Abstract
In this study, we have used glucagon as a model system for analyzing amyloid fibrillogenesis by hydrogen exchange MALDI mass spectrometry (HXMS). The hydrogen exchange mass spectrometry data correlated well with the traditional method based on Thioflavin T fluorescence and provided quantitative information by measuring the fibrillating molecules directly. The hydrogen exchange mass spectrometry data collected during fibrillogenesis revealed that glucagon fibrillation was a two component system showing an on/off type of interaction where only monomeric and fibrils were present without any substantial amount of intermediate species. This was evident by the extensive deuteration of the monomer and protection of the entire 29 residue glucagon peptide upon fibrillation.. The method complements the traditional procedures and has the potential to provide new information with respect to the nature of transient species, the structure of the growing fibrils and the mechanism of formation.
Keywords: hydrogen exchange, MALDI, glucagon, amyloid fibrils
1. Introduction
Several proteins are known to cause diseases by forming amyloid fibril and deposition including, amyloid-β protein in Alzheimer’s disease, α-synuclein related to Parkinson’s disease, TGFBip related to cornea dystrophies and Islet amyloid polypeptide in type 2 diabetes [1–4].
In all cases, amyloids are formed by the stacking of individual protein molecules through intermolecular β-sheet formation, leading to a characteristic cross-β structure where the β-strands are perpendicular to the axis of the resulting fibril. Individual fibrils, each consisting of a single contiguous β-sheet, may stabilize each other by lateral association. These fibrils are often stable due to a combination of backbone hydrogen bonding between the β-strands and specific side-chain interactions [5, 6]. In an attempt to prevent or even dissolve amyloid fibril formation, extensive work has been carried out to understand the structure and formation of these fibrils.
Amyloid fibrils are often characterized by a unique combination of variable characteristics, including Thioflavin T (ThT) staining, Circular Dichroism (CD) spectrum fingerprints, thermostability, heat capacity (that is, the extent to which the fibrils can bind water), fiber diffraction patterns, small angle x-ray scattering, solid state NMR and morphology [1]. Most of these methods are either invasive and/or requires relatively high amount of material. As a consequence of the regularity and stability of the amyloid inter-strand hydrogen bonds, hydrogen exchange is strongly reduced in regions involved in amyloid formation compared to more dynamic areas of the protein. Thus, HXMS provides potentially residue-specific information about the extent of amyloid structure and requires very modest amounts of material typically 5 μg compared to the mg amounts required for more traditional methods mentioned above.
MALDI and ESI based HXMS have been used for more than a decade [7, 8] to study amyloid fibrils including, Aβ protein [9, 10], prion protein [11] and α-synuclein [12]. However, these studies were focused on the comparison of the monomeric forms to the mature forms in an attempt to identify highly protected areas in the fibril in order to understand more about the molecular structures. In the present study, MALDI based HXMS is used as a supplementary method to follow the fibrillogenesis; the process by which the fibrils are formed.
Glucagon was used as the model system, a 29 residue peptide hormone that function as a insulin antagonist and is used for the treatment of hypoglycaemia [13]. Glucagon is under physiological conditions present at low concentrations where it remains monomeric. However, in vitro at low pH (<pH 4) it readily fibrillates in a reproducible manner and has been extensively used as a model system for studies of amyloid fibril formation [14–20]. Notably, the peptide is able to form a number of fibrils differing in secondary, tertiary or quaternary structures depending on the conditions used during fibril formation [14, 16, 18, 19]. In this study we have used hydrogen exchange mass spectrometry to analyze the fibrillogenesis of glucagon fibrils formed at pH 2.5. The data revealed a two-component system showing an on/off type of interaction where only monomers and fibrils were present without any substantial amount of intermediate species.
2. Experimentals
2.1 Materials
Pharmaceutical grade glucagon (GlucaGen®, Novo Nordisk A/S) was a kind gift from Novo Nordisk A/S (Bagsværd, Denmark). D2O (99.9%) was from Euriso-top and Immobilized pepsin was from Thermo Scientific, PlusOne Urea was from GE healthcare, and ThT was from Sigma-Aldrich. Glucagon concentration was determined spectrophotometrically at 280 nm using the theoretical molar extinction coefficients E = 8250 M−1 cm−1 (GPMAW software, lighthouse data). ThT concentration was measured based on the extinction coefficient of 36000 M−1 cm−1 at 412 nm.
2.2 Fibril formation
Monomeric material was generated by filtering glucagon (0.2 μm micro spin filter, Lida Manufacturing Corp) dissolved in 50 mM glycine, pH 2.5. The concentration was adjusted to 2 mg/ml before the fibrillation was allowed to proceed by subjecting the monomeric glucagon to mechanical agitation at 25°C using a Vortex mixer set at 900 rpm. During the course of the fibrillation process aliquots were removed for HXMS and ThT analyses. For the analyses of isolated fibrils the fibrillated material was collected by centrifugation at 10.000×g for 10 minutes and washed 3 consecutive times in 50 mM glycine pH 2.5.
2.3 Thioflavin T analysis
During fibrillation samples were removed for ThT analysis. The fibrillating samples were diluted 10 times in 45 μM ThT, 50 mM glycine pH 2.5 and measured in 96 well black polystyrene microtiter plates (NUNC) using a plate reader (FLUOstar omega from BMG Labtech) capable of measuring fluorescence at 450 nm (excitation) and 485 nm (emission). All values plotted were based on an average of three independent measurements.
2.4 Circular Dichroism (CD) spectroscopy
All the CD spectra were recorded on a Jasco-810 CD spectrophotometer. A 4 mg/ml glucagon fibril solution was diluted 24 times in both 50 mM Glycine (pH 2.5) as well as in 20 mM Tris-HCl buffer (pH 7.4) and then used for far-UV measurements. Before measurements, the usually viscous fibril solution was sonicated briefly in a sonicator to reduce light scattering effects. Far-UV measurements were made between 250 and 200 nm with a step resolution of 0.2 nm using a 1 mm path length quartz cuvette. The scanning speed was set at 50 nm/min and the band width was 0.2 nm. Each far-UV was an average of three measurements, taken under identical conditions and corrected for buffer background signal.
2.5 Hydrogen exchange
The Glucagon samples (2 mg/ml) were deuterated by diluting the sample 24 times in 99% D2O containing 20 mM Tris-HCl pH 7.4 (uncorrected for the isotopic effect on pH glass electrodes) resulting in a glucagon concentration of 83 μg/ml. For the analyses of monomeric or fibrillated glucagon the samples were incubated in D2O for 30 sec, 1, 5, 10 and 15 min. Samples removed during fibrillation were incubated in D2O for 5 minutes. The exchange was quenched by adding ice-cold 1 M glycine pH 2.5 to a final concentration of 100 mM. All samples were made in triplicates.
2.6 Dissolving of fibrils and pepsin digestion
Monomeric glucagon was generated by dissociating the fibrils in 50 mM Glycine pH 2.4, containing 4 M Urea. For analysis of full-length glucagon 10 μl were micro-purified using StageTips (Proxeon) and analyzed by MALDI-TOF MS (see section 2.8). A schematic outline of the protocol is shown in Figure 1. Solubilized sample containing 28 mg/ml glucagon was digested in the buffer described above by adding a slurry of pepsin immobilized on 6% cross-linked agarose beads (Thermo Scientific). The final composition of the buffer during pepsin digestion was 3 M Urea, 50 mM glycine pH 2.5 containing 12.5% (v/v) immobilized pepsin. The samples were digested for 5 minutes at 0°C before the immobilized pepsin was pelleted by centrifugation at 800xg. Aliquots of 10 μl supernatant were micro-purified using StageTips (Proxeon) and analyzed by MALDI-TOF MS using a Q-TOF Ultima Global mass spectrometer (Micromass/Waters Corp.) (see section 2.8). The peptic peptides were identified by MALDI-TOF MS/MS of unlabeled samples, The collected data were analyzed by using a combination of the GPMAW software package (lighthouse data) and the MASCOT search engine (Matrix science). Three of the peptides generated by pepsin corresponding to residue 1–21 (2461.13 Da), 10–21 (1500.73 Da) and 10–29 (2520.22 Da) were used during the hydrogen exchange experiments.
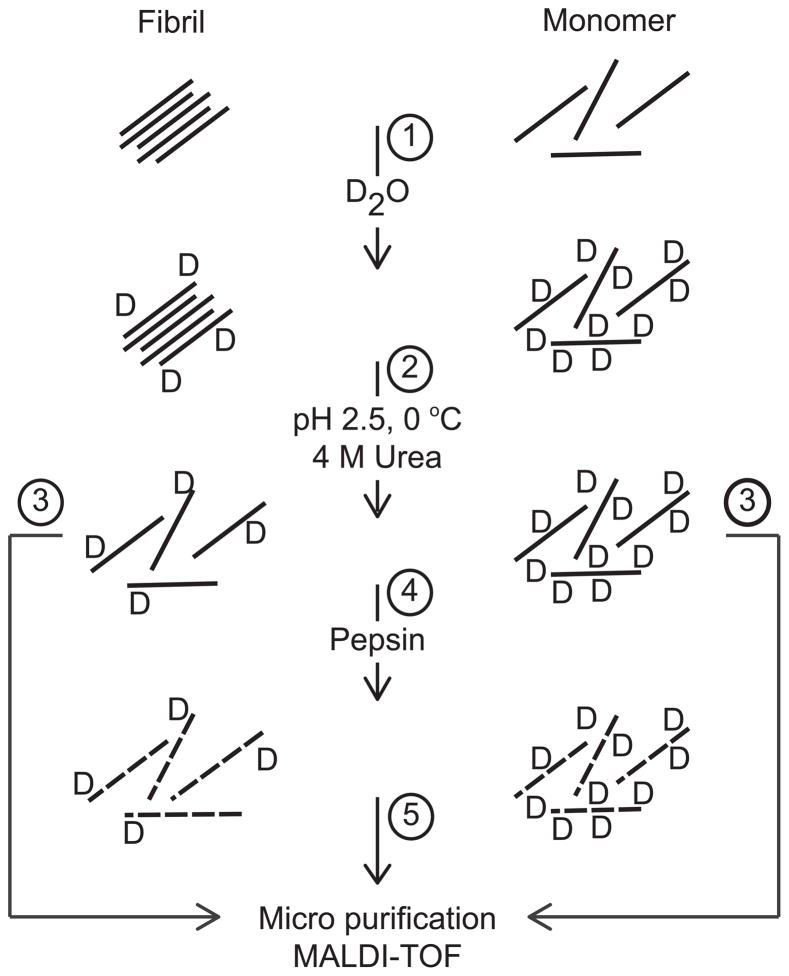
Figure 1. Schematic representation of the utilized protocol.
Monomeric, fibrillated or a mixture of monomeric and fibrillated glucagon were diluted 24 times in D2O (1). The hydrogen deuterium exchange reaction was quench by lowering pH and temperature and fibrillated species were dissolved by urea (2). Full-length glucagon was micro purified and analyzed by MALDI-TOF (3). For the analysis of glucagon fragments the urea treated samples were digested with immobilized pepsin (4). The pepsin generated peptides were micro purified prior to MALDI-TOF analysis (5).
2.7 Hydrogen exchange under denaturing conditions
Purified glucagon amyloid fibrils at 2 mg/ml were deuterated by diluting the sample 24 times in 20 mM Tris-HCl pH 7.4 buffer in 99% D2O (uncorrected for the isotopic effect on pH glass electrodes) containing an increasing concentration of urea ranging from 0.5–8 M. The diluted samples contained 83 μg/ml glucagon. The exchange/unfolding was continued for 5 min before quenching the reaction by adding ice-cold 1 M glycine pH 2.5 to a final concentration of 100 mM. The quenched samples were immediately diluted 3 times in ice-cold 8 M urea, 50 mM glycine pH 2.5 lowering the temperature to 0°C. Prior to MS analysis 10 μl of the samples were desalted by micro-purification using StageTips (Proxeon).
2.8 MALDI-TOF MS
The MALDI-MS sample preparation was based on the method described by J. G. Mandell et al. [21]. To obtain the optimal combination of signal intensity and minimize matrix mediated back-exchange 1.5 mg/ml α-cyano-4-hydroxycinnamic acid, pH 2.5 (adjusted using 2% trifluoroacetic acid, TFA) in 80% acetonitrile/0.1 % TFA was used. The samples (full-length and pepsin treated) were micro-purified using StageTips (Proxeon) equilibrated in chilled 0.1% TFA and spotted directly on to a cooled MALDI target. The target was quickly placed under moderate vacuum in a desiccator at 25°C causing the sample to dry within 2–3 minutes and immediately transferred to the mass spectrometer for analysis. In total, no more than 20 minutes was spent from quenching to the end of the analysis.
MS or MS/MS spectra were collected in a Q-TOF Ultima Global mass spectrometer (Micromass/Waters Corp.) calibrated using a polyethylene glycol mixture. External calibration of each MS spectrum was performed using Glu-fibrinopeptide B (m/z 1570.6774). The obtained MS or MS/MS data were processed using Masslynx 4.0 software package (Micromass).
2.9 HXMS data processing
For deuterium uptake, calculations were based on approximately 75 spectra recorded over 2 min. Deuterium uptake curves were made using HX-Express [22] employing a peak threshold of 2% of base peak intensity (BPI) and the width set at 50% of BPI. No corrections for back-exchange were made, since all measurements were obtained within similar time periods. The back-exchange was thus considered to be similar in all samples measured. Furthermore, the measurements recorded of aliquots removed during the fibrillation were based on the relative proportion of fibril and monomeric glucagon, eliminating the need for correction of back-exchange.
The relative amount of fibril was calculated from smoothed and centred mass spectra (intensity based on the peak area) as the total intensity of the 3482–3485 m/z region (fibril) relative the total intensity from the 3482–3485 m/z region (fibril) and the 3496–3500 m/z region (monomer). These two m/z intervals account for approximately 60% of total intensity of both the monomeric and fibrillated isotope cluster. The intensity from 3481–3485 m/z (fibril) and 3492–3500 m/z (monomer) was used to compensate for the drift in the isotopic cluster caused by the increasing urea concentration during the denaturation studies.
3. Results
3.1 Glucagon is significantly protected upon fibril formation
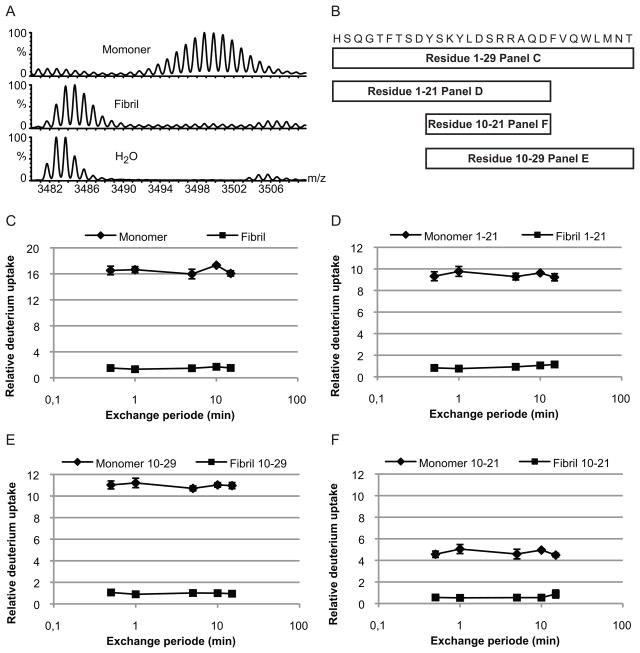
The initial HXMS characterization of the glucagon monomers and fibrils was carried out by increasing the exchange time from 30 sec to 15 min (Figure 2). The analyses were performed on both full-length glucagon (1–29) and three peptides obtained by pepsin cleavage (Figure 1 and 2b). The three peptides (residues 1–21, 10–21 and 10–29) covered the entire sequence. Maximum exchange was observed for the full-length monomer and the individual peptides after 30 seconds of hydrogen exchange with a relative deuterium uptake of approximately 16 out of the 28 available backbone amide hydrogens for full-length glucagon (Figure 2c–f). The lack of complete deuterium exchange is most likely caused by back exchange during sample preparation. Depending on the peptide sequence and specific sample preparation back-exchange, although constant, can be 15–50% during MALDI based HXMS analysis [7].
Figure 2. Relative Deuterium uptake in monomeric and fibrillated glucagon.
Monomeric (◆) and fibrillated (■) glucagon were diluted in D2O and incubated at 25°C for different periods, before the exchange was quenched by lowering the pH to 2.5 and the temperature to 0°C. The samples were either full-length glucagon or peptic peptides covering the entire sequence. (A) Mass spectra of monomeric (top) and fibrillated (middle) full-length glucagon after 10 min hydrogen exchange, with an unlabelled sample shown for comparison (bottom). (B) Sequence and schematic representations of the peptides generated by pepsin treatment of glucagon. The relative deuterium uptakes are shown for (C) full-length glucagon, (D) residue 1–21, (E) residue 10–29 and (F) residue 10–21. Error bars show the standard deviation (non visual error bar indicates standard deviation within the size of the marker).
Following an initial relative deuterium uptake of 1.5 completed after 30 sec, the fibrils did apparently not exchange further over time (figure 2c–f). After 15 minutes of incubation, the deuterium uptake persisted to be approximately 1.5. Even after 24 hrs of hydrogen exchange, the observed deuterium uptake for the fibril remained less than 2 deuterium (data not shown). Since the glucagon fibrils were formed at pH 2.5 and the deuteration at pH 7.4 CD were spectra were recorded of fibrils at both pH values. These data showed that the fibrils were not affected by the change in pH (data not shown). Our glucagon exchange data revealed that HXMS is able to differentiate between monomeric and fibrillated glucagon based on the difference between the isotopic cluster assigned to the fibril and the monomer (Figure 2a). Furthermore, the results indicated that glucagon obtain protection against hydrogen exchange along the entire peptide backbone upon fibrillation. However, it can not be ruled out that the protected population have incorporated several deuterons during isotopic labeling and lost those again as a result of back-exchange.
3.2 Fibril formation followed by HXMS
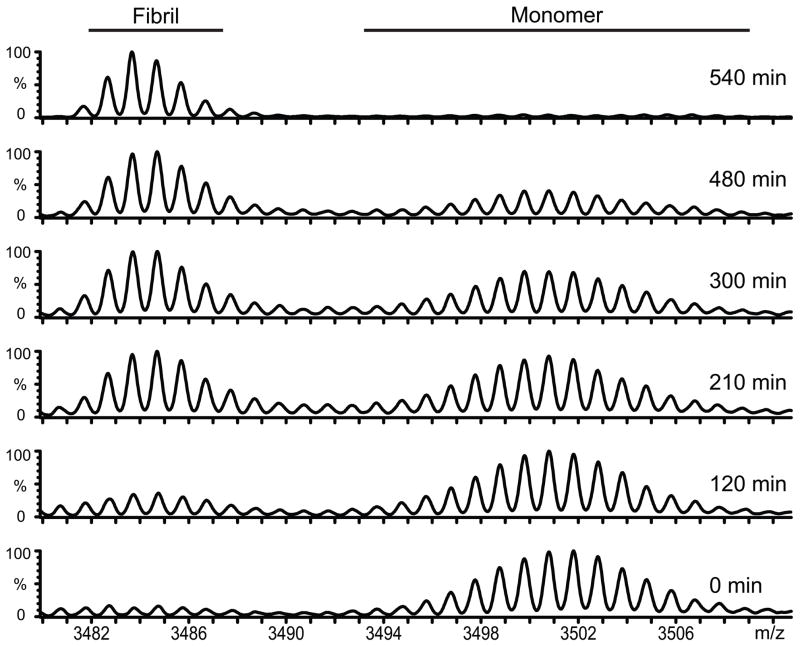
The unambiguous separation of the isotope cluster corresponding to fibrillated glucagon and monomeric glucagon facilitated the analysis of the fibrillation process by HXMS. This was initially demonstrated by removing aliquots during fibrillation and subjecting the sample to HXMS (Figure 3). Interestingly, we only observed isotope clusters corresponding to monomeric or fibrillated glucagon. Accordingly, if other types of glucagon species are present in significant amounts during fibrillation, they show the same type of hydrogen exchange as either monomer or fibril. Intermediate species containing prefibrillar assemblies which have not yet assumed the canonical cross-β structure would be expected to show a hydrogen exchange level midway between monomer and fibril. The simplest explanation for our observations is that glucagon only exists in two states, either monomeric or fibrillar. Glucagon also forms trimers at concentrations of 2 mg/ml and above [15], but our exchange conditions were performed at concentrations around 0.1 mg/ml where the monomer is the only significant species for the soluble species. Since we have a two-component system we can use our HXMS data to quantify the amount of fibril present in the sample to generate a HXMS based fibrillation curve, which can be compared to more traditional methods such as ThT fluorescence.
Figure 3. Fibrillation followed by HXMS.
During the first 12 hrs of glucagon fibrillation aliquots from the same fibrillating sample were withdraw for HXMS analysis. Representative spectra obtain at different time points during the fibrillation are shown. From the bottom its is aliquots removed from a fibrillating sample after 0, 120, 210, 300, 480 and 540 minutes.
3.3 ThT analysis and relative quantification of fibril formation
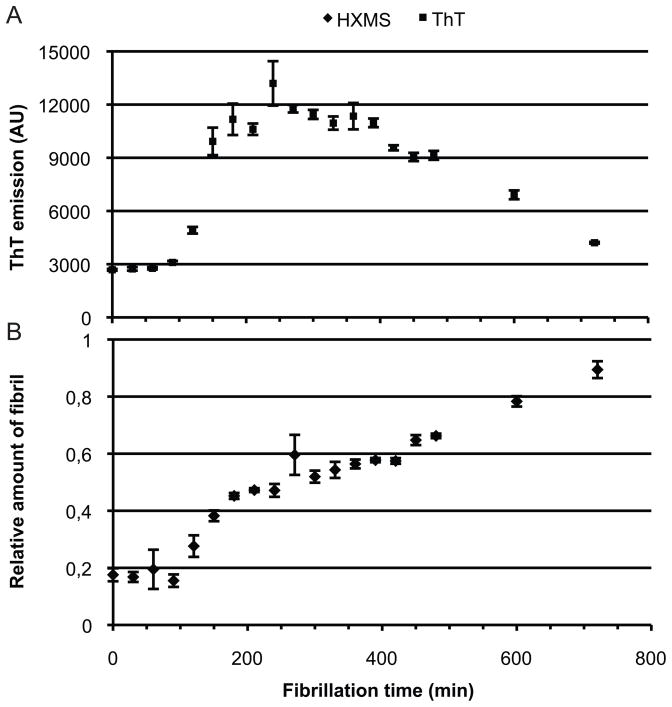
ThT fluorescence is one of the most widely used methods for following amyloid fibril formation [23]. The applicability of HXMS as a tool for monitoring the formation of amyloid fibrils was assessed by measuring ThT fluorescence and deuterium exchange of the same sample undergoing fibrillogenesis (Figure 4). ThT signal and hydrogen exchange were measured on aliquots from the same fibrillating sample. Samples were removed every 30 min followed by incubation in D2O for 5 min. To quantify the proportion of fibril present during the fibrillation process, we calculated the amount of fibril based on the intensity of the isotopic cluster corresponding to the monomer or the fibril (see Figure 3). By using the quantitative relation between the isotopic cluster corresponding to the fibrillated and the monomeric glucagon we were able to minimize the influence of the variations in back exchange observed during MALDI-MS.
Figure 4. Relative amount of fibrillated glucagon.
During the fibrillation process of glucagon aliquots from the same sample were withdraw for either ThT (A) or HXMS (B) analysis. The relative amount of fibrillated glucagon was calculated based on the intensity of the isotopic cluster corresponding to monomeric and fibrillated glucagon. All analysis were made in triplicates with 5 min hydrogen exchange. Error bars indicates the standard deviation.
There was a clear relationship between ThT signal and the amount of fibrillated glucagon observed during the fibrillation (Figure 4). Additionally, when the ThT signal has reached its maximum an indication of a plateau was observed in the HXMS analysis. This plateau indicates that a concentration threshold has been reached where the fibrillation is halted. Further fibrillation appears to dependent on a rearrangement of the fibril, which causes the ThT signal to decrease. The apparent continuous increase in the relative fibril amount can either be due to consumption of monomeric glucagon by the fibril or additional protection of fibril associated glucagon. These results demonstrate a correlation between the ThT assay and HXMS analysis of the fibrillation process suggesting that HXMS is a useful tool for analyzing the kinetics of amyloid fibril formation.
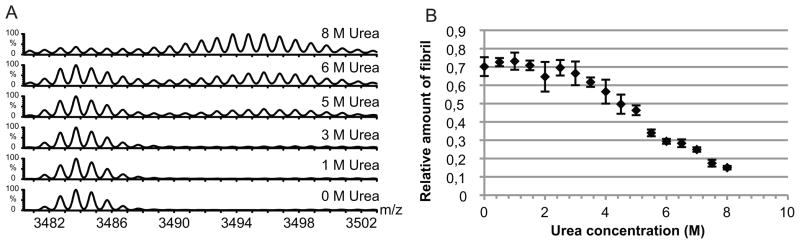
3.3 Urea induced denaturation of glucagon amyloid fibrils
If glucagon fibrils contain a fibrillating core more stable than the remaining fibril, an increase in hydrogen exchange outside the fibril core is likely to occur as the fibril gradually unravels by the addition of incremental concentrations of urea. Glucagon fibrils are known to dissociate at concentrations above 2 M urea [18]. This approach was applied to mature glucagon fibrils using a constant 5 min exchange rate (Figure 5b). Under these experimental conditions is was not possible to demonstrate a gradual increase in deuterium uptake, instead a direct transition from fibril to monomer was observed (Figure 5a). In these experiments, full-length glucagon showed an increased deuterium uptake at urea concentrations above 3.5 M. This increase was caused by the appearance of an isotopic cluster related to monomeric glucagon and not a gradual shift of the isotopic cluster (Figure 5a).
Figure 5. Urea induced dissociation of glucagon fibrils.
Centrifugation purified glucagon fibrils were analyzed with HXMS under increasing urea concentration. Spectra obtain at different urea concentrations are shown (A). (B) The relative amount of fibril during urea dependent unfolding of glucagon fibrils was calculated from the intensity of the isotopic cluster corresponding to monomeric and fibrillated glucagon. Error bars show the standard deviation.
The isotopic cluster corresponding to both fibril and monomeric glucagon did not show the same degree of deuterium uptake as compared to the analyses without urea. The relative deuterium uptake for monomeric glucagon in 8 M urea was approximately 10 compared to approximately 16 without urea. This lower deuterium uptake is not due to a structural change but is most likely caused by the lower relative concentration of deuterium caused by a high concentration of exchangeable hydrogens originating from urea.
The relative amount of fibril can be quantified similarly to the quantification during the fibrillation process (Figure 5b). When the relative amount of fibril is shown in relation to the urea concentration a clear dissociation above 3.5 M is observed. Furthermore, similarly to the fibrillation, the solvation involves two processes. From 0–3.5 M urea a small change of fibril is seen, which is followed by a rapid solvation above 3.5 M urea. While previous results have showed fibril dissociation above 2 M urea [18] our data revealed a continuous dissociation as the urea concentration was increased. This discrepancy is probably due to the short time period in which the unfolding was allowed to proceed before HXMS (5 min) as compared to the previous study measuring dissociation for 48h. Our data indicates that the entire glucagon polypeptide chain becomes unprotected/dissolved supporting the hypothesis of a two-component system during glucagon fibrillation as well as during fibril dissolution. Furthermore, the data supports the involvement of a rearrangement of the fibrils also seen with ThT analysis and HXMS during fibrillation.
We conclude that there is no localized fibril core with a higher protection level than in other regions of glucagon. All parts of the molecule are incorporated into the fibril structure like a magnet snapping in place and are released in the same way upon denaturation. In addition, the urea induced unfolding of the glucagon fibril, was proceeded by a gradual release of glucagon from the fibril rather than a destabilization of different parts of the peptides to giving rise to a more loose fibril.
4. Discussion
In this study we demonstrate the potential of MALDI-MS based HXMS for the analyses of fibril structure, formation and dissociation. MALDI and ESI based HXMS are well-established techniques for the study of amyloid fibrils [7, 8]. In the present study we attempt to evaluate both the fibrillogenesis and the dissolution process exploiting the large difference in exchange rate between the monomer and fibril. The data correlate well with the widely used ThT assay for studying fibril formation. The method described is based on MALDI-MS avoiding the complication of in-line pepsin digestion and a cooled HPLC chromatography set-up [7]. The protocol may produce some variations in back-exchange, however the large difference in the exchange rate of the soluble glucagon monomers and the amyloid fibrils compensate for this discrepancy. MALDI-MS has the additional advantage of being more tolerant towards insoluble samples such as amyloid fibrils and generates simple spectra as compared to ESI-MS, facilitating the data analysis.
Besides the ability to observe the fibrillogenesis (backbone protection), we also obtain structural information of the fibrils. The data show that when glucagon is incorporated into the fibril, it changes from a nearly unprotected to an almost fully protected form, indicating that the entire polypeptide is involved in the amyloid structure. A transition from a nearly unprotected form in the monomeric state to nearly fully protected in the fibrillated form have previously been observed for α-synuclein where a section of approximately 60 residues comprising the central part of the protein obtained a complete protection upon fibrillation [12]. The gain of full protection indicates a fibril structure in which the entire glucagon peptide participate in the β-conformation.
This high degree of protection cannot necessarily be used as evidence for amyloid fibrils formation, since alternative structures can lead to similarly protection. In this case, HXMS analysis of putative novel amyloid systems has to be combined with other methods such as CD spectroscopy, ThT fluorescence [23] or fiber diffraction [24], which will indicate the presence of β-sheet structure.
Amyloid fibrils are often detected by staining methods using either Congo red or ThT [25]. These dyes are excellent all-round dyes providing a simple and relatively robust assessment of the presence of fibrils. Furthermore, the absolute amount of fibrils can under favorable circumstances be estimated from Congo Red data [26]. However, Congo Red is not compatible with acidic conditions due to chromophore protonation, while alkaline conditions can compromise ThT measurements [27]. Furthermore, both Congo Red [28] and ThT [29] may give rise to false positives in the presence of non-fibrillar β-sheet structure, while biopolymers such as DNA and cellulose also bind ThT [30]. Moreover, in the case of glucagon the ThT signal decline after having reached a maximum early on in the fibrillation making it difficult to detect the mature fibrils with ThT. We attribute this to the rearrangement of the fibrils forming a more compact structure, leading to a lower number of available ThT binding sites per glucagon molecules [18, 19]. HXMS represents a more direct method to study amyloid structures circumventing the variability of more traditional staining methods. The use of HXMS for analysis of fibril formation exploits the high sensitivity of mass spectrometry. For comparison, we used 9 nmol glucagon in each well of the microtiter plate during ThT measurement, while the HXMS analysis consumed less than 60 pmol of material. While sensitivity usually is of less important when analyzing recombinant material or model peptides, this is not the case when analyzing material obtained from amyloid fibrils formed in vivo. In combination with the high sensitivity, we also get relative quantitative information using HXMS.
Several studies have used HXMS for the analysis of different amyloid fibrils including Aβ protein [9, 10], prion protein [11] and α-synuclein [12]. However, these studies were focused on the structure of the mature fibril. Here we have used HXMS as an analytical tool for exploring fibrillogenesis of glucagen. By using HXMS to follow fibril formation, it will most likely be possible to identify potential initial fibrillating cores or determine if the fibrillation occurs by a all or nothing mechanism as seen with glucagon. The fibril core can also be probed by systematically destabilizing the fibril through stepwise addition of denaturants as done in this study. In this case the data indicates that the entire glucagon peptides is involved in the fibril formation. For larger fibrillating systems such as α-synuclein, TGFBip and prion protein, the gradual denaturation of mature fibrils in combination with HXMS hold the potential for identifying the fibrillation core and/or difference in stability of the core.
5. Conclusion
In this study, we have used HXMS to analyze the fibrillogenesis of glucagon as a model system for amyloid fibril formation in general. We have used MALDI-MS in combination with micro tip purification resulting in a low-tech easily accessible method. The use of mass spectrometry for analyzing fibril formation is significantly more sensitive than traditional methods based on UV-Vis spectroscopy or NMR. Additionally, mass spectrometry provides quantitative data. Due to the relatively fast analysis time, it is possible to obtain structural information during the actual fibrillogenesis, rather than being limited to examining the end product.
The abbreviations used are
- HXMS
hydrogen exchange mass spectrometry
- ThT
Thioflavin T
- CD
Circular Dichroism
- TFA
trifluoroacetic acid
- BPI
base peak intensity
Footnotes
The work was supported by grants from the Danish Natural Science Research Council (J.J.E.).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Frokjaer S, Otzen DE. Protein drug stability: a formulation challenge. Nat Rev Drug Discov. 2005;4:298–306. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- 3.Kolstoe SE, Wood SP. Drug targets for amyloidosis. Biochem Soc Trans. 2010;38:466–470. doi: 10.1042/BST0380466. [DOI] [PubMed] [Google Scholar]
- 4.Runager K, Enghild JJ, Klintworth GK. Focus on molecules: Transforming growth factor beta induced protein (TGFBIp) Exp Eye Res. 2008;87:298–299. doi: 10.1016/j.exer.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 6.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nazabal A, Schmitter JM. Hydrogen-deuterium exchange analyzed by matrix-assisted laser desorption-ionization mass spectrometry and the HET-s prion model. Methods Enzymol. 2006;413:167–181. doi: 10.1016/S0076-6879(06)13009-8. [DOI] [PubMed] [Google Scholar]
- 8.Kheterpal I, Cook KD, Wetzel R. Hydrogen/deuterium exchange mass spectrometry analysis of protein aggregates. Methods Enzymol. 2006;413:140–166. doi: 10.1016/S0076-6879(06)13008-6. [DOI] [PubMed] [Google Scholar]
- 9.Kheterpal I, Zhou S, Cook KD, Wetzel R. Abeta amyloid fibrils possess a core structure highly resistant to hydrogen exchange. Proc Natl Acad Sci U S A. 2000;97:13597–13601. doi: 10.1073/pnas.250288897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang SS, Tobler SA, Good TA, Fernandez EJ. Hydrogen exchange-mass spectrometry analysis of beta-amyloid peptide structure. Biochemistry. 2003;42:9507–9514. doi: 10.1021/bi0342766. [DOI] [PubMed] [Google Scholar]
- 11.Nazabal A, Hornemann S, Aguzzi A, Zenobi R. Hydrogen/deuterium exchange mass spectrometry identifies two highly protected regions in recombinant full-length prion protein amyloid fibrils. J Mass Spectrom. 2009;44:965–977. doi: 10.1002/jms.1572. [DOI] [PubMed] [Google Scholar]
- 12.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL., Jr Structure and properties of alpha-synuclein and other amyloids determined at the amino acid level. Proc Natl Acad Sci U S A. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang G, Zhang BB. Glucagon and regulation of glucose metabolism. Am J Physiol Endocrinol Metab. 2003;284:E671–678. doi: 10.1152/ajpendo.00492.2002. [DOI] [PubMed] [Google Scholar]
- 14.Andersen CB, Hicks MR, Vetri V, Vandahl B, Rahbek-Nielsen H, Thogersen H, Thogersen IB, Enghild JJ, Serpell LC, Rischel C, Otzen DE. Glucagon fibril polymorphism reflects differences in protofilament backbone structure. J Mol Biol. 2010;397:932–946. doi: 10.1016/j.jmb.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Andersen CB, Otzen D, Christiansen G, Rischel C. Glucagon amyloid-like fibril morphology is selected via morphology-dependent growth inhibition. Biochemistry. 2007;46:7314–7324. doi: 10.1021/bi6025374. [DOI] [PubMed] [Google Scholar]
- 16.Jeppesen MD, Hein K, Nissen P, Westh P, Otzen DE. A thermodynamic analysis of fibrillar polymorphism. Biophys Chem. 2010;149:40–46. doi: 10.1016/j.bpc.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Oliveira CL, Behrens MA, Pedersen JS, Erlacher K, Otzen D. A SAXS study of glucagon fibrillation. J Mol Biol. 2009;387:147–161. doi: 10.1016/j.jmb.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Pedersen JS, Dikov D, Flink JL, Hjuler HA, Christiansen G, Otzen DE. The changing face of glucagon fibrillation: structural polymorphism and conformational imprinting. J Mol Biol. 2006;355:501–523. doi: 10.1016/j.jmb.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 19.Pedersen JS, Flink JM, Dikov D, Otzen DE. Sulfates dramatically stabilize a salt-dependent type of glucagon fibrils. Biophys J. 2006;90:4181–4194. doi: 10.1529/biophysj.105.070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svane AS, Jahn K, Deva T, Malmendal A, Otzen DE, Dittmer J, Nielsen NC. Early stages of amyloid fibril formation studied by liquid-state NMR: the peptide hormone glucagon. Biophys J. 2008;95:366–377. doi: 10.1529/biophysj.107.122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandell JG, Falick AM, Komives EA. Measurement of amide hydrogen exchange by MALDI-TOF mass spectrometry. Anal Chem. 1998;70:3987–3995. doi: 10.1021/ac980553g. [DOI] [PubMed] [Google Scholar]
- 22.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 23.LeVine H., 3rd Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 24.Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, Serpell LC. The common architecture of cross-beta amyloid. J Mol Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 25.Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 26.Klunk WE, Jacob RF, Mason RP. Quantifying amyloid by congo red spectral shift assay. Methods Enzymol. 1999;309:285–305. doi: 10.1016/s0076-6879(99)09021-7. [DOI] [PubMed] [Google Scholar]
- 27.Fodera V, Groenning M, Vetri V, Librizzi F, Spagnolo S, Cornett C, Olsen L, van de Weert M, Leone M. Thioflavin T hydroxylation at basic pH and its effect on amyloid fibril detection. J Phys Chem B. 2008;112:15174–15181. doi: 10.1021/jp805560c. [DOI] [PubMed] [Google Scholar]
- 28.Khurana R, Uversky VN, Nielsen L, Fink AL. Is Congo red an amyloid-specific dye? J Biol Chem. 2001;276:22715–22721. doi: 10.1074/jbc.M011499200. [DOI] [PubMed] [Google Scholar]
- 29.De Ferrari GV, Mallender WD, Inestrosa NC, Rosenberry TL. Thioflavin T is a fluorescent probe of the acetylcholinesterase peripheral site that reveals conformational interactions between the peripheral and acylation sites. J Biol Chem. 2001;276:23282–23287. doi: 10.1074/jbc.M009596200. [DOI] [PubMed] [Google Scholar]
- 30.Ilanchelian M, Ramaraj R. Emission of thioflavin T and its control in the presence of DNA. J Photoch Photobio A. 2004;162:129–137. [Google Scholar]