Activation of RNase L endonuclease activity is part of the mammalian innate immune response to viral infection. The poliovirus RNA genome contains a sequence in its protein-coding region that can act as a competitive inhibitor (ciRNA) of RNase L, the first example of an RNA that directly inhibits the activity of an RNase. By using a combination of biophysical, mutational, and functional studies, the authors have mapped features of the three-dimensional architecture of the ciRNA in its unbound form. Local tight RNA–RNA backbone packing occurs in parts of the structure, but the fold appears to be less stable than many other tightly packed RNAs. This feature may allow the ciRNA to accommodate the translocation of ribosomes and polymerase across this multifunctional region of the viral RNA, but also functions as an RNase L inhibitor.
Keywords: RNA structure, poliovirus, kissing hairpins, RNase L, antiviral countermeasure
Abstract
Activation of RNase L endonuclease activity is part of the mammalian innate immune response to viral infection. The poliovirus RNA genome contains a sequence in its protein-coding region that can act as a competitive inhibitor of RNase L. Mutation, sequence, and functional analysis of this competitive inhibitor RNA (ciRNA) revealed that its activity depends on specific sequences, showed that a loop–loop hairpin interaction forms in the ciRNA, and suggested the presence of a loop E motif. These features lead to the hypothesis that the ciRNA's function is conferred in part by a specific three-dimensional folded RNA architecture. By using a combination of biophysical, mutational, and functional studies, we have mapped features of the three-dimensional architecture of the ciRNA in its unbound form. We show that the loop–loop interaction forms in the free ciRNA and affects the overall structure, perhaps forming long-range tertiary interactions with the loop E motif. Local tight RNA–RNA backbone packing occurs in parts of the structure, but the fold appears to be less stable than many other tightly packed RNAs. This feature may allow the ciRNA to accommodate the translocation of ribosomes and polymerase across this multifunctional region of the viral RNA but also to function as an RNase L inhibitor.
INTRODUCTION
Single-stranded positive sense RNA viruses such as hepatitis A and C, poliovirus, yellow fever, dengue, rubella, coronaviruses, and many others infect a significant percentage of the world's population and are substantial threats to human health (Katz 2006; Martin and Lemon 2006; Rezza 2007; Teixeira and Barreto 2009; Gardner and Ryman 2010). The genomes of single-stranded positive sense RNA viruses vary in size from ∼3–32 kilobases in length, with most <10 kilobases long, and thus are much smaller than the genomes of their hosts. Even the largest single-stranded positive sense RNA viral genomes generally encode less than a dozen genes (Gorbalenya et al. 2006). Thus, it is not surprising that many of these viruses have evolved to use structured regions of their RNA genomes to regulate molecular processes necessary for successful infection. In some cases, these structured RNAs fold into specific, stable three-dimensional structures that interact with components of the host cell, with other parts of the viral genome, or with virally encoded proteins. These interactions manipulate the host cell's machinery or control viral processes, influencing steps as diverse as translation, replication, encapsulation, localization, and others (e.g., Dreher 2009; Filbin and Kieft 2009; Giedroc and Cornish 2009; Steil and Barton 2009; Zoll et al. 2009). It is clear that structured RNA sequences can be a critical component of the replication strategy of these viruses. Therefore, a detailed understanding of these viral RNA structures and their functions is necessary for a complete understanding of single-stranded positive sense RNA viruses. In addition, understanding how structured viral RNAs manipulate cellular machinery reveals aspects of how the machinery functions, providing insight into basic cellular processes (Cullen 2009).
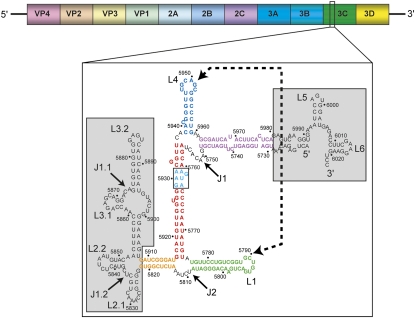
Poliovirus is a single-stranded positive sense RNA virus that is a useful model for understanding fundamental mechanisms of viral infection, including the use of RNA structure. Poliovirus is a human enterovirus in the order of Picornavirales with an RNA genome of ∼7.5 kilobases (Kitamura et al. 1980; Racaniello and Baltimore 1981), capped with the VPg protein (Flanegan et al. 1977; Lee et al. 1977; Nomoto et al. 1977), and packaged into an icosahedral nonenveloped capsid (Hogle et al. 1985). The virus' RNA genome utilizes several different regions of structured RNA to drive important events during infection. This includes an internal ribosome entry site (IRES) and a cloverleaf structure that are both found in the 5′ UTR (Pelletier and Sonenberg 1988; Barton et al. 2001), and the cis-acting replication element (CRE) in the protein coding region (Paul et al. 2000; Goodfellow et al. 2003; Steil and Barton 2009). Recently, another viral RNA sequence of novel function was discovered in the poliovirus genome, in the open reading frame (ORF) that encodes for the viral 3C protease (3CPro) (Fig. 1). This RNA sequence can competitively inhibit the endonuclease activity of RNase L and thus has been termed the poliovirus competitive inhibitor RNA (ciRNA) (Han et al. 2007; Townsend et al. 2008a). The ciRNA is able to operate both in cis and in trans to protect RNAs from cleavage by RNase L, the first identified example of an RNase-inhibiting RNA.
FIGURE 1.
Secondary structure of the ciRNA. At top is a diagram of the poliovirus RNA genome with open reading frames for the viral protein products shown. The ciRNA is found in the 3CPro coding region. (Below) The proposed secondary structure is shown. Regions in gray are not part of the “minimum” functional ciRNA. Various secondary structure elements are colored and labeled, and the kissing interaction is shown with a dashed line and arrows. The WT ciRNA used in this study did not contain any of the RNA in gray.
RNase L is expressed in most human tissues and is a critical downstream molecule in the interferon α and β antiviral pathways (Liang et al. 2006; Chakrabarti et al. 2011). Mutations to, and overexpression of, RNase L has been linked to diseases including prostate cancer, chronic fatigue syndrome, fibrosarcoma, benign prostatic hyperplasia, and others (Casey et al. 2002; Xiang et al. 2003; Nijs and De Meirleir 2005). Inactive RNase L exists as a monomer. Activation of 2′-5′ oligoadenylate synthetase (OAS) by double-stranded RNA (dsRNA) or interferon induces synthesis of 2′-5′ oligoadenylate (2-5A), which then binds to the ankyrin repeats of RNase L (for review, see Liang et al. 2006). 2-5A binding induces dimerization and activation of RNase L, which then cleaves RNA in the cell preferentially at 5′-UpA-3′ or 5′-UpU-3′ and leads to apoptosis. Remarkably, when the poliovirus ciRNA is included in in vitro reactions containing RNase L and activator 2-5A, RNA cleavage by the activated enzyme is inhibited (Han et al. 2007; Townsend et al. 2008a,b). The ciRNA also can block RNA cleavage by the constitutively active isolated endonuclease domain of RNase L (Dong and Silverman 1997), demonstrating that it interacts directly with that domain (Townsend et al. 2008a).
The ciRNA sequence which optimally inhibits RNase L cleavage contains 303 nucleotides (nt; 5720–6023 in the poliovirus 1 Mahoney Strain genome) of the 3CPro coding region of the viral genome (Han et al. 2007). Mutagenesis and truncation of the ciRNA combined with RNase L cleavage inhibition assays have identified that a sequence of ∼170 nt is sufficient to inhibit the enzyme. The proposed secondary structure of this RNA contains five helices and two three-way junctions (Fig. 1). In addition, this proposed secondary structure contains two RNA stem–loops (L1 and L4) that show base-complementarity in their apical loops and thus could form a long-range loop–loop “kissing” interaction. Indeed, functional studies of RNAs with mutations to these loops that disrupt or restore the putative kissing interaction indicate the presence of a kissing interaction (Han et al. 2007; Townsend et al. 2008b). In addition, there is a putative loop E motif in one helix of the proposed secondary structure; mutations to this element decrease the ability of the ciRNA to inhibit RNase L cleavage (Townsend et al. 2008b). The ciRNA sequence is more conserved than is the sequence of the poliovirus genome overall. The fact that the ciRNA sequence is maintained (even in wobble positions) suggests not only that this part of the genome encodes protein but that its RNase L inhibiting function contributes to the success of the virus.
The presence of functionally important secondary structure elements, long-range RNA–RNA interactions, and the high degree of conservation led us to hypothesize that the ciRNA adopts a specific and stably folded three-dimensional structure that is necessary for function. We used a combination of biochemical and biophysical studies to test this idea. Here, we show that the tertiary fold of the ciRNA includes long-range RNA–RNA interactions. Formation of these interactions appears to be important for the inhibitory function of the ciRNA. By using these data, we present a model for the global three-dimensional architecture of the ciRNA that can serve as the basis for additional hypotheses and experimentation.
RESULTS
Functional and structural roles of the kissing hairpins
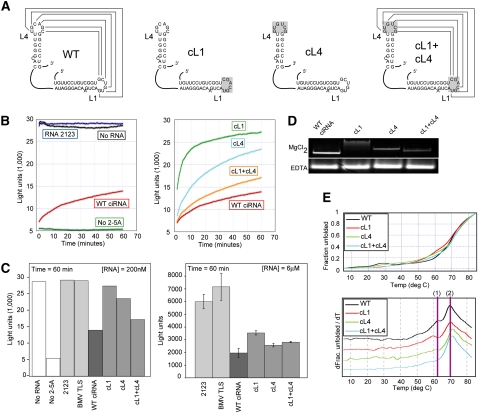
The ability of loops L1 and L4 of the ciRNA to kiss has not been explored directly using structural methods. We therefore generated a set of mutant ciRNAs in which all six of the nucleotides in each loop were mutated (Fig. 2A). These are mutant cL1 (the sequence of L1 replaced by the reverse sequence of L4), mutant cL4 (the sequence of L4 replaced by the reverse sequence of L1), and mutant cL1+cL4 (L1 and L4 reverse sequences are exchanged). RNAs cL1 and cL4 do not have the potential to form a kissing hairpin interaction, while in mutant cL1+cL4 the potential is restored by switching of the two loop sequences. Because our mutants differ from those used in previous studies (which contained mutations to just 3 of the 6 nt in each loop), we measured the ability of cL1, cL4, and cL1+cL4 to inhibit RNase L using the previously described fluorescence-based assay with a doubly labeled substrate RNA (Thakur et al. 2005; Townsend et al. 2008a,b). The substrate consists of an U-A rich oligonucleotide with a 6-carboxyfluorescein (6FAM) fluorophore and Black Hole Quencher-1 (BHQ1) attached to the 5′ and 3′ ends, respectively. Cleavage of the probe by 2-5A–activated RNase L spatially separates the fluorophores, and under this condition, 6FAM can be excited and the resulting emission detected. If the ciRNA inhibits the RNase L, the probe remains intact and the BHQ1 quenches the fluorescence from 6FAM. Thus, emission of light indicates cleavage by RNase L, and a lack of light indicates inhibition of RNase L activity.
FIGURE 2.
Function and structural role of the kissing hairpins. (A) Diagram of the mutants used to study the structural role of the kissing hairpin interaction. For clarity, only the L1 and L4 elements are shown. The loop(s) mutated in each RNA are shaded gray. (B, left) Time course of fluorescence from a representative in vitro RNase L cleavage assay. Controls with no added RNA, with a negative control RNA (2123), or without 2-5A activator are shown. The inhibition effect of WT ciRNA also is included. (Right) Comparison of WT, cL1, cL4, and cL1+cL4 mutant RNAs in the cleavage assay. All concentrations of RNA = 200 nM. (C, left) Bar graph depiction of the experiment shown in panel B. The amount of fluorescence at time = 60 min is shown for each experiment. An additional negative control RNA (BMV TLS, a structured RNA form the 3′ UTR of the brome mosaic virus genomic RNA) also is included. One representative experiment is shown. (Right) Bar graph of an in vitro cleavage assay done at elevated RNA concentrations (6 μM) showing how the mutants inhibit nearly as well as WT at this concentration of RNA. Data are three averaged experiments; error bars, 1 SD from the mean. (D) Native gels of WT ciRNA and the cL1, cL4, and cL1+cL4 mutants. The top gel contained 5 mM MgCl2; the bottom contained 2 mM EDTA. (E) Thermal denaturing curves of WT ciRNA and the three kissing loop mutants. At top is the raw data normalized to reflect fraction RNA folded as a function of temperature; at bottom is the first derivative of the data. The two transitions discussed in the text are indicated.
We assayed the ability of these ciRNAs to inhibit RNase L at two RNA concentrations: 200 nM and 6 μM. Figure 2, B and C, shows that in the absence of 2-5A, no RNase L–catalyzed cleavage is observed, while upon addition of 2-5A, robust cleavage is detected almost immediately. Addition of a negative control RNA of similar size (2123, an RNA sequence from the poliovirus genome) (Han et al. 2007) did not inhibit the RNase. However, addition of the wild-type (WT) ciRNA to the reaction results in inhibition. At concentration of RNA = 200 nM, both mutants cL1 and cL4 show a decreased ability to inhibit RNase L compared with the WT (Fig. 2B,C), consistent with the effect observed with other mutations to these loops (Han et al. 2007; Townsend et al. 2008b). cL1 showed the greatest loss of inhibition activity. The cL1+cL4 double mutant contains the restored potential to form a kissing interaction, and consistent with this, it inhibits RNase L cleavage to a greater degree than does cL1 or cL4 but not as well as does the WT ciRNA. This may indicate that the sequence within each of the loops, and not just their ability to form a kissing interaction, is important for full ciRNA function. At higher concentration of RNA, the negative control RNAs are still unable to inhibit the RNase L cleavage (Fig. 2C, right), but cL1 and cL4 are now fairly efficient inhibitors, although not quite as effective as the WT. This suggests that although the potential to form the kissing interaction contributes to the full inhibitor activity of the ciRNA, it is not absolutely essential at higher concentration of RNA. Overall, our data confirm the presence of a kissing interaction and show that our mutants can be used to study the implications of this interaction for the global structure of the ciRNA.
To explore the ability of the long-range kissing interaction to stabilize a specific three-dimensional ciRNA fold, we compared the global structures of the WT and loop mutants using nondenaturing (native) polyacrylamide gel electrophoresis, which is very sensitive to changes in the global conformation of an RNA. In general, more compact molecules migrate faster, and changes in the migration rate indicate changes in the overall structure of the RNA. We used a gel containing 5 mM MgCl2 to study the RNA in the folded state, and 2 mM EDTA to study the RNA in its unfolded state (although secondary structures may form). As shown in Figure 2D, the cL1 and cL4 mutations that disrupt the potential kissing interaction cause a retardation of the band in the MgCl2 gel, but there is no effect in EDTA. This suggests that these mutations do not radically alter the secondary structure of the RNA, but they alter the Mg2+-induced folded three-dimensional structure, resulting in a less compact structure relative to the WT RNA. The greatest retardation effect is observed for cL1, which was also the mutation with the lowest RNase L inhibition efficiency. cL4 shows less retardation than cL1, but its structure is clearly altered. Furthermore, the double-loop mutant cL1+cL4's migration rate is faster than either of the two single-loop mutants and is nearly as fast as the WT. This suggests that the cL1+cL4 mutant RNA has a structure that is more compact than either of the mutants with a disrupted kiss, consistent with formation of a restored kissing interaction. However, the migration rate is not completely restored to the position of the WT ciRNA, corresponding to the inability of this mutant to fully inhibit the RNase to the same degree as WT ciRNA. We interpret these results as indicating that although the kissing interaction forms and stabilizes the native fold of the molecule, the specific sequences within L1 and L4 are also structurally and functionally important.
Thermal denaturation of kissing hairpin mutants
To examine the structural role of the L1 and L4 loop sequences and kissing interaction in more detail, we thermally denatured WT, cL1, cL4, and cL1+cL4 RNAs and monitored their unfolding by ultraviolet (UV) absorbance as a function of temperature (“melting curve” analysis). If L1 and L4 are involved in stabilization of higher-order structure, we expect to see changes in the denaturation curves as a result of loop mutation. The melting curve data were normalized to reflect the fraction RNA unfolded. To identify specific transitions more readily, the first derivative of each curve also was calculated and plotted (Fig. 2E). Examination of the WT RNA plot shows two major transitions: The first (1) is ∼62°C and the second (2) is ∼70°C. In mutant cL1, transition 1 shifts to a lower temperature by ∼2°C, suggesting that the structural feature associated with this denaturation event is destabilized. In mutant cL4, transition 1 effectively disappears, and transition 2 increases by ∼1°C–2°C. This is a subtle change but is reproducible. The denaturation behavior of cL4 suggests that the structural feature that gives rise to transition 1 is destabilized to a greater degree than in cL1, but other structural features are slightly stabilized. When the double mutant cL1+cL4 is examined, we see that it does not show a melting profile that matches WT despite the fact that the kissing interaction has been restored. In fact, the melting profile of cL1+cL4 more closely resembles that of cL4 in that the transition 1 is not evident and transition 2 is shifted to a slightly higher temperature relative to the WT.
The identity of the structural features that give rise to each transition in the denaturation profiles cannot be determined based on the data presented here. One possibility for the increased temperature of transition 2 is that the cL4 mutation stabilizes a secondary structure in that loop, and this transition reflects the denaturation of this loop. However, these data also suggest that the sequence in the loops is important for the structure of the RNA beyond formation of a kissing interaction. If formation of the kissing interaction were the sole structural role of L1 and L4, we would expect cL1+cL4 to behave as WT; our data show it does not in both the native gel and denaturation experiments. Our results suggest that the loops are thermodynamically coupled to more than one transition in the denaturation curve. This leads to the hypothesis that the loops not only kiss but form long-range interactions with other parts of the ciRNA structure.
Functional and structural roles of the putative loop E motif
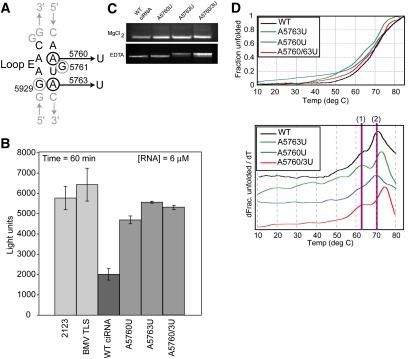
Previous studies reported that mutation of two G bases in the ciRNA's putative loop E motif (G5761 and G5929) decreased the ability of the ciRNA to inhibit RNase, indicating this region is functionally important (Townsend et al. 2008b). To investigate the structural and functional role of the putative loop E motif directly, we made three mutants designed to specifically prevent formation of a proper loop E structure (Fig. 3A). Specifically, A5760 was replaced by a U (A5760U) to disrupt formation of an A•A parallel base-pair and potentially allow alternate base-pairs to form (such as a U-A). Likewise, A5763 was replaced by a U (A5763U) to disrupt formation of an A•G sheared base-pair but allowing an alternate base-pair to form (a U•G wobble). Mutant A5760/3U contains both mutations.
FIGURE 3.
Function and structural role of the putative loop E motif. (A) Secondary structure diagram of the putative loop E motif, with noncanonical base-pairs shown. Nucleotides previously mutated and studied are circled in gray; mutations we generated and studied are circled in black, and the mutation is indicated. (B) Bar graph of in vitro cleavage assays with the loop E mutants, WT, and control RNAs. Graph depicts the fluorescence at time = 60 min, averaged from three experiments. Error bars, 1 SD from the mean. (C) Native gels of WT ciRNA and three loop E mutants. The top gel contained 5 mM MgCl2; the bottom contained 2 mM EDTA. (D) Thermal denaturing curves of WT ciRNA and the three loop E mutants. At top is the raw data normalized to reflect fraction RNA folded as a function of temperature; at bottom is the first derivative of the data. The two transitions discussed in the text are indicated.
The three loop E mutants were tested in the RNase L inhibition assay, again with WT ciRNA as a positive control and the 2123 RNA as negative controls (Fig. 3B). We included an additional negative control, the tRNA-like sequence from the 3′ end of the brome mosaic virus (BMV), which is known to form a compact structure with emerging stem–loops (Hammond et al. 2009). With all three loop E mutations, we observe nearly complete loss of the ciRNA's ability to inhibit RNase L, even at elevated concentrations of RNA. These data confirm that sequences with the loop E motif are critical for function of the ciRNA, more so than the kissing interaction.
To determine if these loop E mutations alter the global structure of the ciRNA, we used native gel analysis identical to that done for the L1 and L4 mutants (Fig. 3C). We observed no differences in the migration rate of WT, A5760U, A5763U, and A5760/3U on the Mg2+-containing gel, and only a slight difference was seen on the EDTA-containing native gels for mutants A5763 and A5760/3U. The most straightforward interpretation of this result is that local changes in the secondary structure occurred when A5763 was mutated (as expected), and these are seen as small changes in migration in the EDTA-containing gel. However, when the molecule adopts its Mg2+-induced global fold, none of these mutations alter the overall architecture of the RNA to a measurable degree or to the degree that the cL1 and cL4 mutants do. Therefore, the putative loop E motif is critical for function but is not a major determinant of the global three-dimensional fold of the ciRNA. Furthermore, the fact that these loop E mutants are deficient in inhibiting RNase L but appear to have a global fold similar to WT suggests that the overall global fold is not the sole determinant of ciRNA function but that important local structures and specific sequences must be present within the fold.
Thermal denaturation of the putative loop E motif mutants
To determine if the loop E mutations could affect these same denaturation transitions as did the kissing loop mutations, we subjected A5760U, A5763U, and A5760/3U to the same analysis (Fig. 3D). Mutant A5760U showed little or no changes in the two observed transitions, with perhaps only a slight change in transition 1. In contrast, both A5763U and A5760/3U showed an increase in the temperature at which transition 2 occurs by several degrees, while the location of transition 1 did not change. As with the L1 and L4 mutants, we did not attempt to derive thermodynamic parameters from these data, and we cannot assign them to specific structural features in the ciRNA. It possible that these mutations increase the stability of the secondary structure in the putative loop E elements by causing this element to become more like a canonical helix, accounting for the increase in the temperature of translation 2. Nonetheless, these data show that mutations to the putative loop E motif can alter the same transitions that are altered by mutations to the kissing hairpin loops.
Probing the backbone solvent accessibility of the folded ciRNA
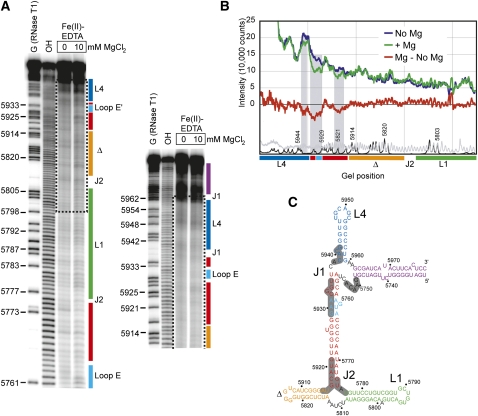
To determine if long-range RNA–RNA interactions in the ciRNA induce a tightly packed higher-order fold similar to other functional RNAs, we used hydroxyl radical (•OH) probing on the “minimal” ciRNA element in the presence and absence of Mg2+ (Fig. 4). •OH probing uses solvent-borne radicals to cleave the RNA backbone wherever it is solvent exposed, independent of sequence and secondary structure. Reduced •OH cleavage is a result of solvent protection due to tight RNA backbone packing within a higher-order folded structure. •OH probing of the ciRNA revealed localized regions of subtle protection that are just visible to the naked eye on the gel (Fig. 4A). We quantified and normalized the probing data (Fig. 4B) and mapped reproducibly protected areas onto the proposed secondary structure of the ciRNA (Fig. 4C). The protections are found in and adjacent to the two three-way junctions, and on one side of the putative loop E motif, suggesting these areas are involved in tight packing within the Mg2+-induced global fold. However, these protections are somewhat more subtle than protections observed for some other folded RNAs. For example, probing studies of Dicistroviridae intergenic region IRESs and the BMV TLS (RNAs of similar size to the ciRNA) conducted in our laboratory exhibit an average of a 1.5-fold decrease in cleavage in folded regions upon the addition of magnesium (Costantino and Kieft 2005; Hammond et al. 2009), while the ciRNA's protection average is 1.2, with only one part of the RNA showing a protection of 1.5-fold. The more subtle protections observed in the ciRNA probing might indicate that although the folded ciRNA has some tightly packed regions, particularly around its junction elements, the overall global fold is less tightly packed than these comparable RNAs or its structure might be “breathing” to a degree that the solvent-borne radicals can partially access the RNA backbone even in folded regions.
FIGURE 4.
Hydroxyl radical (•OH) probing of the WT ciRNA. (A) Representative gel from a •OH probing experiment (using Fe(II)-EDTA) of the WT ciRNA shown in panel C. Two different gel run times are shown. From left to right, the lanes contain an RNase T1 sequencing reaction, a base hydrolysis reaction, the •OH probing in the absence of Mg2+, and the probing in the presence of 10 mM Mg2+. Secondary structural regions are indicated to the right of the gel in colors that match those in Figure 1 and panel C. The dashed box indicates the portion of the gels shown in panel B. (B) Quantification of the portion of the gel shown in panel A with a dashed box, normalized and graphed to show intensity of radiation as a function of location in the gel. The trace in the absence of Mg2+ is blue and in the presence of 10 mM Mg2+ is in green, and the difference is in red. Shaded gray regions indicate regions that show subtle protection in the presence of Mg2+. The trace of the RNase T1 and base hydrolysis lanes are shown in black and gray at the bottom of the graph, respectively. Positions of secondary structure elements are shown under the graph in colors that match panel C. (C) Secondary structure diagram of the WT ciRNA used in the probing, color-coded as in Figure 1. Regions that showed reproducible protection from solvent in three independent probing experiments are shaded dark gray.
Small-angle X-ray scattering of the ciRNA
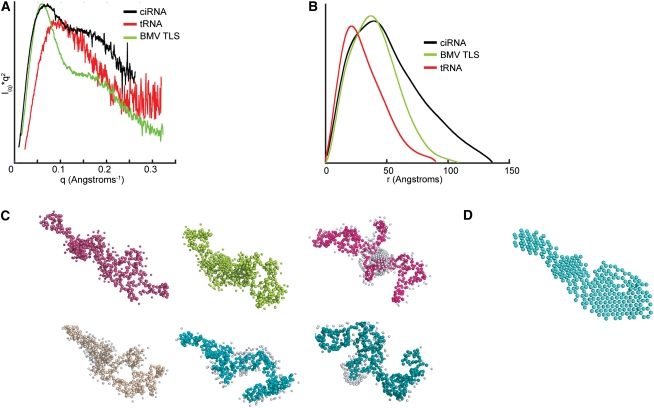
Small-angle-X-ray scattering (SAXS) can be used to query the overall shape of folded RNAs, therefore we applied it to the WT minimum ciRNA. Chemically pure ciRNA was folded in the presence of Mg2+, made conformationally pure by size-exclusion chromatography, and subjected to SAXS analysis in a manner identical to that previously described for tRNA and tRNA mimics (Hammond et al. 2009, 2010). The data were used to construct a Kratky plot, which can be used to determine the degree to which a macromolecule is folded into a compact structure; an inverted parabolic Kratky plot indicates a folded molecule. For comparison, we include previously published data of the BMV TLS and phenylalanine yeast tRNA (Fig. 5A; Hammond et al. 2009). SAXS of the BMV TLS yields a Kratky plot with an inverted parabola that indicates a compact fold. Likewise, SAXS of tRNA yields a plot that indicates a folded molecule but one that is less compact than the BMV TLS as indicated by the “tail” at high q-values. Compared with these, the Kratky plot of the ciRNA shows features consistent with a folded structure but also contains a “tail” that does not drop to lower values, and thus the molecule is best described as partially folded, comprising a structure that is not as compact as those adopted by the BMV TLS or tRNA.
FIGURE 5.
Ab initio shape reconstructions of WT ciRNA from SAXS. (A) Kratky plots constructed from SAXS data of the ciRNA (black), phenyalanine yeast tRNA (red), and the BMV TLS RNA (green). The tRNA and BMV data and curves were previously published (Hammond et al. 2009). (B) Plots of the pairwise distribution functions for the same RNAs shown in panel A. Colors match panel A. (C) Six independent representative shape reconstructions. (D) Average of 10 independent reconstructions.
The scattering data were used to obtain information about the size and shape of the ciRNA in the presence of Mg2+. The measured radius of gyration of the ciRNA is 40.9 Å compared with 31.9 Å for the BMV TLS, although the two RNAs are nearly identical in terms of length (169 nt for BMV TLS and 168 for the ciRNA RNA used in SAXS). This indicates that the ciRNA is a more extended molecule overall. The plot of the pairwise distribution function shows that the ciRNA has more scattering pairs that are distant from each other, indicated by the great P(r) at higher r-values (Fig. 5B). The calculated Dmax value that is derived from this curve is 136 Å for the ciRNA (compared with 108 Å for the BMV TLS), which again indicates that the ciRNA is a less compact structure. However, the maxima of the ciRNA P(r) curve is shifted toward larger distances compared with tRNA, similar to what is seen for the BMV TLS. The shift to ∼40 Å in the BMV TLS curve has been interpreted as evidence of helices packing together; thus, there is evidence in the P(r) curve of higher-order packing within the folded ciRNA but also for a structure that is more extended than is the BMV TLS. Taken together, the SAXS data are consistent with the Fe(II)-EDTA data, indicating a molecule that is partially folded or may be sampling both compact and extended conformations and thus its compact fold is relatively unstable in the unbound form.
SAXS data can be used to generate ab initio reconstructions that are consistent with the scattering data. These reconstructions lack detail but can be used to gain a general ideal of the overall shape of the molecule that gives rise to the scattering. This method has been used successfully in our laboratory to examine the structures of tRNA-mimicking RNAs (Hammond et al. 2009, 2010), and others have used it to examine other folded RNAs, such as riboswitches and catalytic RNAs (Garst et al. 2008; Lipfert et al. 2008; Stoddard et al. 2010). Figure 5C contains six representative members of the family of reconstructions of the ciRNA that were generated and are consistent with the scattering data. Comparison of these solutions shows that there is no one single structure that is consistent with the scattering data, but rather several structures that could give rise to the observed data. However, all of the SAXS structures show a fairly elongated molecule with extensions that are either packed together or extended away from one another to some degree. The rough similarity of these solutions led us to average the structures (Fig. 5D). Although this average structure cannot be viewed as any more “correct” than any of the individual structures, it provides a means to observe the average solution consistent with the scattering data. This average structure is largely elongated but contains a thicker bulge region in the center that is consistent with RNA helices packed side by side.
DISCUSSION
In order to be a competitive inhibitor, the ciRNA must bind to RNase L, compete for binding of the substrate to the active site, and not present a viable substrate. We hypothesize that this ability is due to a specific ciRNA structure that contacts the enzyme using the same surfaces used to bind substrate RNA, but the structure of the ciRNA prevents it from being cleaved. Here, we tested the first part of this hypothesis, that being the presence of a ciRNA higher-order fold. Our data show that the ciRNA folds into a higher-order structure and that this structure is important for function. Furthermore, the fold of the RNA involves secondary structural elements that come together in three-dimensional space. Long-range tertiary interactions give rise to a structure that includes a kissing hairpin interaction and a loop E motif that must work together to drive function, perhaps through long-range interactions that involve all these structural elements.
Our data structurally confirm the presence of a loop–loop interaction within the ciRNA fold that is not essential for ciRNA inhibition of RNase L but increases this function. In contrast, mutations to the loop E motif are extremely detrimental to the function of the inhibitor (Townsend et al. 2008b). Loop E motifs are known to interact with proteins and to be involved in RNA–RNA interactions (Hausner et al. 1987; Moazed et al. 1988; Leontis and Westhof 1998; Lancaster et al. 2008). Both types of interactions may play a role in the ciRNA's function. The kissing loops likely act to stabilize the active structure of the RNA, an effect reported in other folded RNAs to include the Neospora VS ribozyme (Rastogi et al. 1996; DeAbreu et al. 2011) and the aptamer domain of the lysine riboswitch (Blouin and Lafontaine 2007; Garst et al. 2008; Blouin et al. 2011). In both of these structured RNAs, a loop–loop kissing interaction contributes to stabilization of the active conformation and thus to the structure-dependent biological activity of the RNA. Our structural and functional data support the idea that role of the kissing interaction is to shift the conformational equilibrium of the ciRNA toward the active inhibiting structure that is critically dependent on the loop E motif, while not being absolutely essential for the RNase L inhibition activity.
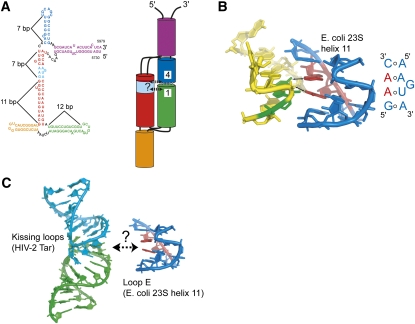
To help guide further experiments aimed at understanding the structure-function relationship of the ciRNA, we constructed a model for the overall architecture of the folded but unbound ciRNA that reconciles available data (Fig. 6A). In constructing this model, we did not initially make use of the SAXS data due to the fact that multiple solutions were obtained. Rather, we based the model on the behavior of the mutant RNAs on native gels, •OH probing, denaturation curves, and the propensity of RNA helices to coaxially stack in junctions. In this model, the global architecture of the RNA comprises two helical stacks packed side by side, a common feature of folded RNAs. The model structure contains two three-way junctions that we are depicting with stacked helices. For J2 (the junction from which stem–loop L1 emerges), we modeled this using the proposed three-way junction rules of Lescoute and Westhof (2006). The orange helix will stack coaxially with the red helix owing to the fact that there no nucleotides are in the junction between these two helices. This configuration may then allow the green helix (which contains the L1 kissing loop) to swing into position to pair with L4. For junction J1, we modeled the purple helix stacked with the blue helix (which contains the L1 kissing loop), but it seems equally plausible for the purple helix and red helix to stack. Either stacking arrangement is consistent with our data.
FIGURE 6.
Model of the global architecture of the ciRNA. (A) At left is the proposed secondary structure of the WT ciRNA with elements color coded and labeled. The lengths of several helices in the structure (in term of base-pairs) is indicated. To the right of the secondary structure diagram is a model of the global architecture of the ciRNA, with cylinders representing the helical regions, black lines indicating junction regions, base-pairs that form the kissing interactions shown with gray lines, and L1 and L4 labeled. Hypothetical interactions between the kissing loops and the loop E motif (cyan) are shown with dashed arrows and a question mark. (B) Interaction between the loop E motif found in helix 11 of 23S rRNA from E. coli (blue and red) and other parts of the rRNA (yellow and green) (Zhang et al. 2009). The secondary structure of the loop E motif is shown alongside. Two adenosines in the loop E (red) make contact through the minor groove (arrows) to a G nucleotide (green). (C) At left is the NMR structure of a kissing loop interaction based on the HIV-2 Tar RNA that contains six paired bases in the loop (Chang and Tinoco 1997). At right is the loop E motif from panel B. The dashed arrow indicates a speculative interaction that could form between RNA motifs of this type.
Within the model, L1 and L4 form the kissing interaction, and counting the number of base-pairs in the relevant helices suggests that the kissing loops and the loop E are spatially proximal in the folded structure (Fig. 6A). By extension, they could interact directly, and this could be how they work together to allow RNase L inhibition. To our knowledge, a direct tertiary interaction between kissing loops and a loop E motif has not been reported in a structured RNA (the lysine riboswitch contains both elements, but they do not directly interact) (Garst et al. 2008), but one can speculate how such an interaction could occur. The loop E sequence of ciRNA is identical to a loop E sequence in helix 11 of Escherichia coli 23S ribosomal RNA to include an adjacent C-A base-pair (Leontis and Westhof 1998). Within the ribosome, this loop E forms tertiary contacts to the minor groove of a distorted adjacent helix (Fig. 6B), suggesting the ciRNA loop E is capable of a similar interaction. To illustrate one possibility, Figure 6C shows the NMR structure of a kissing loop interaction derived from the HIV-2 Tar loop that contains six paired nucleotides in the “kiss” (Chang and Tinoco 1997), matching the six predicted to form in the ciRNA. The kissing interface provides a relatively open minor groove that could be recognized by the loop E motif; distortion of the minor groove by the kiss might facilitate this interaction. It is important to note that this mode of interaction between the kissing loops and the loop E motif is speculative, but it does provide one possibility for how this putative tertiary contact could occur.
Although the global model shown in Figure 6A (including the possible tertiary contact) remains to be fully tested, it is consistent with data presented here. First, if the predicted interactions between L1 and L4 and loop E form, mutation of each of the loops (kissing and loop E) would give rise to different behavior on native gels, as we observe. Also, this model predicts that there should be solvent-protected regions of RNA backbone consistent with close helical packing and structuring of the junction regions, and that parts of the putative loop E motif should be protected from solvent due to their involvement in interactions with the kissing loops. Our probing data exhibit these effects. This model also explains how the kissing interactions could enhance the inhibitor function of the ciRNA, which seems to be critically dependent on the loop E motif. Finally, although the SAXS data were not initially considered when building our model, the model is similar to the average solution obtained from the shape reconstruction. Complete testing of this hypothetical model will likely require higher-resolution structural studies.
One interesting aspect of the poliovirus ciRNA's structure is the fact that the compact folded state appears to be less stable than are folded RNAs of similar size, an observation based on the •OH probing and the SAXS analysis. While this effect may be due to the elimination of the portions of the RNA removed from the full-length sequence (Fig. 1), this is unlikely in that removal of some or all of this nonessential RNA had little effect on the ability of the ciRNA to inhibit RNase L (Han et al. 2007; Townsend et al. 2008a). If these sequences substantially stabilized the active fold of the ciRNA, we would expect a greater functional effect upon their removal. The lower stability of the ciRNAs is intriguing because the poliovirus ciRNA is an example of a viral RNA sequence that can perform several roles: It can bind to and inhibit RNase L; it is a template for protein synthesis; and it is a template for RNA replication. The ciRNA is therefore multifunctional and responds to several distinct evolutionary pressures. The first is to maintain the correct protein-encoding sequence; the second is to be readily unwound by the ribosome and the replication machinery; and the third is to fold into a structure capable of competitively inhibiting RNase L. These functions of the ciRNA depend on very different molecular conformations. When acting as a substrate for translation or replication, the ciRNA sequence must unfold to allow passage of the ribosome or polymerase, and when acting as an inhibitor, the RNA must adopt a specific compact fold. We speculate that the nature of the ciRNA's fold allows it to adopt both structures readily.
To date, the poliovirus ciRNA is the only reported example of an RNA that competitively inhibits an RNase. However, other types of structured and functional RNA sequences that manipulate the host's cellular components are found in the coding sequences of diverse viruses. The ability of RNA viruses to mutate rapidly and also to share genetic material supports the idea that new RNA sequences capable of folding into specific structures and performing novel functions may be hidden in the coding regions of many viruses.
MATERIALS AND METHODS
DNA/plasmid construction
Plasmids containing various ciRNA sequences were generated using standard molecular biology and cloning methods. Briefly, DNA inserts containing the ciRNA sequence between two ribozymes (hepatitis δ ribozyme at the 3′ end and a hammerhead ribozyme at the 5′ end) and under control of a T7 promoter were generated by PCR, using a template plasmid provided by David Barton (University of Colorado Denver School of Medicine). The resultant plasmids were inserted into the EcoR1/BamH1 sites of the pUC19 vector, amplified in E. coli strain DH5α, and verified by sequencing. Plasmids encoding mutant RNAs were generated by QuickChange mutagenesis (Stratagene).
RNA transcription and purification
DNA templates for in vitro transcription by T7 polymerase were generated by PCR from these plasmids. Reaction conditions were as follows: 40 mM Tris-HCl (pH 8.1), 1 mM spermidine, 5 mM dithiothreitol, 0.01% Triton X-100, 60 mM MgCl2, 8 mM each ribonucleotide triphosphate (pH adjusted to 8.0), and 0.04 mg/mL T7 RNA polymerase. The reactions were incubated overnight at 37°C and then were placed for 5 min at 65°C to promote complete cleavage of the ribozymes. The reaction was then precipitated by adding 4 volumes of ice-cold 100% ethanol and the RNA collected by centrifugation. The resulting RNA was purified by gel electrophoresis on a 10% denaturing polyacrylamide gel in 1× TBE buffer. The band containing the RNA was visualized by UV-shadowing, excised from the gel, crushed, and allowed to elute into RNase-free water overnight, with shaking at 4°C. The gel pieces were filtered out of the solution using a 0.22-μm filter (Nalgene). The RNA then was concentrated using a centrifugal filter with a 10,000 molecular weight cutoff (Amicon). Purity was assessed by polyacrylamide gel electrophoresis.
Thermal denaturation (melting) analysis
RNA sample dilutions for melting curves were calculated so as to have an OD260 absorbance of between 0.8 and 1 by using the equation: Dilution = ([RNA in mg/mL] × (1000))/((40) × (1 to 0.8)). It was found that an RNA final concentration of 0.25 μg/μL gave a good signal. Using that concentration, 50 μg of RNA was added to a tube along with ddH2O to 194 μL. RNA was heated to 85°C and then allowed to cool to room temperature for 10 min. Four microliters of 100 mM MgCl2 and 2 μL 1 M NaCacodylate (pH 7.5) were added to each sample. Samples were then placed in a 4°C cold block until they were ready to run. To remove any aggregates, the samples were spun down in Eppendorf spin filters at 14,000 rpm for 2 min in a benchtop microfuge. Samples were loaded into a 10-mm cuvette and loaded into a Jasco Circular Dichroism machine. The temperature was increased at 0.5°C/min with a measurement range of 4°C–85°C. Measurements were taken every second. Jasco software provided the first derivatives of the data, after which all data were exported to either Excel or KaleidaGraph.
Radioactive end labeling
First, 100–150 μg of RNA was added to 1× PNK buffer (70 mM Tris-HCl at pH 7.6, 10 mM MgCl2, 5 mM DTT), 20 units PNK enzyme, and 1 μL concentrated P32 containing γ-labeled ATP (MP Biomedicals) in a 10 μL solution. The reaction was then incubated for 1 h at 37°C, followed by desalting using a BioRad Micro Bio-Spin 30 Chromatography Column. Resulting solution was then added to equal volume RNA loading buffer and purified on 10% denaturing polyacrylamide gel and eluted in buffer (500 mM NaOAc at pH 5.2, 0.1% SDS) using the crush and soak method. Samples were then ethanol precipitated and resuspended in DEPC-treated water.
•OH probing
•OH probing and generation of RNase T1 and hydrolysis ladders were performed as previously described (Kieft et al. 2007). Briefly, to anneal the RNA the sample was heated in DEPC-treated water for 1 min at 85°C and cooled for 20 min at room temperature to reach folding equilibrium and then placed in 30 mM HEPES-NaOH (pH 7.5), 0 or 10 mM MgCl2, and 0.2 mg/mL total yeast tRNA. The RNA was then moved to 4°C and allowed to cool for 15 min. Cleavage reactions were then conducted for 2 min at 4°C with the addition of 1 μL of Fe(II)-EDTA solution (20 mM sodium EDTA, 10 mM ferrous ammonium sulfate), 1 μL of 50 mM sodium ascorbate, and 1 μL of 1% hydrogen peroxide were added sequentially to the reaction tube, and the reaction was incubated for 2 min at 37°C. Reaction was quenched with 1 μL of 100 mM thiourea and 10 μL of 9 M urea/1 mM EDTA and resolved on 10% sequencing denaturing polyacrylamide gels (1× TBE, 7 M urea, 29:1 acrylamide:bis-acrylamide). Gels were dried and visualized with a phosphorimager. Quantitation of the data was performed using the program ImageQuant, and the data were analyzed using Excel and Kaleidagraph.
Nondenaturing (native) gels
RNA was heated in DEPC-treated water for 1 min at 85°C and cooled for 20 min at room temperature and then added to nondenaturing RNA loading dye (2× TH Buffer, 0.5 mg/mL bromophenol blue and xylene cyanol, and 10% glycerol) with either MgCl2 or EDTA. Reactions were run on 10% polyacrylamide gels in 1× TRIS-HEPES buffer with either MgCl2 or EDTA.
FRET-based RNase L activity assay
A dual-labeled RNA FRET probe was used as a substrate to monitor RNase L activity (Thakur et al. 2005). The substrate was derived from a 36-nt intergenic sequence of respiratory syncytial virus (RSV; UUAUCAAAUUCUUAUUUGCCCCAUUUUUUUGGUUUA) with a fluorophore (6FAM or 6-carboxyfluorescein) at the 5′ terminus and a BHQ1 at the 3′ terminus (Sigma-Proligo). Reactions (50 μL) contained 25 mM Tris-HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 50 μM ATP, 7 mM β-mercaptoethanol, 100 nM FRET probe, 100 nM or 6 μM ciRNA, and full-length RNase L and 2–5A (20 nM each). Reactions were incubated at room temperature in a black 96-well microtiter round bottom plate. Fluorescence was activated using 485-nm wavelength excitation; 535-nm wavelength emissions were detected at 1-min intervals for 60 min in a Bio-Tek Synergy HT Multi-Detection Microplate Reader, according to the method described by Townsend et al. (2008a,b).
Small-angle X-ray scattering
SAXS was performed as previously described (Hammond et al. 2009, 2010). Briefly, purified RNA was refolded by heating for 3 min to 85°C, then for 5 min at 55°C, and then for 2 min at 37°C. The RNA was then diluted into a buffer containing 20 mM MPS (pH 6.5), 50 mM KCl, 7.6 mM MgCl2 and purified using a Superose 6 PC 3.2 gel-filtration column on an Ettan LC liquid chromatography system configured with a Rheodyne in-line vacuum degasser and a 0.01-μM Millipore solvent filter. Data were collected on samples at ∼5–10 mg/mL RNA concentration with exposures of 10, 30, and 100 sec. Scattering data collected from matching buffer were subtracted to generate the ciRNA's SAXS scattering profile. A dilution series (2/3 dilution) was used to check for interparticle interference. The 100-sec exposure was analyzed using PRIMUS; the GNOM was used to estimate Dmax. Output from GNOM was used in GASBOR to generate ab initio shape reconstructions (Svergun et al. 2001). The number of residues was set at nucleotides × 2.6.
ACKNOWLEDGMENTS
We thank Dr. Robert Rambo (Lawrence Berkeley National Lab) for SAXS data collection and advice. We also thank Drs. David Barton and Hannah Townsend (UC Denver) for many useful discussions and sharing of reagents and protocols, Dr. Robert Silverman (Cleveland Clinic) for sharing purified RNase L, and the members of the Kieft Lab for many useful discussions. This manuscript was critically read by R. Batey, B. Silverman, D. Barton, J. Whitehurst, D. Costantino, and T. Colussi, and we thank them for their comments. J.S.K. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030007.111.
REFERENCES
- Barton DJ, O'Donnell BJ, Flanegan JB 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J 20: 1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin S, Lafontaine DA 2007. A loop–loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13: 1256–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin S, Chinnappan R, Lafontaine DA 2011. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic Acids Res 39: 3373–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey G, Neville PJ, Plummer SJ, Xiang Y, Krumroy LM, Klein EA, Catalona WJ, Nupponen N, Carpten JD, Trent JM, et al. 2002. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet 32: 581–583 [DOI] [PubMed] [Google Scholar]
- Chakrabarti A, Jha BK, Silverman RH 2011. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res 31: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KY, Tinoco I Jr 1997. The structure of an RNA “kissing” hairpin complex of the HIV TAR hairpin loop and its complement. J Mol Biol 269: 52–66 [DOI] [PubMed] [Google Scholar]
- Costantino D, Kieft JS 2005. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA 11: 332–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR 2009. Viral RNAs: lessons from the enemy. Cell 136: 592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAbreu DM, Olive JE, Collins RA 2011. Additional roles of a peripheral loop–loop interaction in the Neurospora VS ribozyme. Nucleic Acids Res 39: 6223–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Silverman RH 1997. A bipartite model of 2-5A-dependent RNase L. J Biol Chem 272: 22236–22242 [DOI] [PubMed] [Google Scholar]
- Dreher TW 2009. Role of tRNA-like structures in controlling plant virus replication. Virus Res 139: 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin ME, Kieft JS 2009. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 19: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan JB, Petterson RF, Ambros V, Hewlett NJ, Baltimore D 1977. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc Natl Acad Sci 74: 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner CL, Ryman KD 2010. Yellow fever: a reemerging threat. Clin Lab Med 30: 237–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT 2008. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 283: 22347–22351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc DP, Cornish PV 2009. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res 139: 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow IG, Kerrigan D, Evans DJ 2003. Structure and function analysis of the poliovirus cis-acting replication element (CRE). RNA 9: 124–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117: 17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JA, Rambo RP, Filbin ME, Kieft JS 2009. Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA 15: 294–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JA, Rambo RP, Kieft JS 2010. Multi-domain packing in the aminoacylatable 3′ end of a plant viral RNA. J Mol Biol 399: 450–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JQ, Townsend HL, Jha BK, Paranjape JM, Silverman RH, Barton DJ 2007. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J Virol 81: 5561–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner TP, Atmadja J, Nierhaus KH 1987. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie 69: 911–923 [DOI] [PubMed] [Google Scholar]
- Hogle JM, Chow M, Filman DJ 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229: 1358–1365 [DOI] [PubMed] [Google Scholar]
- Katz SL 2006. Polio: new challenges in 2006. J Clin Virol 36: 163–165 [DOI] [PubMed] [Google Scholar]
- Kieft JS, Costantino DA, Filbin ME, Hammond J, Pfingsten JS 2007. Structural methods for studying IRES function. Methods Enzymol 430: 333–371 [DOI] [PubMed] [Google Scholar]
- Kitamura N, Adler C, Wimmer E 1980. Structure and expression of the picornavirus genome. Ann N Y Acad Sci 354: 183–201 [DOI] [PubMed] [Google Scholar]
- Lancaster L, Lambert NJ, Maklan EJ, Horan LH, Noller HF 2008. The sarcin–ricin loop of 23S rRNA is essential for assembly of the functional core of the 50S ribosomal subunit. RNA 14: 1999–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YF, Nomoto A, Detjen BM, Wimmer E 1977. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci 74: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 1998. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J Mol Biol 283: 571–583 [DOI] [PubMed] [Google Scholar]
- Lescoute A, Westhof E 2006. Topology of three-way junctions in folded RNAs. RNA 12: 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SL, Quirk D, Zhou A 2006. RNase L: its biological roles and regulation. IUBMB Life 58: 508–514 [DOI] [PubMed] [Google Scholar]
- Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DM 2008. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure 16: 1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Lemon SM 2006. Hepatitis A virus: from discovery to vaccines. Hepatology 43: S164–S172 [DOI] [PubMed] [Google Scholar]
- Moazed D, Robertson JM, Noller HF 1988. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334: 362–364 [DOI] [PubMed] [Google Scholar]
- Nijs J, De Meirleir K 2005. Impairments of the 2-5A synthetase/RNase L pathway in chronic fatigue syndrome. In Vivo 19: 1013–1021 [PubMed] [Google Scholar]
- Nomoto A, Kitamura N, Golini F, Wimmer E 1977. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc Natl Acad Sci 74: 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol 74: 10359–10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325 [DOI] [PubMed] [Google Scholar]
- Racaniello VR, Baltimore D 1981. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci 78: 4887–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi T, Beattie TL, Olive JE, Collins RA 1996. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J 15: 2820–2825 [PMC free article] [PubMed] [Google Scholar]
- Rezza G 2007. Prevention and control of emerging infections: a challenge for the 3rd millennium. New Microbiol 30: 358–361 [PubMed] [Google Scholar]
- Steil BP, Barton DJ 2009. Cis-active RNA elements (CREs) and picornavirus RNA replication. Virus Res 139: 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT 2010. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH 2001. Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MG, Barreto ML 2009. Diagnosis and management of dengue. BMJ 339: b4338 doi: 10.1136/bmj.b4338 [DOI] [PubMed] [Google Scholar]
- Thakur CS, Xu Z, Wang Z, Novince Z, Silverman RH 2005. A convenient and sensitive fluorescence resonance energy transfer assay for RNase L and 2′,5′ oligoadenylates. Methods Mol Med 116: 103–113 [DOI] [PubMed] [Google Scholar]
- Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ 2008a. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA 14: 1026–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend HL, Jha BK, Silverman RH, Barton DJ 2008b. A putative loop E motif and an H-H kissing loop interaction are conserved and functional features in a group C enterovirus RNA that inhibits ribonuclease L. RNA Biol 5: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Z, Murakami J, Plummer S, Klein EA, Carpten JD, Trent JM, Isaacs WB, Casey G, Silverman RH 2003. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res 63: 6795–6801 [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, Cate JH 2009. Structures of the ribosome in intermediate states of ratcheting. Science 325: 1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoll J, Heus HA, van Kuppeveld FJ, Melchers WJ 2009. The structure–function relationship of the enterovirus 3′-UTR. Virus Res 139: 209–216 [DOI] [PubMed] [Google Scholar]