Fig. 1A). In another RNA strand bound to the second protein molecule present in the asymmetric unit of the crystal only one ribonucleotide is ordered and visible in clear electron density (not further discussed). In the Bs-CspB·rC7 complex one of the two protein molecules present in the asymmetric unit interacts with an RNA molecule (Fig. 1B). Here, only the five pyrimidines out of the seven nucleotides of the strand are observed. For the terminal purine nucleotides no electron density is detectable.
FIGURE 1.
Crystal structures of Bs-CspB in complex with RNA. The ribbon (Bs-CspB) and stick (RNA) representation of the complexes (A) Bs-CspB·rU6 and (B) Bs-CspB·rC7 are labeled according to the secondary structure elements, the loops (L), and the nucleotides.
Ligand binding surface and protein–ligand interactions
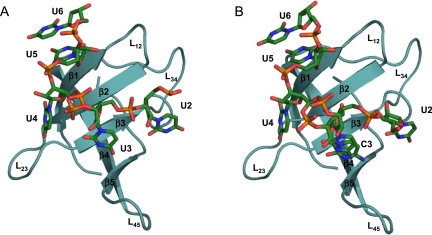
In the following we shall focus on structural aspects of the protein and the ligands and on those interactions between Bs-CspB and RNA that are observed in both crystal structures unless stated otherwise. The surface of the CSP is separated into two areas with different properties. One side of the protein is decorated by acidic sidechains, which give rise to a prominent negative surface potential that does not favor RNA binding. On the opposite side, several aromatic amino acids surrounded by basic and polar groups form an amphipathic surface. These amino acids originate mainly from the first three β strands of Bs-CspB and establish the ligand binding site (Fig. 2A,B). Further protein groups participating in ligand binding are located in β strand 5.
FIGURE 2.
Electrostatic surface potential of the Bs-CspB ligand binding site. The electrostatic potential is projected onto the molecular surface of the protein of (A) Bs-CspB·rU6 and (B) Bs-CspB·rC7. The electrostatic potential was calculated with APBS (Baker et al. 2001) for pH 7.5, with a range from −5 kT (red) to 5 kT (blue).
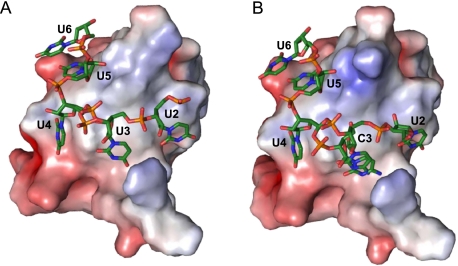
Oligonucleotide binding is dominated by base stacking and involves the sidechains of His29, Phe27, Phe17, Trp8, and the RNA bases of U2 to U5 (Fig. 3). The equally solvent-exposed aromatic sidechain of Phe15 is not involved in the binding of the RNA ligand. A symmetry-related Bs-CspB molecule forms a base stack between the sidechain of Phe38 and the base of U3 in rU6 and C3 in rC7, respectively. Several hydrogen bonds mediate the interaction with the ligand. One hydrogen bond connects the sidechain of Gln59 to the N3 of U2 in Bs-CspB·rC7. This contact is water-mediated in Bs-CspB·rU6. Further hydrogen bonds are formed between the sidechain of Asp25 and N3 of U4 and between the sidechains of Lys7 as well as Trp8 and the O2 of U4. Asp10 contacts O2 of U5 directly in Bs-CspB·rU6 and via a water molecule in Bs-CspB·rC7. Additional water-mediated contacts are formed between the Pro58 backbone carbonyl group and the O4 of U3 and the N4 of C3, respectively. Moreover, an interaction between the backbone carbonyl group of Asp25 and O4 of U4 as well as the backbone carbonyl group of Phe9 and the N3 of U5 is observed. Finally we could identify only one interaction between the protein and the sugar-phosphate backbone. In both crystal structures the 2′-OH group of U5 forms a hydrogen bond with the sidechain of Asn10. The central residue of loop L45 Arg56 forms intermolecular contacts with symmetry-related molecules instead of forming a hydrogen bond with a phosphate group as observed in the Bs-CspB·dT6 and Bc-Csp·dT6 crystal structures.
FIGURE 3.
Stacking and polar interactions between Bs-CspB and the ligands (A,C) rC7 and (B) rU6. (A) A stereo representation of the Bs-CspB molecule with the longest observed nucleotide sequence of rC7 is shown. The backbone of the protein molecule is depicted as gray object; protein groups involved in stacking interactions and water-mediated or direct hydrogen bonding are colored according to the CPK scheme with the exception of carbon, which is in gray. The RNA is colored according to the CPK scheme with the exception of carbon, which is green. Direct hydrogen bonds between protein and RNA are shown as dotted lines. In a schematic depiction of ligand binding the RNA strands (B) rU6 and (C) rC7 are depicted in black and groups of Bs-CspB in dark gray (bb, protein backbone). The longest observed nucleotide sequence of rC7 is presented. A group belonging to a symmetry-related Bs-CspB is displayed in light gray. Stacking interactions between aromatic sidechains and RNA bases are shown as solid gray lines. Hydrogen bonds are displayed as dotted lines. The numbers of the contact subsites for individual bases are given at the bottom, using the numbering scheme introduced earlier (Max et al. 2006).
Structural organization of the ligands
In both crystal structures the RNA is in contact with one protein molecule in contrast to the crystal structure of Bs-CspB in complex with hexathymidine where the DNA strand connects two protein molecules and forms in this way a continuous arrangement of protein and DNA in the crystal (Max et al. 2006). In the crystal structure of Bs-CspB·rU6 as well as in Bs-CspB·rC7 five nucleotides could be built into the electron density (Fig. 4A,B). In the case of the oligoribonucleotide rC7, the five central pyrimidine nucleotides with sequence U2–C3–U4–U5–U6 are resolved in the structure whereas the 5′- and 3′-terminal purine nucleotides are not represented by electron density. The cytidine C3 and the phosphodiester group of the following nucleotide adopt two alternative conformations. The first conformation permits base stacking of U2 onto the imidazole group of His29. In the second conformation this binding is sterically disallowed and, presumably due to high flexibility, no electron density can be detected for nucleotide U2. In the crystal structure of Bs-CspB·rU6 only the phosphodiester group connecting nucleotide U3 with U4 exists in two conformations. The 5′-to-3′ polarity is the same as described for CSP–DNA complexes (Max et al. 2006, 2007) and for most other OB-fold proteins (Theobald et al. 2003). The 5′-end of the oligonucleotide points toward strands β5 of the CSP and the 3′-end toward β1. In both structures the U6 base engages in a stacking interaction with nucleotide U5. Apart form this all RNA bases are oriented toward the hydrophobic protein surface and the sugar-phosphate backbone is exposed to the solvent. In Bs-CspB·rU6 the sugar ring puckers from U2 to U6 are as follows: C3′-exo, C2′-endo, C2′-endo, C3′-endo, and the energetically unfavorable O4′-endo conformation. In Bs-CspB·rC7 the sugar puckering for U2–U6 is C4′-exo, C2′-endo, C2′-endo, C2′-exo, C4′-exo. The C2′-endo pucker, which is strongly avoided in double-stranded A-form RNA (Saenger 1984), is frequently observed along with standard C3′-endo pucker. In the structures, C2′-endo does not lead to steric clashes, however, because the RNA is single-stranded. All nucleotides are in anti conformation except for nucleotide U2 of Bs-CspB·rC7 which is in syn conformation but still in an energetically favorable conformer.
FIGURE 4.
RNA ligands (A) rU6 and (B) rC7 surrounded by their electron density. The ssRNA is colored according to the CPK scheme with the exception of carbon, which is green. Their composite omit electron densities are contoured corresponding to 1.0 σ. (A) Five of six uridine nucleotides of rU6 could be built into the electron density. The phosphodiester group connecting nucleotides 2 and 3 adopts a double conformation. (B) Nucleotides U2 to U6 of rC7 could be built into the density. Nucleotide C3 and the phosphodiester group of the following nucleotide exist in two conformations. The occupancy of U2 is 0.5 such that a sterical clash with the C3 nucleotide which would collide in one of its two conformations is avoided.
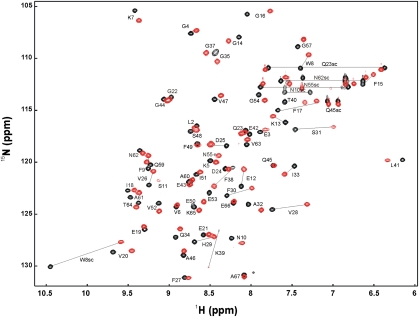
NMR titration of Bs-CspB by rU6 and rC7
Two NMR titrations of a 15N-enriched Bs-CspB sample with unlabeled rU6 and rC7 were performed. A superposition of the first and the last HSQC spectrum of the experiment for Bs-CspB·rC7 is depicted in Figure 5. Most amide protons showed a fast exchange on the NMR chemical shift time scale allowing a direct assignment of amide resonances in the complexes Bs-CspB·rU6 and Bs-CspB·rC7 from gradual shifts during the titration. The following residues stand out from the mean weighted change in chemical shift of their backbone resonances ΔδMW(1HN,15N) with a value >0.13 ppm.
FIGURE 5.
2D 1H/15N HSQC spectrum of free (black) and rC7-bound (red) Bs-CspB in 20 mM HEPES, 50 mM NaCl, pH 7.5, 10% D2O at 20°C. The spectrum of rC7-bound Bs-CspB was recorded at a 1.5-fold molar excess of rC7.
Bs-CspB titrated with rU6: β strand 1 (Lys7), β1–β2 loop L12 (Glu12), RNP motif I (Gly14, Phe15, Gly16), β strand 3 (Asp25), RNP motif II (Val26, Val28, His29, Phe30), β3–β4 loop L34 (Ser31, Ile33, Phe38, Lys39, Thr40):
Bs-CspB titrated with rC7: β strand 1 (Lys7, Trp8, Phe9), β1–β2 loop L12 (Glu12), RNP motif I (Lys13, Phe15, Gly16, Val20), β strand 3 (Asp25), RNP motif II (Val26, Val28, His29, Phe30), β3–β4 loop L34 (Ser31, Ala32, ILe33, Phe38, Lys39), β strand 4 (Glu53) (Fig. 6A).
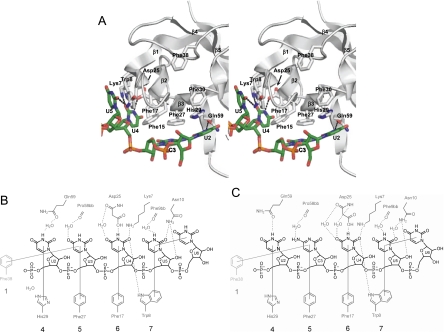
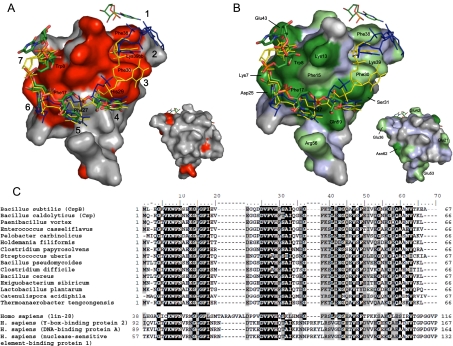
FIGURE 6.
Ligand binding surface of Bs-CspB and its phylogenetic conservation. (A) The interaction of the ssRNA rC7 with Bs-CspB as monitored by NMR titration experiments is depicted and compared with published CSP ssDNA complexes. For the surface representation, the crystal structure of the Bs-CspB·rC7 complex has been used. Residues standing out from chemical shift changes of the backbone amides (ΔδMW > 0.13 ppm) are shown in red. Amino acid residues stacked against RNA or DNA bases in the crystals of Bs-CspB·rC7, Bs-CspB·rU6, and the ssDNA complexes Bs-CspB·dT6 and Bc-Csp·dT6 as well as the backbone of Lys39 are labeled. The numbers of the respective contact subsites for individual bases are also indicated. The ssRNA rC7 of the present work is presented as sticks and colored according to the CPK scheme with the exception of carbon, which is in green. The symmetry-related C3 base contacting subsite 1 is also indicated. The ssDNA molecule of the Bs-CspB·dT6 complex is depicted in blue and the ssDNA molecule of the Bc-CspB·dT6 complex in yellow. The small surface depicts the protein backside. (B) Same representation as in A with the Bs-CspB surface colored according to sequence conservation. Most residues forming the ligand interaction site are conserved at the level of at least 75% identity (dark green) or similarity (light green). Invariant regions which originate from the protein backbone are colored light blue. (C) Sequence alignment of bacterial CSP (upper) and CSD from human proteins (lower panel). Residues that are conserved at a level of at least 75% identity or similarity are highlighted in black or gray, respectively.
The backbone resonances of these residues are sensitive to binding of rU6 or rC7 either by direct interaction or by conformational rearrangements remote from the binding interface and are in excellent agreement with those revealed by Bs-CspB titrated with the DNA fragment dT7 (Zeeb et al. 2006). The residue Phe17, which is involved in binding DNA in previously described Bs-CspB·dT6 and Bc-Csp·dT6 complex structures, shows slow exchange on the NMR chemical shift time scale. A striking difference between the experiment with rU6 and rC7 was observed. Whereas in the titration with rC7 the backbone and the sidechain peaks of Trp8 shift, in the titration with rU6 the backbone resonance changes its position only marginally and the sidechain resonance disappears due to intermediate exchange.
Thus, the same amino acids forming the binding platform in the crystal structures described in this report could be also identified by a substantial chemical shift perturbation during NMR titration. A remarkable feature is that additional amino acids, which are involved in binding of ssRNA, could be identified by the NMR experiment. This may be taken to indicate subtle differences in complex architectures in solution and in the crystal.
Binding affinity of different ssRNAs to Bs-CspB
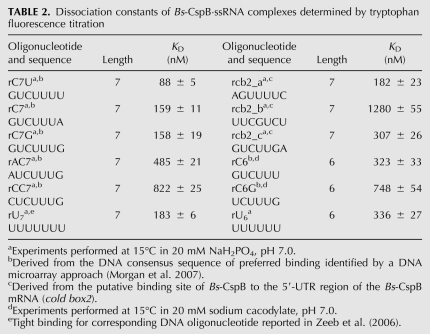
Different oligonucleotides were tested by tryptophan fluorescence titration experiments to assess the sequence specificity of RNA binding to Bs-CspB and to identify suitable ssRNA fragments for cocrystallization. Some fragments were derived from the putative binding site of Bs-CspB to the 5′-UTR region of the Bs-CspB mRNA (cold box2) (Graumann et al. 1997) and others from the DNA consensus sequence of preferred binding identified by a DNA microarray-based approach (Morgan et al. 2007). The preference of Bs-CspB for T-rich stretches of ssDNA (Lopez et al. 2001) and the reported tight binding for the DNA fragment heptathymidine (Zeeb et al. 2006) prompted us to analyze the interaction of Bs-CspB with the corresponding RNA analogs hexa- and heptauridine (Table 2). The observed dissociation constants (KD) are in the medium to high nanomolar range. The KD values for the ssRNA fragment rC7G and its analogs missing a nucleotide at the 5′ or 3′ end, rC6G and rC6, as well as a comparison of the binding affinities of rU7 and rU6 demonstrate the preference of Bs-CspB for heptanucleotides over hexanucleotides. We identified the tightest binding for an ssRNA fragment with the sequence 5′-GUCUUUU-3′ (rC7U). This RNA fragment corresponds to the DNA consensus sequence for high-affinity binders (5′-GTCTTTG/T-3′) revealed by a DNA microarray approach. Related oligonucleotides that differ from the best binder only in the first and the last nucleotide increased the KD value up to a factor of 9. ssRNA sequences derived from the putative binding site of Bs-CspB to the 5′ UTR of its own mRNA showed similar binding affinities (rcb2_a, rcb2_c). The remarkably decreased affinity of the oligonucleotide rcb2_b containing a purine base in the middle of the sequence points toward the preference of Bs-CspB for pyrimidine-rich stretches.
TABLE 2.
Dissociation constants of Bs-CspB·ssRNA complexes determined by tryptophan fluorescence titration
Kinetics and thermodynamics of oligonucleotide binding to Bs-CspB
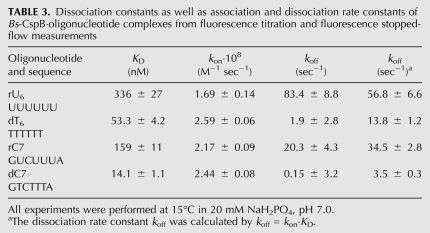
The dissociation constant at equilibrium reflects the ratio between the association (kon) and the dissociation (koff) rate constants. To gain deeper insight into complex formation, both rates have been determined experimentally. The kon and koff rate constants of the ssRNA fragments rU6 and rC7 as well as their ssDNA analogs dT6 and dC7 are shown in Table 3. Both rates were determined by rapid mixing of the protein solution with various concentrations of oligonucleotide. Under pseudo-first-order conditions kon is the slope of the observed rate (kobs) dependent on the oligonucleotide concentration and is better determined than koff which results from an extrapolation to 0 M oligonucleotide and represents the offset of the linear function. Therefore, koff is often determined by koff = kon · KD. The kon of the studied oligonucleotides varied only marginally between 1.69 · 108 M−1 sec−1 and 2.59 · 108 M−1 sec−1. In contrast, koff covered a wider range between 3.5 sec−1 and 56.8 sec−1, revealing a correlation between the variation of koff and the KD derived from equilibrium fluorescence titration experiments. Consequently, reduced affinity results from an increased dissociation rate.
TABLE 3.
Dissociation constants as well as association and dissociation rate constants of Bs-CspB·oligonucleotide complexes from fluorescence titration and fluorescence stopped-flow measurements
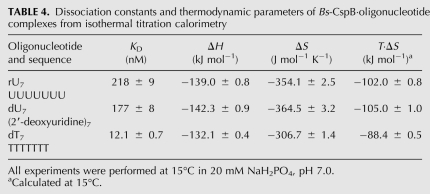
For further investigations we considered the two chemical properties that differentiate RNA from DNA. First, RNA contains ribose instead of deoxyribose, and second the base thymine is substituted by uracil. To investigate the influence of the chemical nature of the nucleic acid, binding affinities were determined by ITC measurements with rU7 (ssRNA fragment), dT7 (ssDNA fragment), and dU7, an oligodeoxyribonucleotide with uracil bases instead of thymine. dU7 differs from rU7 only in the absence of a sugar 2′ hydroxyl and from dT7 only in the absence of a methyl group at the pyrimidine 5 position. The dissociation constants for rU7 and dU7 are almost identical but the KD of dT7 is lowered compared to its RNA analog by a factor of 18 (Table 4). The complex formation is characterized by an exothermic reaction. The individual enthalpy (ΔH) values for all three oligonucleotides vary only 4% from the mean. The free energy (ΔG) of binding was determined from ΔG = −RT ln(1/KD). Hence the entropy (ΔS) of binding at 15°C could be calculated using the Gibbs-Helmholtz relation ΔG = ΔH – TΔS. As expected, it decreases upon binding for all nucleotides. Interestingly, the values for rU7 and dU7 are almost identical, whereas the change in entropy for dT7 is significantly lower.
TABLE 4.
Dissociation constants and thermodynamic parameters of Bs-CspB·oligonucleotide complexes from isothermal titration calorimetry
DISCUSSION
We determined the crystal structure of Bs-CspB in complex with two different single-stranded RNA oligonucleotides. In both Bs-CspB·RNA complexes one RNA molecule binds to one protein molecule, which is in contrast to previously determined crystal structures of Bs-CspB and Bc-Csp in complex with the DNA fragment hexathymidine (Max et al. 2006, 2007). In the Bs-CspB·dT6 structure the DNA strands bridge adjacent protein molecules in the crystal lattice, and in the Bc-Csp·dT6 structure a domain-swapped protein dimer was observed. Nevertheless, all four structures revealed a common mode of oligonucleotide binding characterized by a conserved set of nucleotide-binding subsites.
The RNA binding site is formed by an amphipathic protein surface. The interactions with the ligand are dominated by stacking interactions between RNA bases and aromatic protein sidechains. Although the RNA ligands comprise six nucleotides in the Bs-CspB·rU6 complex and seven in the Bs-CspB·rC7 complex only five are resolved by sufficient electron density in both structures.
Structural characteristics of the ligand binding and assignment of binding subsites
The ligand binding site of Bs-CspB·RNA complexes can be divided in several contact subsites. From the previously assigned seven binding subsites based on the crystal structures of CSP in complex with DNA fragments (Max et al. 2006, 2007) four were also observed in the complexes with RNA (Fig. 3B,C). Interaction subsites 1 to 3 remain empty. Remarkably, a stacking interaction of Phe38 from a symmetry-related protein molecule to the nucleobase of U3/C3 is reminiscent of an occupied subsite 1. At subsite 4 the U2 base forms a stack with His29. The base of U3 in Bs-CspB·rU6 and the base of C3 in Bs-CspB·rC7 stack with the aromatic ring of Phe25, forming subsite 5. In subsite 6 the U4 base stacks with Phe17. This position stands out from the other subsites because of the number of direct contacts involving the base. Three hydrogen bonds are formed between the head groups of the pyrimidine ring and amino acid sidechains. All other bases are fixed with fewer contacts in their respective binding pockets. The last interaction subsite, numbered 7, is characterized by a stacking interaction of the U5 base and Trp8. It is worth mentioning that Asn10 participates in a hydrogen bond with the 2′-OH group of the ribose ring of nucleotide U5 in subsite 7. This interaction could not be formed with a DNA backbone and is thus specific for binding of RNA. In all other binding subsites the ribose rings are exposed to the solvent and do not contact the protein surface. This arrangement prevents the CSP to clearly distinguish between ribose and deoxyribose. The nucleotide U5 and the last nucleotide represented by sufficient electron density, U6, form a base stack. The position of nucleotide U6 in the crystal is stabilized by contacts to symmetry-related protein molecules, indicating that base stacking in Bs-CspB RNA complexes in solution is unlikely.
The ligand binding site thus shields the polar base edges in the nucleoprotein complex. This enables Bs-CspB to counteract double-strand formation of RNA, which may be essential for the presumed RNA-chaperoning activity or transcription antitermination function of Bs-CspB.
Bs-CspB·RNA assemblies in solution
In the present study, RNA nucleotide binding to only four subsites of Bs-CspB was observed, whereas up to six subsites were occupied in the crystal structures of DNA single strands bound to Bs-CspB or Bc-Csp (Max et al. 2006, 2007). This prompted us to analyze the binding of Bs-CspB to RNA fragments in solution. The change in protein chemical shifts during NMR titration experiments allowed us to localize the binding site of rU6 and rC7 (red surface in Fig. 6A). Thereby we obtained evidence that the protein surface covered by the ligand is more expanded in solution than in the crystal structure. The amide proton cross peaks of the amino acids Phe38 and Phe30, in particular, experience chemical shift changes that are among the highest of all protein residues. In comparison, the sidechains of these two phenylalanines are involved in interactions with the bases of two nucleotides bound to subsites 1 and 2, which form a four-membered stack of aromatic rings in the Bc-Csp·dT6 crystal structure (yellow stick representation in Fig. 6) and the bridge to the subsequent oligonucleotide in the Bs-CspB·dT6 crystal structure (blue stick representation in Fig. 6). Moreover, the cross peak of Lys39 experiences a substantial chemical shift perturbation upon binding (Fig. 5). In the Bs-CspB·dT6 structure the backbone atoms of Lys39 form hydrogen bonds with the edge of the base-bound subsite 2. These findings demonstrate that in solution rU6 and rC7 occupy binding subsite 2 of Bs-CspB. The absence of ligand-protein interactions in subsite 2 in the crystal structure can be explained by crystal packing hindering the access of the ligand to this subsite. Based on these findings we can define an interaction interface for rC7 and rU6 in solution. The heptanucleotide rC7 covers all seven binding subsites and the hexanucleotide rU6 occupies subsites 2 to 7. Note that NMR chemical shift mapping cannot per se differentiate between changes induced by direct contacts or by remotely induced conformational changes. The phylogenetic conservation in prokaryotes of residues building all seven binding subsites further confirms the NMR chemical shift mapping results (green in Fig. 6B; Fig. 6C; Max et al. 2007). In human proteins with CSD one or both the phenylalanine of subsites 1 and 2 are missing.
In case of rU6 it is also conceivable that a shift in register causes the ligand to bind to subsite 1–6. This could occur either by Bs-CspB gliding on the oligonucleotide or by a dissociation/rebinding mechanism. Despite the apparent discrepancy between the solution-state RNA binding and the crystal structures of Bs-CspB·rU6 and Bs-CspB·rC7, the crystallographic analysis presented here provides a clear picture of the recognition of RNA substrates by Bs-CspB in binding subsite 4 to 7.
Comparison between ssRNA and ssDNA binding to Bs-CspB
The affinities of several oligonucleotides to Bs-CspB were tested by tryptophan-fluorescence titration. We observed a tighter binding for heptanucleotides compared to hexanucleotides. This confirms the expectation of seven binding subsites also suggested by the NMR titration experiments. Nevertheless, the affinity of ssRNA is significantly lower than the affinity of ssDNA for which a low to subnanomolar range was reported (Max et al. 2006). A direct comparison of the single-stranded ribonucleic acids used for crystallization and their deoxyribonucleic acid analogs revealed 6 to 11 times higher KD values. To elucidate whether the discrepancy in affinity of ssRNA and ssDNA originates from differences in the rate of association or dissociation we measured the kinetics of complex formation by stopped-flow fluorescence. The similar kon values indicate low specificity of Bs-CspB association to all oligonucleotides examined regarding the base composition and the type of ribose rings, whereas koff varies according to the respective KD determined at equilibrium. In this sense a low binding affinity of Bs-CspB, because of unfavorable or missing interactions in the protein–ligand interface, is reflected only in an increased koff. This raises the question as to what causes the increased koff in ssRNA compared to its ssDNA analog.
All structures of oligonucleotides bound to CSPs now available demonstrate that the binding between the protein and the ligand is dominated by the base stacking on aromatic protein sidechains. The ribose or deoxyribose rings are oriented toward the solvent and do not directly contact the protein surface. Therefore, the substitution of the thymine bases by uracil in RNA should be the reason for the lower affinity of ssRNA to Bs-CspB and not sugar-phosphate backbone interactions. To prove this hypothesis we measured the binding constants of the heptanucleotides rU7 (ssRNA), dT7 (ssDNA), and dU7 (chimera) by isothermal titration calorimetry. The chimera shows a KD value in the medium nanomolar range. The substitution of its deoxyribose ring by ribose in rU7 changed the KD only marginally, but the substitution of the uracil base by thymine as in dT7 increased the affinity dramatically. Therefore, it can be concluded that the additional methyl group of thymine can be responsible for the higher affinity of dT7. The observed decrease in entropy for rU7, dU7 and dT7 is due to conformational restrictions in the complex and contributes unfavorably to ΔG. The entropy change of dT7 stands out from the two other oligonucleotides with a significant lower value. In this sense the increased binding affinity of dT7 is mainly a consequence of the lower entropy change upon binding. This lower entropy change might be a consequence of the hydrophobic effect of the additional methyl group of thymine in the unbound form.
The here observed lower affinity of Bs-CspB for ssRNA compared to ssDNA determined by biophysical methods in vitro renews a controversial discussion in the literature about the in vivo function of the CSPs. In E. coli, ssDNA binding of CSPs has been reported as transcription stimulation after cold shock (La Teana et al. 1991; Jones et al. 1992; Brandi et al. 1994). However, in E. coli CspA is one of the most abundant proteins at 37°C during early exponential growth (Brandi et al. 1999) and B. subtilis CspB and CspC are profoundly induced in the stationary phase (Graumann and Marahiel 1999). Therefore, induction upon cold shock, amino acid starvation (Fraser et al. 2006), and antibiotic treatment (VanBogelen and Neidhardt 1990) might indicate numerous other cellular functions of CSPs, especially because knockout of single and multiple CSPs did not result in clear phenotypes in E. coli (Xia et al. 2001) and in B. subtilis (Graumann et al. 1997). For example, an RNA chaperone function (Jiang et al. 1997) or a transcription antitermination function of Bs-CspB (Phadtare et al. 2002a,b; Phadtare 2004) has been suggested and later contested in cell-free translation and transcription experiments (Hofweber et al. 2005). Our present in vitro findings revealed that Bs-CspB shows generally a higher affinity to ssDNA compared to the respective ssRNA fragments up to a factor of 18. In vivo, additionally the cellular concentration of target RNA and DNA has to be considered. For a rather unspecific binding such as chaperoning, the expected ratio of ssDNA compared to ssRNA is much smaller than a factor of 18 and therefore ssDNA binding of Bs-CspB is very unlikely. In the case of a specific event such as antitermination or binding to the 5′ UTR of a specific mRNA the factor of 18 becomes relevant and then the preferred target will be the ssDNA.
MATERIALS AND METHODS
Bs-CspB overexpression and purification
The gene encoding the CSP Bs-CspB in pET 11a was overexpressed in E. coli BL21(DE3) using the T7 RNA promoter system. Bs-CspB was purified as described previously (Schindelin et al. 1992; Schindler et al. 1995) with the following modifications. The cell suspension was centrifuged for 10 min and 8300g at 4°C. The pellet was suspended in 50 mM Tris/HCl pH 8.0, and the suspension obtained was sonified in intervals for a total of 3 min. The broken cells were centrifuged for 30 min at 48,000g, and the supernatant was applied to a Fractogel EMD TMAE (M) (Merck) anion exchange column. Bound protein was eluted with a NaCl gradient ranging from 0 to 0.6 M in 50 mM Tris/HCl pH 7.8. Fractions containing Bs-CspB were pooled, and (NH4)2SO4 was added to a final concentration of 2.3 M. The solution was applied to a Butyl Sepharose 4 Fast Flow (GE Healthcare) aliphatic hydrophobic interaction column equilibrated with 50 mM Tris/HCl pH 7.6, 2.3 M (NH4)2SO4. After washing, elution was performed by reducing (NH4)2SO4 concentration to 0 M. Fractions containing protein were dialyzed against 50 mM Tris/HCl pH 7.2, 100 mM KCl. Subsequent gel filtration (HiLoad Superdex 75 16/60) in the same buffer led to pure Bs-CspB as judged from SDS-polyacrylamide gels. Finally the protein solution was dialyzed against 20 mM (NH4)HCO3 and freeze-dried.
Complex formation
The HPLC-purified RNA fragments were purchased from Thermo Fisher Scientific GmbH. A buffer with 20 mM HEPES pH 7.5, 50 mM NaCl, was prepared and 0.1% (by volume) diethylpyrocarbonate was added and stirred for 12 h followed by autoclaving at 121°C for 20 min. For complex formation, protein and RNA were dissolved in this buffer and mixed in a 1:1.25 molar ratio. Excess RNA was separated using a Superdex 75 16/60 prep grade (GE Healthcare) gel-filtration column, and the complexes were concentrated using Vivaspin 3 kDa concentrators (Vivascience). The complex solution was checked for sample homogeneity by dynamic light scattering.
Crystallization
Crystallization experiments were carried out at 293 K using the sitting-drop vapor-diffusion method. The protein–RNA complex solution was mixed in equal volumes with the reservoir solution containing 31% (w/v) PEG 3350, 0.25 M MgCl2, 0.1 M Tris pH 8.5 for the Bs-CspB·rU6 complex, and 30% (w/v) PEG 4000, 0.2 M MgCl2, 0.1 M Tris pH 8.5 for the Bs-CspB·rC7 complex.
X-ray diffraction data collection and processing
Crystals were harvested from the drops and directly flash-frozen in liquid nitrogen. X-ray diffraction data were collected for the Bs-CspB·rU6 complex at a wavelength of 1.5418 Å at an in-house X-ray generator using a MAR345 image plate detector and for the Bs-CspB·rC7 complex at a wavelength of 0.9184 Å at beamline BL 14.2 of Freie Universität Berlin at BESSY (Berlin) (Heinemann et al. 2003) using a Rayonix MX-225 CCD detector. Complete data sets were collected to maximal resolutions of 1.68 Å and 1.38 Å, respectively. The XDS package (Kabsch 1993) was used to integrate reflection intensities. The quality of the data is summarized in Table 1.
Structure determination, model building, and refinement
Structure factor phases were determined via molecular replacement by Phaser version 2.1.4 (McCoy et al. 2007) and free Bs-CspB (1CSP) (Schindelin et al. 1993) as a template. The structure was refined using REFMAC5 version 5.5.0072 (Murshudov et al. 1997). Five percent of the reflections were set aside for cross-validation, and Rfree was used to adjust the refinement strategy and monitor progress while the correlation between models and experimental data was analyzed with SFCHECK (Vaguine et al. 1999). SFCHECK was also used to illustrate unbiased electron densities of the RNA ligands as displayed in Figure 4. The resulting structural models and experimental data for Bs-CspB·rU6 and Bs-CspB·rC7 were deposited in the Protein Data Bank under entry codes 3PF5 and 3PF4, respectively.
Fluorescence titration of Bs-CspB with oligonucleotides
For the determination of dissociation constants (KD) of protein-oligonucleotide complexes proteins at concentrations of 0.5–6 μM were used depending on the dissociation constant and titrated with DNA or RNA oligonucleotides. Experiments were performed at 15°C in 20 mM NaH2PO4 pH 7.0. The samples were gently stirred during titration. After 2 min of equilibration the intrinsic fluorescence of Bs-CspB Trp8 was excited at 280 nm and monitored at 343 nm by using a Jasco FP6500 spectrofluorimeter. The sample volume was 1.7 mL, and the increase in volume during titration was <5%. The fluorescence was corrected for buffer fluorescence and dilution. The binding isotherms were analyzed according to the binding equation:
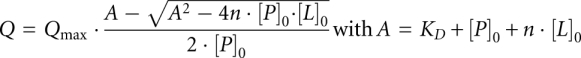 |
where Q is the quenching of the intrinsic fluorescence intensity of Trp8 after each addition of oligonucleotide. Qmax represents the maximal quenching upon complete saturation of the protein with nucleic acid. [P]0 and [L]0 are the protein and oligonucleotide concentrations. n is the number of Bs-CspB molecules bound to one oligonucleotide strand and KD the dissociation constant of the complex.
Measurement of association kinetics
Association kinetics were measured using an Applied Photophysics SX-20MV stopped-flow instrument. An excitation wavelength of 280 nm was used, and the emission was monitored using a 320 nm cut-off filter. All experiments were carried out at 15°C in 20 mM sodium phosphate buffer pH 7.0. One volume of 0.12 μM protein solution was mixed with one volume of 0.6–3.2 μM oligodeoxyribonucleotide or 0.8–2.2 μM oligoribonucleotide. Data collected from six to eight shots were averaged and fitted using the software supplied with the stopped-flow instrument.
Isothermal titration calorimetry
Isothermal titration calorimetry was carried out using a MicroCal ITC calorimeter (MicroCal, Inc.). Into a Bs-CspB solution (6 μM) aliquots of dU7 (60 μM) or dT7 (60 μM) were injected. The measurements were performed at 15°C in 20 mM sodium phosphate pH 7.0. The data were analyzed with the ORIGIN software (Microcal Software).
NMR titration experiments
All oligonucleotides were purchased from Thermo Fisher Scientific GmbH except for dU7, which was purchased from biomers.net GmbH. All NMR titration experiments were performed at 20°C on a Bruker Avance III 600 spectrometer. The initial protein concentration was 100 μM of uniformly 15N enriched Bs-CspB dissolved in 20 mM HEPES pH 7.5, 50 mM NaCl, 10% D2O. The titration experiments were carried out by successive addition of aliquots of unlabeled rU6 or rC7 stock solution dissolved in the same buffer as the protein until a 1:1.5 molar excess of RNA was reached. Complex formation was monitored by recording 2D 1H/15N-HSQC spectra with WATERGATE solvent suppression after each ssRNA addition. The mean weighted change in chemical shift was calculated by the formula: ΔδMW(1HN,15N) = ([Δδ(1HN)2 + (Δδ(15N)/25)2]/2)0.5 (Grzesiek et al. 1996).
Alignment of CSD sequences
A BLAST search was performed using the NCBI's Reference Sequence Database (RefSeq) and the PSI-BLAST algorithm to find homologous sequences to Bs-CspB. The 400 closest hits were used to define a profile, which was trained by a further iteration cycle. The 400 closest hits reported after the third search were used for a multiple sequence alignment. Thirteen sequences with the highest degree of divergence from Bs-CspB were added to the sequences of Bs-CspB and Bc-Csp. A second BLAST search was performed similar to that described above to find homologous sequences in Homo sapiens. The four closest hits were added to the list. All sequences were aligned using CLUSTAL W (Thompson et al. 1994) and the level of conservation was calculated for each sequence position and shaded at a level of 75% sequence identity or similarity using BioEdit (Hall 1999).
ACKNOWLEDGMENTS
This work was supported by the Fonds der Chemischen Industrie, the Deutsche Forschungsgemeinschaft, and the initiative ProNet-T3 of the German Federal Ministry of Education and Research (BMBF). Significant investments into the NMR facility of the University Halle-Wittenberg from the European Regional Development Fund (ERDF) by the European Union are also gratefully acknowledged.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.02809212.
REFERENCES
- Bae W, Jones PG, Inouye M 1997. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J Bacteriol 179: 7081–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W, Xia B, Inouye M, Severinov K 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc Natl Acad Sci 97: 7784–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA 2001. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi A, Pon CL, Gualerzi CO 1994. Interaction of the main cold shock protein CS7.4 (CspA) of Escherichia coli with the promoter region of hns. Biochimie 76: 1090–1098 [DOI] [PubMed] [Google Scholar]
- Brandi A, Spurio R, Gualerzi CO, Pon CL 1999. Massive presence of the Escherichia coli ‘major cold-shock protein’ CspA under non-stress conditions. EMBO J 18: 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs K, Karplus PA 1997. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol 4: 269–275 [DOI] [PubMed] [Google Scholar]
- Fang L, Hou Y, Inouye M 1998. Role of the cold-box region in the 5′ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J Bacteriol 180: 90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser KR, Tuite NL, Bhagwat A, O'Byrne CP 2006. Global effects of homocysteine on transcription in Escherichia coli: Induction of the gene for the major cold-shock protein, CspA. Microbiology 152: 2221–2231 [DOI] [PubMed] [Google Scholar]
- Giaquinto L, Curmi PM, Siddiqui KS, Poljak A, DeLong E, DasSarma S, Cavicchioli R 2007. Structure and function of cold shock proteins in archaea. J Bacteriol 189: 5738–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliodori AM, Di Pietro F, Marzi S, Masquida B, Wagner R, Romby P, Gualerzi CO, Pon CL 2010. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol Cell 37: 21–33 [DOI] [PubMed] [Google Scholar]
- Graumann PL, Marahiel MA 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci 23: 286–290 [DOI] [PubMed] [Google Scholar]
- Graumann PL, Marahiel MA 1999. Cold shock proteins CspB and CspC are major stationary-phase-induced proteins in Bacillus subtilis. Arch Microbiol 171: 135–138 [DOI] [PubMed] [Google Scholar]
- Graumann P, Schroder K, Schmid R, Marahiel MA 1996. Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol 178: 4611–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P, Wendrich TM, Weber MH, Schröder K, Marahiel MA 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol Microbiol 25: 741–756 [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Stahl SJ, Wingfield PT, Bax A 1996. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry 35: 10256–10261 [DOI] [PubMed] [Google Scholar]
- Hall TA 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98 [Google Scholar]
- Heinemann U, Bussow K, Mueller U, Umbach P 2003. Facilities and methods for the high-throughput crystal structural analysis of human proteins. Acc Chem Res 36: 157–163 [DOI] [PubMed] [Google Scholar]
- Hofweber R, Horn G, Langmann T, Balbach J, Kremer W, Schmitz G, Kalbitzer HR 2005. The influence of cold shock proteins on transcription and translation studied in cell-free model systems. FEBS J 272: 4691–4702 [DOI] [PubMed] [Google Scholar]
- Horn G, Hofweber R, Kremer W, Kalbitzer HR 2007. Structure and function of bacterial cold shock proteins. Cell Mol Life Sci 64: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Fang L, Inouye M 1996. The role of the 5′-end untranslated region of the mRNA for CspA, the major cold-shock protein of Escherichia coli, in cold-shock adaptation. J Bacteriol 178: 4919–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hou Y, Inouye M 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem 272: 196–202 [DOI] [PubMed] [Google Scholar]
- Jones PG, Krah R, Tafuri SR, Wolffe AP 1992. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J Bacteriol 174: 5798–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kremer W, Schuler B, Harrieder S, Geyer M, Gronwald W, Welker C, Jaenicke R, Kalbitzer HR 2001. Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima. Eur J Biochem 268: 2527–2539 [DOI] [PubMed] [Google Scholar]
- La Teana A, Brandi A, Falconi M, Spurio R, Pon CL, Gualerzi CO 1991. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc Natl Acad Sci 88: 10907–10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MM, Yutani K, Makhatadze GI 2001. Interactions of the cold shock protein CspB from Bacillus subtilis with single-stranded DNA. Importance of the T base content and position within the template. J Biol Chem 276: 15511–15518 [DOI] [PubMed] [Google Scholar]
- Max KE, Zeeb M, Bienert R, Balbach J, Heinemann U 2006. T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB. J Mol Biol 360: 702–714 [DOI] [PubMed] [Google Scholar]
- Max KE, Zeeb M, Bienert R, Balbach J, Heinemann U 2007. Common mode of DNA binding to cold shock domains. Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus. FEBS J 274: 1265–1279 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitta M, Fang L, Inouye M 1997. Deletion analysis of cspA of Escherichia coli: Requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol Microbiol 26: 321–335 [DOI] [PubMed] [Google Scholar]
- Morgan HP, Estibeiro P, Wear MA, Max KE, Heinemann U, Cubeddu L, Gallagher MP, Sadler PJ, Walkinshaw MD 2007. Sequence specificity of single-stranded DNA-binding proteins: A novel DNA microarray approach. Nucleic Acids Res 35: e75 doi: 10.1093/nar/gkm040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HP, Wear MA, McNae I, Gallagher MP, Walkinshaw MD 2009. Crystallization and X-ray structure of cold-shock protein E from Salmonella typhimurium. Acta Crystallogr Sect F Struct Biol Cryst Commun 65: 1240–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller U, Perl D, Schmid FX, Heinemann U 2000. Thermal stability and atomic-resolution crystal structure of the Bacillus caldolyticus cold shock protein. J Mol Biol 297: 975–988 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Phadtare S 2004. Recent developments in bacterial cold-shock response. Curr Issues Mol Biol 6: 125–136 [PubMed] [Google Scholar]
- Phadtare S, Inouye M, Severinov K 2002a. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J Biol Chem 277: 7239–7245 [DOI] [PubMed] [Google Scholar]
- Phadtare S, Tyagi S, Inouye M, Severinov K 2002b. Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J Biol Chem 277: 46706–46711 [DOI] [PubMed] [Google Scholar]
- Ren J, Nettleship JE, Sainsbury S, Saunders NJ, Owens RJ 2008. Structure of the cold-shock domain protein from Neisseria meningitidis reveals a strand-exchanged dimer. Acta Crystallogr Sect F Struct Biol Cryst Commun 64: 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger W 1984. Principles of nucleic acid structure. Springer-Verlag, New York [Google Scholar]
- Schindelin H, Herrler M, Willimsky G, Marahiel MA, Heinemann U 1992. Overproduction, crystallization, and preliminary X-ray diffraction studies of the major cold shock protein from Bacillus subtilis, CspB. Proteins 14: 120–124 [DOI] [PubMed] [Google Scholar]
- Schindelin H, Marahiel MA, Heinemann U 1993. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature 364: 164–168 [DOI] [PubMed] [Google Scholar]
- Schindelin H, Jiang WN, Inouye M, Heinemann U 1994. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc Natl Acad Sci 91: 5119–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler T, Herrler M, Marahiel MA, Schmid FX 1995. Extremely rapid protein folding in the absence of intermediates. Nat Struct Biol 2: 663–673 [DOI] [PubMed] [Google Scholar]
- Schnuchel A, Wiltschek R, Czisch M, Herrler M, Willimsky G, Graumann P, Marahiel MA, Holak TA 1993. Structure in solution of the major cold-shock protein from Bacillus subtilis. Nature 364: 169–171 [DOI] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS 2003. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct 32: 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaguine AA, Richelle J, Wodak SJ 1999. SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr 55: 191–205 [DOI] [PubMed] [Google Scholar]
- VanBogelen RA, Neidhardt FC 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci 87: 5589–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MH, Marahiel MA 2002. Coping with the cold: The cold shock response in the Gram-positive soil bacterium Bacillus subtilis. Philos Trans R Soc Lond B Biol Sci 357: 895–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Ke H, Inouye M 2001. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol Microbiol 40: 179–188 [DOI] [PubMed] [Google Scholar]
- Xia B, Ke H, Jiang W, Inouye M 2002. The Cold Box stem-loop proximal to the 5′-end of the Escherichia coli cspA gene stabilizes its mRNA at low temperature. J Biol Chem 277: 6005–6011 [DOI] [PubMed] [Google Scholar]
- Zeeb M, Max KE, Weininger U, Löw C, Sticht H, Balbach J 2006. Recognition of T-rich single-stranded DNA by the cold shock protein Bs-CspB in solution. Nucleic Acids Res 34: 4561–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]