A majority of SNPs (single nucleotide polymorphisms) map to noncoding and intergenic regions of the genome. Noncoding SNPs are often identified in genome-wide association studies (GWAS) as strongly associated with human disease. Two such disease-associated SNPs in the 5′ UTR (Untranslated Region) of the human FTL (Ferritin Light Chain) gene are predicted to alter the ensemble of structures adopted by the mRNA. High-accuracy single nucleotide resolution chemical mapping reveals that these SNPs result in substantial changes in the structural ensemble in agreement with the computational predictions. The data confirm that the FTL 5′ UTR is a “RiboSNitch,” an RNA that changes structure if a particular disease-associated SNP is present.
Keywords: RiboSNitch, SNP, mRNA, partition function, transcript
Abstract
A majority of SNPs (single nucleotide polymorphisms) map to noncoding and intergenic regions of the genome. Noncoding SNPs are often identified in genome-wide association studies (GWAS) as strongly associated with human disease. Two such disease-associated SNPs in the 5′ UTR of the human FTL (Ferritin Light Chain) gene are predicted to alter the ensemble of structures adopted by the mRNA. High-accuracy single nucleotide resolution chemical mapping reveals that these SNPs result in substantial changes in the structural ensemble in agreement with the computational prediction. Furthermore six rescue mutations are correctly predicted to restore the mRNA to its wild-type ensemble. Our data confirm that the FTL 5′ UTR is a “RiboSNitch,” an RNA that changes structure if a particular disease-associated SNP is present. The structural change observed is analogous to that of a bacterial Riboswitch in that it likely regulates translation. These data further suggest that specific pairs of SNPs in high linkage disequilibrium (LD) will form RNA structure-stabilizing haplotypes (SSHs). We identified 484 SNP pairs that form SSHs in UTRs of the human genome, and in eight of the 10 SSH-containing transcripts, SNP pairs stabilize RNA protein binding sites. The ubiquitous nature of SSHs in the transcriptome suggests that certain haplotypes are conserved to avoid RiboSNitch formation.
INTRODUCTION
Noncoding but nonetheless transcribed regions of the human genome often play important regulatory functions in the cell (Griffiths-Jones et al. 2005; Rana 2007; Chu and Herschlag 2008; Tseng et al. 2009; Halvorsen et al. 2010). In particular, the 5′ and 3′ UTRs of genes are central components of the regulatory machinery (Lai et al. 1998; Pesole et al. 1999; Boffa et al. 2008; Halvorsen et al. 2010; Kilty et al. 2010). In bacteria, Riboswitches in 5′ UTRs will bind small molecules (often metabolite precursors) that alter their conformation to regulate protein expression levels (Tucker and Breaker 2005; Edwards and Ferre-D'Amare 2006; Weinberg et al. 2007; Stoddard et al. 2008; Wang et al. 2008a). Riboswitches exist because certain RNA sequences will adopt multiple, near isoenergetic conformations; small molecule binding stabilizes one conformation over the other (Wilkinson et al. 2005; Edwards and Ferre-D'Amare 2006; Stoddard et al. 2008; Wang et al. 2008a; Tseng et al. 2009). Analogously, we expect that specific mutations will significantly alter the RNA conformational ensemble (Halvorsen et al. 2010). If the RNA plays a regulatory role in the cell, the consequences of these structural changes can be a contributor to human disease (Teresi et al. 2007; Pezzolesi et al. 2008).
A RiboSNitch is a functional RNA regulatory element (generally in the 5′ or 3′ UTR of a gene or a noncoding RNA) where one or more single nucleotide polymorphisms (SNPs) or mutations cause a large conformational rearrangement in the RNA structural ensemble (Halvorsen et al. 2010). Of particular interest are RiboSNitches where the SNP or mutation is associated with a human disease (Chappell et al. 2006; Kimchi-Sarfaty et al. 2007; Garcia-Barcelo et al. 2009; Glinskii et al. 2009; Fujimoto et al. 2010; Kilty et al. 2010). Using the SNPfold algorithm (Halvorsen et al. 2010), we predicted that two SNPs in the 5′ UTR of the human FTL (Ferritin Light Chain) gene associated with hyperferritinemia cataract syndrome (a rare but dominant hereditary disorder resulting in early onset cataracts) alter the mRNA structural ensemble (Ferrari et al. 2006; Sanchez et al. 2006; Burdon et al. 2007; Halvorsen et al. 2010). Our data reveal the ensemble of conformations adopted by mRNA is exquisitely sensitive to specific mutations despite being described as “unstructured.” Furthermore, these structural changes can be rescued with multiple secondary compensating mutations. This raises the possibility that certain SNPs in high linkage disequilibrium (LD) stabilize critical regions in transcripts. A comprehensive analysis of common human genetic variation reveals that such “structure-stabilizing haplotypes” are ubiquitous in mRNA and in a majority of cases stabilize RNA binding protein (RBP) sites.
RESULTS
Disease-associated SNPs repartition the RNA conformational ensemble
Certain noncoding RNAs (for example the Ribosome and self splicing introns) have evolved to fold into unique three-dimensional structures with highly specific function (Woodson and Leontis 1998; Silverman et al. 2000; Weikl et al. 2004; Sykes and Levitt 2005; Laederach et al. 2007; Waldsich 2008). However, many regions in the transcriptome are under selective pressure that will not necessarily favor a single conformation (Peng et al. 1995; Rana 2007; Chu and Herschlag 2008; Regulski and Breaker 2008; Tseng et al. 2009; Halvorsen et al. 2010). Instead, they adopt an ensemble of conformations that cannot be described by a single structure (Russell and Herschlag 2001; Mathews 2004; Bernhart et al. 2006; Tyagi and Mathews 2007; Waldispuhl and Clote 2007). This is the case for a majority of human mRNA transcripts, including the 5′ UTR of the human FTL gene.
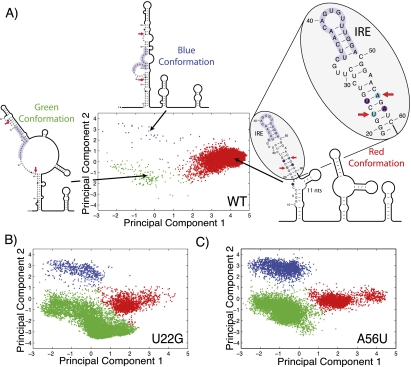
To visualize the conformational diversity of the FTL 5′ UTR, we performed Boltzmann sampling of suboptimal secondary structures and projected these onto a principal component decomposition of the ensemble (Ding et al. 2004, 2005). Unlike traditional minimum free energy methods for predicting RNA structure (e.g., mFold), Boltzmann sampling allows us to visualize the conformational ensemble of an mRNA (Halvorsen et al. 2010). Each dot in Figure 1A represents one suboptimal RNA structure, and the distance between them is correlated to the difference in structure. What is apparent from Figure 1 is that the human FTL 5′ UTR is predicted to adopt three major conformations. The wild-type (WT) sequence (Fig. 1A) adopts primarily one conformation (red), while two disease-associated mutations (U22G and A56U, Fig. 1B,C, respectively) favor the alternative blue and green conformations. The FTL 5′ UTR includes an Iron Response Element (IRE) which is indicated in light purple (Fig. 1A). Only for the WT sequence does a majority of structures adopt a conformation where the IRE is in the correct hairpin conformation. These calculations suggest a putative mechanism for the disease association. The Iron Responsive Element-Binding Protein (IREBP) binds to the IRE and represses translation (Sanchez et al. 2006). When the disease-associated SNP shifts the equilibrium away from the WT conformation (Fig. 1A), FTL translation is enhanced leading to the hyperferritinemia phenotype (Burdon et al. 2007).
FIGURE 1.
(A) Projection of 10,000 Boltzmann sampled suboptimal structures of the human FTL 5′ UTR onto a principal component decomposition of the structural space (Ding et al. 2005). The projection allows us to visualize the conformational ensemble of the noncoding RNA. We identify three distinct conformations illustrated by the representative secondary structures (blue, green, and red conformations). The FTL 5′ UTR includes an IRE, which is a regulatory motif known to repress translation when bound by the IREBP (Ferrari et al. 2006). In the two alternative conformations (blue and green), the IRE does not adopt a functional hairpin conformation. (B,C) Visualizations of the structural ensemble for the U22G and A56U SNP-containing FTL 5′ UTRs, respectively, identified in patients suffering from hyperferritinemia cataract syndrome (Cremonesi et al. 2001, 2003; Ferrari et al. 2006; Burdon et al. 2007). Both SNPs repartition the ensemble favoring the blue and green conformations. We propose that the FTL 5′ UTR is a “RiboSNitch,” which much like a bacterial Riboswitch (Tucker and Breaker 2005; Grundy and Henkin 2006; Gilbert et al. 2008) undergoes an important structural rearrangement that controls translation. On the secondary structure diagrams the two hyperferritinemia cataract syndrome SNPs (U22G and A56U) are indicated with arrows while SNPfold predicted structure-stabilizing mutations for U22G are indicated in purple.
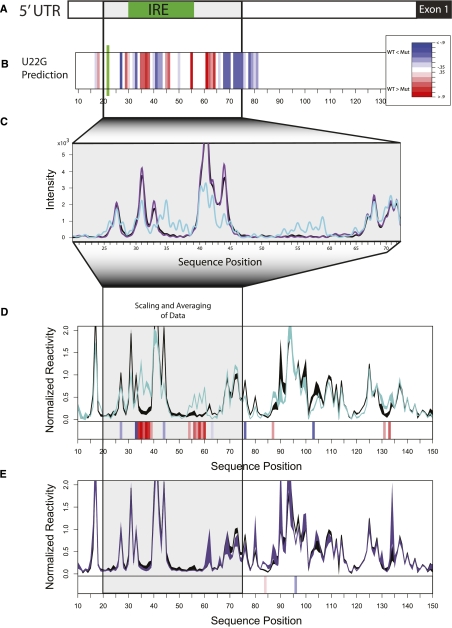
A particularly important feature of computing RNA ensembles such as those illustrated in Figure 1 is that they also predict the probability of base-pairing for each nucleotide (Mathews 2004; Bernhart et al. 2006; Waldispuhl and Clote 2007). The frequency of formation for a base pair in the ensemble is proportional to the probability of that base pair (Ding et al. 2004, 2005). Figure 2A schematically represents the FTL 5′ UTR, and Figure 2B is a heatmap of the predicted difference between the WT and U22G base-pair probabilities. We immediately observe another important aspect of this system. Even though the disease-associated mutation is not in the IRE, the largest predicted changes in base-pair probability are within the functional element.
FIGURE 2.
(A) Schematic of the human FTL 5′ UTR and Exon 1; the IRE is indicated in green. (B) SNPfold prediction of the changes in the probability of a base being paired caused by the U22G hyperferritinemia-associated SNP (Cremonesi et al. 2001, 2003; Ferrari et al. 2006; Burdon et al. 2007) as a heatmap, with red indicating the disease-allele results in higher base-pair probability and blue indicating a lower probability. (C) Raw capillary traces of SHAPE analysis of the WT (black), U22G (cyan, disease-associated), and G4A (purple, not disease-associated) FTL 5′ UTRs. (D) Quantification, averaging, and scaling of multiple repeats for WT (black) and U22G (cyan) SHAPE data. The thickness of the line indicates one standard deviation over six repeats of the experiment. The heatmap below the data is orthologous to the predictions made in B since changes in SHAPE reactivity correlate with changes in base-pair probability (Badorrek and Weeks 2005; Badorrek et al. 2006; Wilkinson et al. 2008). (E) Scaled and averaged repeats for G4A construct indicating the control mutation has no effect on base-pair probability as predicted. These experiments validate the SNPfold prediction and indicate that the FTL 5′ UTR is indeed a RiboSNitch.
Structure mapping confirms the presence of a RiboSNitch in the FTL 5′ UTR
Changes in base-pair probability are experimentally assayed using chemical mapping probes (Mitra et al. 2008; Quarrier et al. 2010). We transcribed the human FTL 5′ UTR and performed selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) chemical mapping experiments (Wilkinson et al. 2005, 2008, 2009). These experiments probe RNA structure with single nucleotide resolution and report the flexibility of the backbone, which is directly correlated with base-pair probability (Wilkinson et al. 2006). Figure 2C illustrates raw capillary (+) SHAPE traces for the WT, U22G, and a control G4A mutation (which is not predicted to affect the RNA structural ensemble, and is also not disease-associated). From these raw traces alone, we visually discern the effect of the U22G mutation on the structure of the FTL 5′ UTR. As predicted by Boltzmann sampling, the IRE nucleotides (C33-A49) have higher SHAPE reactivity for the U22G construct, indicative of a significant decrease in base-pairing. The full extent of the disease-associated SNP's effect on the RNA ensemble is observed when the peak areas are integrated, scaled, and averaged over multiple repeats (Fig. 2D; Mitra et al. 2008). Analogous experiments on the A56U construct reveal a similar effect, consistent with the ensemble calculation prediction (Supplemental Fig. 1). Furthermore the control SNP G4A has only minimal effect on the RNA structure, as predicted (Fig. 2E).
RiboSNitch rescue through double mutation
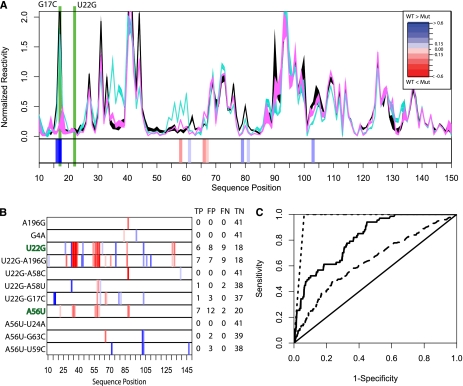
To further validate our ensemble model for the FTL 5′ UTR (Fig. 1) we performed a comprehensive SNPfold analysis to identify secondary mutations that restore the ensemble to WT partitioning. These mutations are reported in Supplemental Table 1. We experimentally validated the top three mutations for both constructs. The SHAPE data for the U22G-G17C double mutant shows that this mutation restores IRE structure to WT (Fig. 3A). This prediction is particularly interesting as it does not simply restore the U22-A58 base pair (Fig. 1, red conformation) through isosteric replacement of a canonical Watson-Crick pair (Lescoute et al. 2005). Instead it further destabilizes the first stem in the blue and green conformations favoring the WT structure.
FIGURE 3.
(A) SHAPE chemical mapping data scaled and averaged over six repeats for the WT (black), U22G (cyan), and U22G-G17C (magenta) FTL 5′ UTRs. The heatmap below the data shows that the U22G-G17C haplotype restores base-pair probability to near WT conformation, as predicted by SNPfold. (B) SHAPE data heatmaps indicating experimentally determined SNP-induced changes in probability of a base being paired for hyperferritinemia (U22G and A56U) and nondiseased genotypes. We made orthologous predictions using SNPfold and report the True Positive, False Positive, and True Negative, and False Negative (TP, FP, TN, and FN, respectively) rates when comparing SNPfold predictions to SHAPE chemical data. (C) ROC analysis of the SNPfold algorithm for predicting changes in IRE base-pairing probability (solid line, AUC = 0.86) and identifying RiboSNitches (dashed line, AUC = 0.97), and using traditional minimum free energy (e.g., mFold) calculations (long dashes, AUC = 0.62). ROC analysis includes SHAPE data collected on the additional 10 FTL 5′ UTR mutants reported in Supplemental Figure 1C.
We systematically collected SHAPE data on the WT, single and double mutants predicted to affect the IRE structure to evaluate the performance of the SNPfold algorithm. Figure 3B illustrates a subset of these data (the additional data are summarized in Supplemental Fig. 1C). The SHAPE data were used to calculate the True Positive, False Positive, True Negative, and False Negative rates of our SNPfold predictions for each base. We compared traditional minimum free energy predictions to our SNPfold algorithm using a Receiver Operator Curve (ROC) analysis (DeLong et al. 1985). The SNPfold algorithm performs better with an area under the curve of 0.86 (solid line, Fig. 3C) compared to that of the minimum free energy predictions (long dashes, Fig. 3C) with an area under the curve of 0.62. Furthermore, we evaluated SNPfold for identifying SNPs that cause a SHAPE reactivity change in more than eight nucleotides in the IRE of FTL. The performance of SNPfold for RiboSNitch detection is high (Fig. 3C, short dashes, area under the curve 0.97).
Structure-stabilizing haplotypes
Our results on the FTL 5′ UTR reveal that specific SNPs can have profound effects on the structural ensemble of noncoding regions of RNA and that the SNPfold algorithm accurately predicts these (Fig. 3C). It is important to realize that a majority of SNPs have little or no effect on the ensemble partitioning of RNA. An analysis of all possible FTL 5′ UTR mutations previously showed that the U22G and A56U SNPs are in the top 5% in terms of their predicted structural effect (Halvorsen et al. 2010). Our experimental results predict that an individual with the U22G-G17C (Fig. 3A) haplotype will not have the hyperferritinemia phenotype since this double mutation stabilizes the IRE in the FTL 5′ UTR. The U22G and A56U mutations are extremely rare, as is hyperferritinemia cataract syndrome. We performed a comprehensive search of HapMap haplotypes and did not find any individuals with the U22G or A56U SNPs (Stranger et al. 2007; Morton 2008).
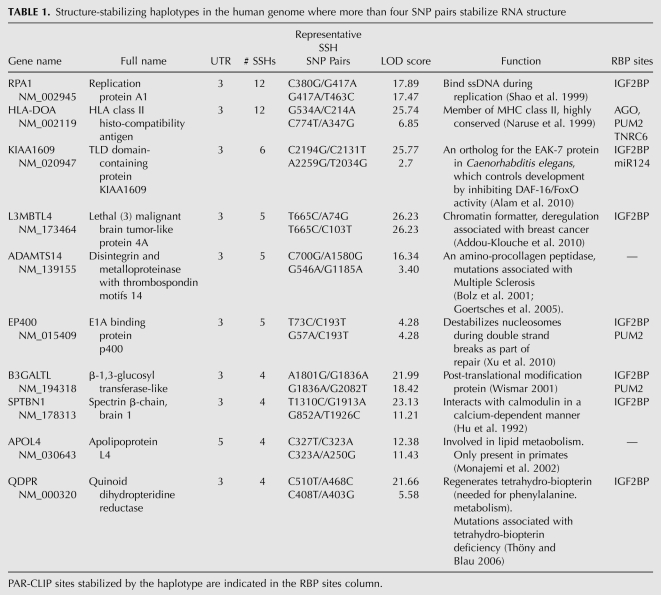
We nonetheless expect to find pairs of SNPs in high LD that stabilize mRNA structure. We call these pairs of SNPs “structure-stabilizing haplotypes,” or SSHs. Our experimental results demonstrate that the SNPfold algorithm is highly predictive of SSHs (area under the curve 0.97) (Fig. 3C), having correctly identified six pairs in the FTL 5′ UTR. We comprehensively analyzed LD SNP pairs in the human genome that map to 5′ and 3′ UTRs to identify potential SSHs. A complete list of the 484 SSH pairs we identified in human UTRs is provided in the supplement (Supplemental Table 2). In Table 1, we report UTRs where more than four SSH pairs were found. These are particularly noteworthy, since observing multiple SSH pairs in the same UTR is unlikely by chance (<1 in 100,000).
TABLE 1.
Structure-stabilizing haplotypes in the human genome where more than four SNP pairs stabilize RNA structure
Figure 4A illustrates the subset of LD SNP pairs that are predicted SSHs in the 3′ UTR of RPA1 (replication protein A1) (Shao et al. 1999). In Figure 4B we plot the mean change in base-pair probability caused by all individual SNPs in the haplotype. This allows us to visualize the regions in the RNA that are stabilized by the SSH and we observe that these coincide with experimentally determined IGF2BP binding sites (Hafner et al. 2010a,b). We performed a similar analysis on the SSHs reported in Table 1, and find that, in eight of the 10 UTRs, the predicted regions of the UTR that are stabilized coincide with RBP or miRNA binding sites determined by photoactivatable-ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) (Hafner et al. 2010a,b).
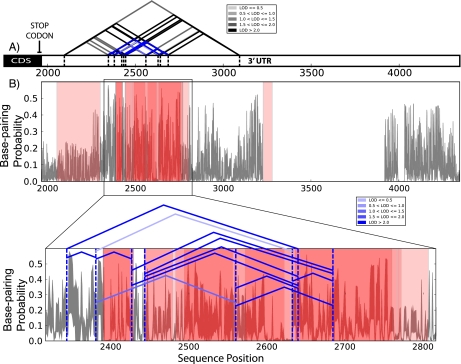
FIGURE 4.
SSH identified in the 3′ UTR of the RPA1 using SNPfold analysis. (A) Schematic representation of RPA1 3′ UTR including the coding sequence (CDS) and all currently known LD SNPs in the region, indicated as a triangle. The LOD scores of the SNPs are indicated in variable shades of gray and the subset of SNP pairs that stabilize structure are indicated in blue. (B) Mean change in base-pair probability based on a SNPfold analysis of the effects of individual SNPs that together stabilize structure. Red translucent bars indicate experimentally determined IGF2BP (insulin-like growth factor binding protein) binding sites (Hafner et al. 2010a; Rodriguez et al. 2010) on the 3′ UTR using the PAR-CLIP approach. The SSH (indicated with a series of solid blue angled lines) stabilizes a majority of IGF2BP binding sites, suggesting an important structural role for this region in the RPA1 3′ UTR. We report in Table 1 an additional 10 SSHs, eight of which stabilize known RBP sites.
DISCUSSION
The SHAPE data we collected validate the presence of a disease-associated RiboSNitch in the FTL 5′ UTR as computationally predicted by SNPfold. Our results illustrate the extent to which specific disease-associated SNPs can impact mRNA transcript ensemble partitioning. The two hyperferritinemia-associated SNPs (U22G and A56U) cause similar large rearrangements in the ensemble partitioning of the RNA. The rearrangements favor alternative conformations where the IRE does not adopt a hairpin conformation, a prerequisite for IREBP binding (Kaygun and Marzluff 2005). We find that the performance of the SNPfold algorithm is better than traditional minimum free energy approaches at identifying SNPs that alter mRNA structure (Fig. 3C) most likely because it takes into account the structural heterogeneity of the transcript (Mathews 2004; Waldispuhl and Clote 2007). Most importantly, our results indicate that transcripts, which are generally thought of as “unstructured,” are nonetheless highly sensitive to specific mutations.
The SNP-induced structural change in the FTL 5′ UTR is analogous in magnitude to that observed upon ligand binding in bacterial Riboswitches (Tucker and Breaker 2005). Furthermore, the effects on FTL protein expression are likely post-transcriptional, explaining the hyperferritinemia phenotype. Indeed, IREBP binding represses translation, such that, if the IRE is indirectly disrupted by SNP-induced ensemble repartitioning, excess protein will be translated. More importantly, secondary compensating mutations exist that restore WT partitioning, and the SNPfold algorithm correctly predicted six of them for both diseased alleles (Fig. 3B). Interestingly, these compensating mutations are not necessarily standard isosteric replacements of canonical base pairs (e.g., U22G-A58C), but also include more complex structural mechanisms (e.g., U22G-G17C) (Fig. 3A).
These results suggest that certain SNP pairs are conserved in the human population because they stabilize specific ensembles of conformations in mRNAs. Of particular interest are genes where multiple SNP pairs stabilize structure (Table 1; Fig. 4). Combined with PAR-CLIP data that identify transcriptome-wide RBP binding sites, an intriguing interplay between population genetics, RBP binding, and mRNA structure is revealed. We have thus far found eight transcript UTRs where multiple SSHs stabilize RBP and/or miRNA binding sites (Table 1). This likely represents only a subset of the genes where SNPs stabilize RNA structure, since we have limited human genetic variation data and PAR-CLIP data for four RBPs and one miRNA. Furthermore, to maintain computational tractability, we analyzed only SNP pairs in LD that map to the same UTR.
To identify pairs of SNPs in high linkage that stabilize RNA structure (i.e., SSHs) we defined a new metric, the Structure Recovery Ratio (SRR, Equation 1 below). Conceptually, the SRR measures the extent to which a pair of SNPs restores structure relative to the SNP that has the largest effect on that structure. Because it is a ratio, the numerical value of the SRR is not dependent on length of the RNA. We show this empirically in Supplemental Figure 2A, where there is no correlation between the SRR and length. We empirically chose an SRR cutoff value of two based on an analysis of the number of SSHs as a function of this threshold (Supplemental Fig. 2B). The mRNAs we identify in Table 1 are particularly interesting as four or more SSHs were found in either the 5′ or 3′ UTRs. We therefore expect that RNA structure likely plays an important regulatory role in these gene's regulation.
Covariation is common in large structured RNAs such as the Ribozyme, tRNAs, and even bacterial Riboswitches (Michel and Westhof 1990; Gutell et al. 2002; Leontis et al. 2002; Wang et al. 2008b). In fact, covariation models are often used to identify structured RNAs in genomic data (Griffiths-Jones et al. 2005). Traditionally, programs such as CMfinder (Yao et al. 2006; Benjamin et al. 2007) or FOLDALIGN (Havgaard et al. 2005) are used to predict structure. These techniques require significant covariation to constrain a structure and make an accurate prediction. Linkage data do not provide sufficient information to determine a structure in this way, which is why we chose to combine our structural predictions with PAR-CLIP data (Fig. 4) to make biological predictions. Our data suggest that certain haplotypes in the human population are conserved because they stabilize specific ensemble partitioning of functional mRNA regions.
We conclude that RiboSNitches are likely ubiquitous in the human genome, and that specific haplotypes are conserved because they preserve mRNA ensemble partitioning. We expect that SSHs in mRNA play an important regulatory role, and that our strategy of combining common genetic variation data with SNPfold analysis and PAR-CLIP data can pinpoint important new regulatory regions in the transcriptome. It is also likely that SNP-induced structure change affects mRNA stability. As more eQTL (expressed Quantitative Trait Loci) data become available, it will be possible to correlate structural change and mRNA stability, greatly expanding our understanding of post-transcriptional regulatory mechanisms (Majewski and Pastinen 2011).
MATERIALS AND METHODS
Computational methods
The SNPfold algorithm was used to predict how RNA structure is modified by SNPs (Halvorsen et al. 2010). The FTL SNPs were originally selected for their disease association. Secondary correcting mutations were predicted by identifying the SNP pair containing the primary mutant that resulted in the highest correlation coefficient to the WT base-pair probability plot as computed by the partition function (McCaskill 1990; Mathews 2004). The control SNPs were selected because SNPfold calculations showed they had the highest Pearson correlation coefficient (CC) in relation to the WT base-pairing. Rescue SNPs were identified by using SNPfold to calculate the CCs of the disease-associated SNP and a secondary SNP pair to the WT base-pairing and selecting those pairs with the highest CC.
From the hg18 genome assemble and hg18 “refgene” datafile, as downloaded from the UCSC dbSNP, 44,186 SNPs were identified. These SNPs satisfied the conditions that they were located in the untranslated region of genes and mapped to fully assembled chromosomes. Of these SNPs, 15,491 are located on the same spliced UTR and have linkage disequilibrium values (LOD scores) greater than or equal to one. The D′ values were extracted from LD flatfiles (available at the Hapmap webpage, http://hapmap.ncbi.nlm.nih.gov/downloads/ld_data/2009-04_rel27/) corresponding to the CEU population of Hapmap (release #27, Phase II and III).
The SRR is defined as
where CCboth is the correlation coefficient of the pairing probability of the RNA with both SNPs present, and CCmin is the correlation coefficient of the pairing probability to WT for the SNP in LD that has the largest effect on RNA structure (smallest CC). In both cases the correlation coefficients are computed with respect to the pairing probability of WT.
Principal component analysis
Principal components were calculated (as described previously; Halvorsen et al. 2010) from a total of 10,000 Boltzmann sampled structures generated equally from the five sequences: WT, C10U, C14G, U22G, and A56U. The projection of the binary representation of these 10,000 structures onto the first two principal components revealed three distinct clusters as identified by the k-means clustering algorithm. The centroid structure of each cluster was drawn using R2R. For a given SNP of interest, we project 10,000 structures, generated just from that sequence, onto the first two principal components with the projections colored according to which cluster they belong to.
SHAPE experiments
The WT FTL 5′ UTR sequence (GCAGTTCGGCGGTCCCGCGGGTCTGTCTCTTGCTTCAACAGTGTTTGGACGGAACAGATCCGGGGACTCTCTTCCAGCCTCCGACCGCCCTCCGATTTCCTCTCCGCTTGCAACCTCCGGGACCATCTTCTCGGCCATCTCCTGCTTCTGGGACCTGCCAGCACCGTTTTTGTGGTTAGCTCCTTCTTGCCAACC) was inserted between the SgfI and MluI sites of the pCMV6-AC nontagged precision shuttle vector (Origene). The modified plasmid was transformed into Escherichia coli DH5α electro competent cells and grown up over night at 37°C. The resulting plasmids were extracted using the Qiagen Miniprep Kit. A T7 promoter (TAATACGACTCACTATAGG) was introduced to the 5′ end of the FTL 5′ UTR during PCR amplification. Overlap PCR was used to create mutants by introducing a point mutation on oligonucleotides used during the first round of amplification with a second round of amplification used to join the PCR fragments. The resulting PCR product was transcribed into RNA using MegaScript followed by MegaClear according to Ambion's protocol.
Selective 2′-Hydroxyl Acylation and Primer Extension (SHAPE) experiments were performed in the following way: A total of 10 pmol RNA was suspended in 12 μL of 0.5× TE buffer and then heated to 95°C for 2 min. Immediately after, the mixture was placed on ice for 2 min. With gentle pipetting, 6 μL of folding mix was added (3.3× folding mix; 333 mM HEPES, pH 8.0, 20 mM MgCl2, 333 mM NaCl) to the mixture and then incubated at 37°C for 20 min. Following incubation, the mixture was split equally to obtain a negative control. The negative control was treated with 1 μL of dimethyl sulfoxide (DMSO) while the experimental mixture was treated with 1 μL of DMSO containing 65 mM N-methylisatoic anhydride (NMIA). Both mixtures were incubated for 45 min at 37°C, or roughly five half-lives of NMIA hydrolysis. RNA incubated at −80°C for 30 min following the addition of 90 μL of water, 4 μL of 5 M NaCl, 4 μL of 5 mg/mL glycogen, 2 μL of 100 mM EDTA pH 8.0, and 350 μL of 100% ethanol. RNA was precipitated by centrifugation at 14,000 rpm at 4°C for 30 min. The RNA was rinsed with 70% ethanol after the removal of the supernatant. After being spun down and dried by speed-vac for 10 min, the RNA was suspended in 10 μL of 0.5× TE.
The resulting mixtures had 1 μL of 5 μM of Cy5-labeled primer solution added and mixed by pipetting. The primers were annealed to the RNA by incubation at 65°C for 5 min, then 35°C for 5 min, and finally placed on ice for 1 min. The solutions with the annealed primers had 7 μL of the reverse transcriptase buffer mix which contained 250 mM KCl, 167 mM Tris-HCl, pH 8.3, 1.67 mM each dNTP, 17 mM DTT, 10 mM MgCl2, and 40 U of Invitrogen's RNaseOUT. The reverse transcriptase buffer mix was also added to the sequencing ladder reactions and, for the reactions lacking NMIA, 1–2 μL of one ddNTP (5mM) was also added. The samples were heated at 52°C for 11 min with 1 μL of Superscript III (Invitrogen) added after the first minute and mixed by pipetting. The resulting cDNA was cleaned up with the addition of 2 μL of 2 M NaOH and incubated at 95°C for 5 min. Following the incubation, the samples were treated with a solution containing 2 μL of 2 M HCl, 3 μL of 3 M Na-acetate, 1 μL of 100 mM MgCl2, and 90 μL of 100% ethanol. The mixture was then centrifuged at 14,000 rpm at 4°C for 30 min and rinsed with 70% ethanol. The resulting cDNA pellets were dried, suspended in 40 μL of Sample Loading Solution (Beckman), and subjected to capillary electrophoresis on a Beckman CEQ 8000.
Taking advantage of the data independent Beckman ladders and using custom written Matlab software (available at http://ribosnitch.bio.unc.edu/The_Laederach_Lab/Software.html), the data from the CEQ were aligned. The ShapeFinder (Vasa et al. 2008) software package was used to analyze the aligned SHAPE data. The reproducibility was determined by calculating the product of the mean SHAPE value for each nucleotide and multiplying it by the square root of the number of sets used divided by the standard deviation of that nucleotide. Our data were normalized using outlier analysis as previously reported resulting in the majority of the reactivities being between 0 and 1.5. SHAPE experiments were carried out under standard RNA folding conditions (10 mM MgCl2 and 100 mM KCl) as described in separate publications on the technique (Wilkinson et al. 2005). CAFA analysis of the SHAPE capillary traces was performed with CAFA and Shapefinder as previously described (Mitra et al. 2008; Vasa et al. 2008). The SHAPE data were normalized using standard outlier analysis (Vasa et al. 2008). ROC analysis is based on thresholding the SHAPE data (DeLong et al. 1985), a Welch's t-test was used to determine significance over a minimum of three repeats per experiment. Raw SHAPE data were made available using the SNRNASM standard (Rocca-Serra et al. 2011) and the data are available at http://snrnasm.bio.unc.edu.
Heatmap graphs
With the exception of the G4A mutant having only three data sets, a minimum of four data sets per mutant were used to perform statistical tests. The Shapiro-Wilkes test comparing the data set to the normal distribution with the same mean and standard deviation was performed individually for each position to confirm the appropriateness of using a Welch's t-test. The Welch's t-test for mean difference was performed at each nucleotide with the null hypothesis being that that the mutant and WT data sets were equal. The degree of mean difference for positions with P-values <0.001 are color-coded red when the mutant has the higher value and blue when the WT has a higher value with the depth of the color indicating the degree of difference in 0.15 steps and a maximum value of 0.6. Even in our most extreme cases, <8% of the positions were non-normal with a P-value <0.05. All statistical tests were performed using version 2.8.1 of R (R development Core Team).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work is supported by grants R00 GM079953 (NIGMS) and R21 MH087336 (NIMH) to A.L. L.D.N. received support from NIH R37-GM39422.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029900.111.
REFERENCES
- Addou-Klouche L, Adélaïde J, Finetti P, Cervera N, Ferrari A, Bekhouche I, Sircoulomb F, Sotiriou C, Viens P, Moulessehoul S, et al. 2010. Loss, mutation and deregulation of L3MBTL4 in breast cancers. Mol Cancer 9: 213 doi: 10.1186/1476-4598-9-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam H, Williams TW, Dumas KJ, Guo C, Yoshina S, Mitani S, Hu PJ 2010. EAK-7 controls development and life span by regulating nuclear DAF-16/FoxO activity. Cell Metab 12: 30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badorrek CS, Weeks KM 2005. RNA flexibility in the dimerization domain of a gamma retrovirus. Nat Chem Biol 1: 104–111 [DOI] [PubMed] [Google Scholar]
- Badorrek CS, Gherghe CM, Weeks KM 2006. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization. Proc Natl Acad Sci 103: 13640–13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin EJ, Dupuis J, Larson MG, Lunetta KL, Booth SL, Govindaraju DR, Kathiresan S, Keaney JF Jr, Keyes MJ, Lin JP, et al. 2007. Genome-wide association with select biomarker traits in the Framingham Heart Study. BMC Med Genet Suppl 1 8: S11 doi: 10.1186/1471-2350-8-S1-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Tafer H, Muckstein U, Flamm C, Stadler PF, Hofacker IL 2006. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol Biol 1: 3 doi: 10.1186/1748-7188-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffa MB, Maret D, Hamill JD, Bastajian N, Crainich P, Jenny NS, Tang Z, Macy EM, Tracy RP, Franco RF, et al. 2008. Effect of single nucleotide polymorphisms on expression of the gene encoding thrombin-activatable fibrinolysis inhibitor: A functional analysis. Blood 111: 183–189 [DOI] [PubMed] [Google Scholar]
- Bolz H, Ramírez A, von Brederlow B, Kubisch C 2001. Characterization of ADAMTS14, a novel member of the ADAMTS metalloproteinase family. Biochim Biophys Acta 1522: 221–225 [DOI] [PubMed] [Google Scholar]
- Burdon KP, Sharma S, Chen CS, Dimasi DP, Mackey DA, Craig JE 2007. A novel deletion in the FTL gene causes hereditary hyperferritinemia cataract syndrome (HHCS) by alteration of the transcription start site. Hum Mutat 28: 742 doi: 10.1002/humu.9501 [DOI] [PubMed] [Google Scholar]
- Chappell S, Daly L, Morgan K, Guetta Baranes T, Roca J, Rabinovich R, Millar A, Donnelly SC, Keatings V, MacNee W, et al. 2006. Cryptic haplotypes of SERPINA1 confer susceptibility to chronic obstructive pulmonary disease. Hum Mutat 27: 103–109 [DOI] [PubMed] [Google Scholar]
- Chu VB, Herschlag D 2008. Unwinding RNA's secrets: Advances in the biology, physics, and modeling of complex RNAs. Curr Opin Struct Biol 18: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremonesi L, Fumagalli A, Soriani N, Ferrari M, Levi S, Belloli S, Ruggeri G, Arosio P 2001. Double-gradient denaturing gradient gel electrophoresis assay for identification of L-ferritin iron-responsive element mutations responsible for hereditary hyperferritinemia-cataract syndrome: Identification of the new mutation C14G. Clin Chem 47: 491–497 [PubMed] [Google Scholar]
- Cremonesi L, Paroni R, Foglieni B, Galbiati S, Fermo I, Soriani N, Belloli S, Ruggeri G, Biasiotto G, Cazzola M, et al. 2003. Scanning mutations of the 5′UTR regulatory sequence of L-ferritin by denaturing high-performance liquid chromatography: Identification of new mutations. Br J Haematol 121: 173–179 [DOI] [PubMed] [Google Scholar]
- DeLong ER, Vernon WB, Bollinger RR 1985. Sensitivity and specificity of a monitoring test. Biometrics 41: 947–958 [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE 2004. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res 32: W135–W141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE 2005. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 11: 1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Ferre-D'Amare AR 2006. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure 14: 1459–1468 [DOI] [PubMed] [Google Scholar]
- Ferrari F, Foglieni B, Arosio P, Camaschella C, Daraio F, Levi S, Garcia Erce JA, Beaumont C, Cazzola M, Ferrari M, et al. 2006. Microelectronic DNA chip for hereditary hyperferritinemia cataract syndrome, a model for large-scale analysis of disorders of iron metabolism. Hum Mutat 27: 201–208 [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Ikeda S, Arai T, Tanaka N, Kumasaka T, Ishii T, Kida K, Muramatsu M, Sawabe M 2010. Polymorphism of SERPINE2 gene is associated with pulmonary emphysema in consecutive autopsy cases. BMC Med Genet 11: 159 doi: 10.1186/1471-2350-11-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barcelo MM, Tang CS, Ngan ES, Lui VC, Chen Y, So MT, Leon TY, Miao XP, Shum CK, Liu FQ, et al. 2009. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung's disease. Proc Natl Acad Sci 106: 2694–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT 2008. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol 15: 177–182 [DOI] [PubMed] [Google Scholar]
- Glinskii AB, Ma J, Ma S, Grant D, Lim CU, Sell S, Glinsky GV 2009. Identification of intergenic trans-regulatory RNAs containing a disease-linked SNP sequence and targeting cell cycle progression/differentiation pathways in multiple common human disorders. Cell Cycle 8: 3925–3942 [DOI] [PubMed] [Google Scholar]
- Goertsches R, Comabella M, Navarro A, Perkal H, Montalban X 2005. Genetic association between polymorphisms in the ADAMTS14 gene and multiple sclerosis. J Neuroimmunol 164: 140–147 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A 2005. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res 33: D121–D124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM 2006. From ribosome to riboswitch: Control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol 41: 329–338 [DOI] [PubMed] [Google Scholar]
- Gutell RR, Lee JC, Cannone JJ 2002. The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct Biol 12: 301–310 [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010a. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. 2010b. PAR-CliP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp 2010: 2034 doi: 10.3791/2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A 2010. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet 6: e1001074 doi: 10.1371/journal.pgen.1001074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havgaard JH, Lyngso RB, Stormo GD, Gorodkin J 2005. Pairwise local structural alignment of RNA sequences with sequence similarity less than 40%. Bioinformatics 21: 1815–1824 [DOI] [PubMed] [Google Scholar]
- Hu RJ, Watanabe M, Bennett V 1992. Characterization of human brain cDNA encoding the general isoform of β-spectrin. J Biol Chem 267: 18715–18722 [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF 2005. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol 25: 6879–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilty SJ, Bosse Y, Cormier C, Endam LM, Desrosiers MY 2010. Polymorphisms in the SERPINA1 (Alpha-1-Antitrypsin) gene are associated with severe chronic rhinosinusitis unresponsive to medical therapy. Am J Rhinol Allergy 24: e4–e9 [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525–528 [DOI] [PubMed] [Google Scholar]
- Laederach A, Chan JM, Schwartzman A, Willgohs E, Altman RB 2007. Coplanar and coaxial orientations of RNA bases and helices. RNA 13: 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW 1998. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development 125: 4077–4088 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E 2002. Motif prediction in ribosomal RNAs Lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie 84: 961–973 [DOI] [PubMed] [Google Scholar]
- Lescoute A, Leontis NB, Massire C, Westhof E 2005. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res 33: 2395–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Pastinen T 2011. The study of eQTL variations by RNA-seq: From SNPs to phenotypes. Trends Genet 27: 72–79 [DOI] [PubMed] [Google Scholar]
- Mathews DH 2004. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 10: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill JS 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 29: 1105–1119 [DOI] [PubMed] [Google Scholar]
- Michel F, Westhof E 1990. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol 216: 585–610 [DOI] [PubMed] [Google Scholar]
- Mitra S, Shcherbakova IV, Altman RB, Brenowitz M, Laederach A 2008. High-throughput single-nucleotide structural mapping by capillary automated footprinting analysis. Nucleic Acids Res 36: e63 doi: 10.1093/nar/gkn267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJ 2002. The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics 79: 539–546 [DOI] [PubMed] [Google Scholar]
- Morton NE 2008. Into the post-HapMap era. Adv Genet 60: 727–742 [DOI] [PubMed] [Google Scholar]
- Naruse TK, Kawata H, Anzai T, Takashige N, Kagiya M, Nose Y, Nabeya N, Isshiki G, Tatsumi N, Inoko H 1999. Limited polymorphism in the HLA-DOA gene. Tissue Antigens 53: 359–365 [DOI] [PubMed] [Google Scholar]
- Peng CK, Buldyrev SV, Goldberger AL, Havlin S, Mantegna RN, Simons M, Stanley HE 1995. Statistical properties of DNA sequences. Physica A 221: 180–192 [DOI] [PubMed] [Google Scholar]
- Pesole G, Liuni S, Grillo G, Ippedico M, Larizza A, Makalowski W, Saccone C 1999. UTRdb: A specialized database of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 27: 188–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzolesi MG, Platzer P, Waite KA, Eng C 2008. Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in Cowden syndrome. Am J Hum Genet 82: 1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrier S, Martin JS, Davis-Neulander L, Beauregard A, Laederach A 2010. Evaluation of the information content of RNA structure mapping data for secondary structure prediction. RNA 16: 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana TM 2007. Illuminating the silence: Understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 8: 23–36 [DOI] [PubMed] [Google Scholar]
- Regulski EE, Breaker RR 2008. In-line probing analysis of riboswitches. Methods Mol Biol 419: 53–67 [DOI] [PubMed] [Google Scholar]
- Rocca-Serra P, Bellaousov S, Birmingham A, Chen C, Cordero P, Das R, Davis-Neulander L, Duncan CD, Halvorsen M, Knight R, et al. 2011. Sharing and archiving nucleic acid structure mapping data. RNA 17: 1204–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez S, Eiriksdottir G, Gaunt TR, Harris TB, Launer LJ, Gudnason V, Day IN 2010. IGF2BP1, IGF2BP2 and IGF2BP3 genotype, haplotype and genetic model studies in metabolic syndrome traits and diabetes. Growth Horm IGF Res 20: 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R, Herschlag D 2001. Probing the folding landscape of the Tetrahymena ribozyme: Commitment to form the native conformation is late in the folding pathway. J Mol Biol 308: 839–851 [DOI] [PubMed] [Google Scholar]
- Sanchez M, Galy B, Dandekar T, Bengert P, Vainshtein Y, Stolte J, Muckenthaler MU, Hentze MW 2006. Iron regulation and the cell cycle: Identification of an iron-responsive element in the 3′-untranslated region of human cell division cycle 14A mRNA by a refined microarray-based screening strategy. J Biol Chem 281: 22865–22874 [DOI] [PubMed] [Google Scholar]
- Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y 1999. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J 18: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman SK, Deras ML, Woodson SA, Scaringe SA, Cech TR 2000. Multiple folding pathways for the P4–P6 RNA domain. Biochemistry 39: 12465–12475 [DOI] [PubMed] [Google Scholar]
- Stoddard CD, Gilbert SD, Batey RT 2008. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA 14: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. 2007. Population genomics of human gene expression. Nat Genet 39: 1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes MT, Levitt M 2005. Describing RNA structure by libraries of clustered nucleotide doublets. J Mol Biol 351: 26–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teresi RE, Zbuk KM, Pezzolesi MG, Waite KA, Eng C 2007. Cowden syndrome–affected patients with PTEN promoter mutations demonstrate abnormal protein translation. Am J Hum Genet 81: 756–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny B, Blau N 2006. Mutations in the BH4-metabolizing genes GTP cyclohydrolase I, 6-pyruvoyl-tetrahydropterin synthase, sepiapterin reductase, carbinolamine-4a-dehydratase, and dihydropteridine reductase. Hum Mutat 27: 870–878 [DOI] [PubMed] [Google Scholar]
- Tseng HH, Weinberg Z, Gore J, Breaker RR, Ruzzo WL 2009. Finding non-coding RNAs through genome-scale clustering. J Bioinform Comput Biol 7: 373–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BJ, Breaker RR 2005. Riboswitches as versatile gene control elements. Curr Opin Struct Biol 15: 342–348 [DOI] [PubMed] [Google Scholar]
- Tyagi R, Mathews DH 2007. Predicting helical coaxial stacking in RNA multibranch loops. RNA 13: 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC 2008. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 14: 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldispuhl J, Clote P 2007. Computing the partition function and sampling for saturated secondary structures of RNA, with respect to the Turner energy model. J Comput Biol 14: 190–215 [DOI] [PubMed] [Google Scholar]
- Waldsich C 2008. Dissecting RNA folding by nucleotide analog interference mapping (NAIM). Nat Protoc 3: 811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Lee ER, Morales DR, Lim J, Breaker RR 2008a. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell 29: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Barr I, Guo F, Lee C 2008b. Evidence of a novel RNA secondary structure in the coding region of HIV-1 pol gene. RNA 14: 2478–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikl TR, Palassini M, Dill KA 2004. Cooperativity in two-state protein folding kinetics. Protein Sci 13: 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. 2007. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 35: 4809–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2005. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J Am Chem Soc 127: 4659–4667 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96 doi: 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM 2009. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA 5: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wismar J 2001. Molecular characterization of h-l(3)mbt-like: A new member of the human mbt family. FEBS Lett 507: 119–121 [DOI] [PubMed] [Google Scholar]
- Woodson SA, Leontis NB 1998. Structure and dynamics of ribosomal RNA. Curr Opin Struct Biol 8: 294–300 [DOI] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD 2010. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol 191: 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Weinberg Z, Ruzzo WL 2006. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics 22: 445–452 [DOI] [PubMed] [Google Scholar]