Abstract
Chromosomal translocations arise from the misjoining of DNA breaks, but the identity of the DNA repair factors and activities involved in their formation has been elusive. Here we show that depletion of CtIP, a DNA end-resection factor, results in a substantial decrease in chromosomal translocation frequency in mouse cells. Moreover, microhomology usage, a signature of the alternative nonhomologous end-joining pathway (alt-NHEJ), is significantly lower in translocation breakpoint junctions recovered from CtIP-depleted cells than in those from wild-type cells. Thus, we directly demonstrate that CtIP-mediated alt-NHEJ has a primary role in translocation formation. CtIP depletion in Ku70−/− cells reduces translocation frequency without affecting microhomology, indicating that Ku70-dependent NHEJ generates a fraction of translocations in wild-type cells. Translocations from both wild-type and Ku70−/− cells have smaller deletions on the participating chromosomes when CtIP is depleted, implicating the end-resection activity of CtIP in translocation formation.
Recurrent chromosomal translocations have been well documented as the hallmark and driving force in various types of cancers1,2. In agreement with the notion that chromosomal translocations originate from the misjoining of DNA breaks, agents known to cause DNA double-strand breaks (DSBs), including ionizing radiation, chemotherapeutic drugs and, in the immune system, recombination-activating gene (RAG) proteins and activation-induced cytidine deaminase (AID), are associated with chromosomal translocations leading to oncogenesis in humans and mice3–8. Thus, identifying the cellular DSB repair factors and pathways required for translocation formation is important for understanding tumor formation and developing strategies for tumor prevention.
Translocation breakpoint junctions rarely occur at homologous sequences, an observation that strongly supports a homology-independent DSB repair mechanism for their formation2,9,10. Consistent with these findings, mechanistic studies have demonstrated that chromosomal translocation formation is highly suppressed during homology-dependent DSB repair mechanisms11,12 and instead arises from some type of nonhomologous end joining (NHEJ)9.
A set of factors, including the DNA end-binding protein complex Ku70–Ku80 (Ku) and the DNA ligase complex ligase IV–Xrcc4, defines the canonical NHEJ pathway13. Unexpectedly, the elimination of Ku or DNA ligase IV–Xrcc4 does not abolish, but rather promotes, chromosomal translocations in a reporter system14,15, in accord with the frequent incidence of oncogenic immunoglobulin H (IgH)-Myc translocations in mice lacking these factors6,7,16. These results implicate an alternative NHEJ (alt-NHEJ, also called A-NHEJ) pathway, rather than canonical NHEJ (also known as C-NHEJ), as the primary mediator of translocation formation in mammalian cells. Consistent with their derivation from an alt-NHEJ pathway, translocation breakpoint junctions from wild-type, Ku70−/− and Xrcc4−/− cells are indistinguishable in many aspects14,15. However, direct evidence linking alt-NHEJ and translocation formation is still lacking, mainly because of the poor understanding of alt-NHEJ.
Alt-NHEJ differs from canonical NHEJ in that its repair products have longer deletions and a greater dependence on short patches of perfectly matched sequences known as microhomologies17,18, both of which are also observed at translocation junctions9,15, further emphasizing the involvement of alt-NHEJ in translocation formation. A mechanistic model of alt-NHEJ has been proposed, in which 5′-to-3′ nucleolytic degradation at a DSB exposes microhomology on the 3′ single-strand DNA tails, and annealing at the microhomology results in loss of the internal sequences after repair19. Notably, 5′-to-3′ end processing, known as resection, is also the initial step of homologous recombination. This similarity underscores the possibility of common resection factor(s) shared between alt-NHEJ and homologous recombination. Indeed, the homologous recombination resection factor CtIP (also called Rbbp8)20 and its yeast homolog Sae2 (ref. 21) have recently been characterized as components of alt-NHEJ22,23.
To study the role of alt-NHEJ and CtIP in the formation of chromosomal translocations, we examined the influence of CtIP knockdown on translocation frequency and the resulting breakpoint junctions in both wild-type and Ku70−/− backgrounds. Our results clearly demonstrate that CtIP is a crucial factor in generating chromosomal translocations, and they support a model whereby CtIP-mediated alt-NHEJ and Ku-dependent canonical NHEJ play major and minor roles in translocation formation, respectively.
RESULTS
CtIP knockdown reduces chromosomal translocations
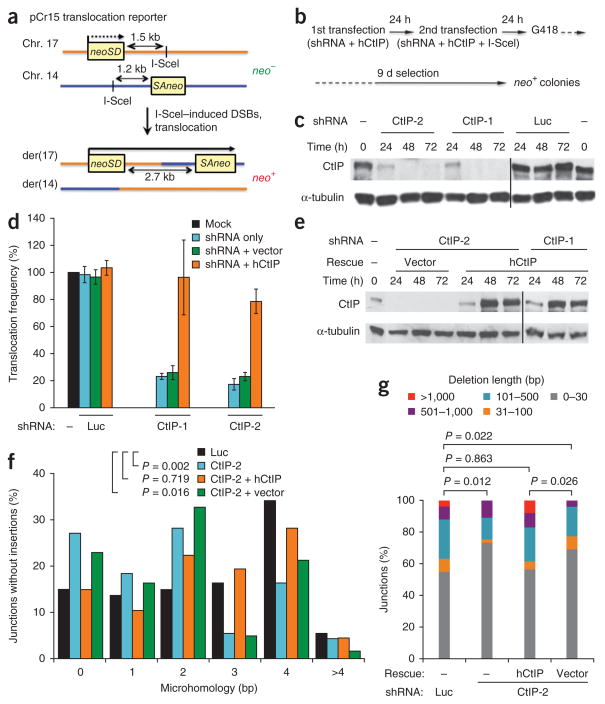
We used a small hairpin RNA (shRNA)-mediated gene knockdown approach to examine the influence of CtIP depletion using our pCr15 translocation reporter, in which two exons of a neomycin phosphotransferase gene (neo) are located on mouse chromosomes 17 and 14, each with an I-SceI endonuclease site demarcating an intron (Fig. 1a)9,14. Expression of I-SceI endonuclease in pCr15 mouse embryonic stem cells leads to DSBs on both chromosomes, and repair by NHEJ associated with translocation restores a neo+ gene on derivative chromosome 17 (der17), such that the translocation frequency can be quantified by counting neo+ colonies after G418 selection.
Figure 1.
CtIP is essential for efficient chromosomal translocation formation by microhomology-prone alt-NHEJ. (a) Diagram of the pCr15 translocation reporter. NHEJ of DSBs induced on chromosomes 17 and 14 by the I-SceI endonuclease generates a chromosomal translocation with a neo+ gene on der(17) in mouse cells. neoSD, neo splice donor; SAneo, splice acceptor neo. (b) Flow chart of shRNA-mediated depletion of CtIP to quantify translocation efficiency. In rescue experiments, an expression vector bearing human CtIP (hCtIP) is included. At the time of the second shRNA transfection, the I-SceI expression vector is introduced to generate DSBs. (c) Representative western blots showing CtIP knockdown with shRNAs CtIP-1 and CtIP-2 or control shRNA to the luciferase gene (Luc). The times relative to the first shRNA transfection are indicated. Black line indicates lanes merged from separate gels. (d) Translocation frequencies of wild-type pCr15 cells treated with the indicated shRNAs alone (blue bars), with an empty expression vector (green bars) and with an hCtIP expression vector (orange bars), as compared to that of mock-treated cells (black bars). All translocation frequencies are normalized for colony survival after shRNA transfection (Supplementary Fig. 1c). Data represent the mean ± 1 s.d. from three or more independent experiments. (e) Representative western blots showing mouse CtIP knockdown rescued by hCtIP expression. Black line indicates lanes merged from separate gels. (f) Distribution of microhomology lengths at der(17) breakpoint junctions from pCr15 cells treated with the control Luc shRNA (black bars), CtIP-2 shRNA (blue bars) or CtIP-2 shRNA rescued by an hCtIP expression vector (orange bars) or an empty vector (green bars). (g) Distribution of deletion lengths for der(17) breakpoint junctions from pCr15 cells treated as in f. P values were calculated with a two-tailed Mann-Whitney test.
To maximally deplete CtIP, we developed a double-transfection protocol in which small hairpin RNA (shRNA) constructs were introduced into cells twice within a 24-h interval (Fig. 1b). Using two different shRNA sequences, denoted CtIP-1 and CtIP-2, efficient CtIP depletion was achieved in a 24- to 72-h window after the first transfection (Fig. 1c). To induce translocations in the pCr15 cells, the I-SceI expression vector was included in the second transfection, and then G418 was added 24 h later, when CtIP depletion was still robust (Fig. 1b,c). Notably, translocation frequencies were 80% lower in cells treated with either CtIP shRNA than in cells treated with a control luciferase (Luc) shRNA (P < 0.001; Fig. 1d and Supplementary Table 1). This reduction was not due to decreased I-SceI expression, decreased cell survival or an altered cell cycle profile upon CtIP depletion (Supplementary Fig. 1a,c,d). To verify the specificity of the knockdowns, we subjected CtIP-depleted pCr15 mouse cells to complementation with a vector expressing human CtIP (Fig. 1e) and found that translocation frequencies were largely restored (Fig. 1d and Supplementary Table 1). These results demonstrate that CtIP is an essential factor for efficient translocation formation in mammalian cells.
Shorter microhomologies and deletions with CtIP knockdown
To gain more insight into the joining events leading to chromosomal translocations, we isolated individual neo+ clones and sequenced the breakpoint junctions. In agreement with previous reports14,15, we observed a range of microhomologies and deletions, as well as a smaller number of insertions (Table 1 and Fig. 1f,g). Microhomologies typically ranged from 0 to 4 bp, with a few longer ones extending up to 7 bp (Fig. 1f). In pCr15 cells treated with control shRNA, the mean length of microhomology at junctions was 2.59 bp, with 56% of the junctions having microhomologies >2 bp (Table 1). Notably, in cells depleted for CtIP the mean length of microhomology was significantly shorter (1.99 bp for shRNA CtIP-1, P = 0.024; 1.80 bp for shRNA CtIP-2, P = 0.002; two-tailed Mann-Whitney test), with substantially fewer junctions having >2-bp microhomology (33% and 26%, respectively; Table 1). Complementation with the human CtIP expression vector (mean 2.54 bp, 52% >2 bp, P = 0.719), but not with the empty vector (mean 1.92 bp, 28% >2 bp, P = 0.016), completely offset the effect of CtIP depletion on microhomology usage (Table 1 and Fig. 1f). Taken together, the concomitant reduction of translocation frequency and microhomology usage upon CtIP depletion provides direct evidence that the formation of chromosomal translocations strongly depends on microhomology-prone alt-NHEJ.
Table 1.
Summary of microhomologies and deletions at translocation junctions
| Treatment | Microhomology
|
Deletion
|
Clones with insertion | Total clones | ||
|---|---|---|---|---|---|---|
| Mean (bp) | >2 bp | Mean (bp) | Median (bp) | |||
| Wild type | ||||||
| Luc | 2.59 | 40/73 (56%) | 191.6 | 17.0 | 12 (14%) | 85 |
| CtIP-1 | 1.99 | 24/73 (33%) | 92.7 | 16.5 | 13 (15%) | 86 |
| CtIP-2 | 1.80 | 24/92 (26%) | 116.6 | 13.5 | 10 (10%) | 102 |
| CtIP-2 + hCtIP | 2.54 | 35/67 (52%) | 252.2 | 17.0 | 9 (12%) | 76 |
| CtIP-2 + vector | 1.92 | 17/61 (28%) | 83.0 | 15.0 | 14 (19%) | 75 |
| Ku70−/− | ||||||
| Luc | 2.56 | 35/71 (49%) | 265.7 | 22.0 | 16 (18%) | 87 |
| CtIP-1 | 2.33 | 37/77 (48%) | 119.7 | 15.0 | 15 (16%) | 92 |
| CtIP-2 | 2.39 | 32/72 (44%) | 116.4 | 15.0 | 17 (19%) | 89 |
| CtIP-2 + hCtIP | 2.55 | 34/62 (55%) | 249.7 | 28.0 | 9 (13%) | 71 |
| CtIP-2 + vector | 2.45 | 34/65 (52%) | 83.6 | 15.0 | 8 (11%) | 73 |
Microhomologies are from junctions without insertions, as the length of microhomologies cannot always be determined in the presence of insertions.
CtIP promotes DNA end resection at DSBs to generate single-stranded DNA for homologous recombination20. A similar role of CtIP in alt-NHEJ would potentially increase deletion lengths as microhomology becomes exposed for annealing. To determine whether CtIP loss affects degradation of DNA ends during translocation formation, we compared overall deletion lengths at the breakpoint junctions. CtIP knockdown caused a significant shift toward shorter deletions, as seen when the deletions were grouped into size classes (Fig. 1g) or analyzed overall for mean (191.6 bp in Luc controls; 92.7 bp for CtIP-1 and 116.6 bp for CtIP-2) and median (17.0 bp in Luc controls; 16.5 bp for CtIP-1 and 13.5 bp for CtIP-2) deletion lengths (Table 1 and Supplementary Fig. 2a). Rescue with the human CtIP expression vector (mean 252.2 bp, median 17.0 bp), but not the empty vector (mean 83.0 bp, median 15.0 bp), restored the deletion length, in all parameters, to a level similar to that in controls (Fig. 1g, Table 1 and Supplementary Fig. 2a). These data indicate that CtIP promotes resection at DNA ends participating in translocation formation.
Ku70−/− cells have reduced translocations with CtIP knockdown
A question arising from our results is which NHEJ pathway is responsible for the remaining translocations recovered with CtIP knockdown. Although we cannot rule out a contribution from residual alt-NHEJ activity due to incomplete CtIP depletion, the significant difference in microhomology seen with CtIP knockdown strongly suggests the involvement of the canonical NHEJ pathway, as the relative bias of canonical NHEJ toward forming junctions with little or no microhomology is in accord with the reduced microhomology found at translocation junctions in the CtIP-depleted cells.
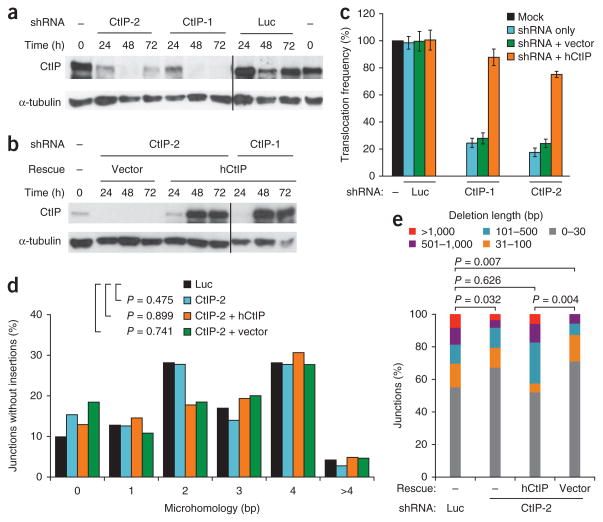
To investigate the role of the canonical NHEJ pathway, we examined translocation formation in the absence of Ku. As seen previously14,15, Ku70−/− pCr15 cells have a higher translocation frequency than wild-type cells (Supplementary Table 1). Given the role for CtIP in translocation formation described above, the higher translocation frequency in the absence of Ku can now be interpreted as follows: Ku normally suppresses chromosomal translocations by inhibiting the resection step of alt-NHEJ (see Discussion). Consistent with this interpretation, CtIP depletion in the Ku70−/− cells led to an 80% reduction in translocations, similar to the reduction seen in wild-type cells, which was substantially restored by expression of human CtIP (P < 0.001; Fig. 2a–c, Supplementary Fig. 1b,c,e and Supplementary Table 1). Further support for this model comes from the finding of increased CtIP binding in Ku70-deficient cells at regions undergoing class-switch recombination (CSR)24.
Figure 2.
In Ku70−/− cells, CtIP is essential for efficient chromosomal translocation formation but does not affect microhomology at breakpoint junctions. (a,b) Representative western blots showing CtIP knockdown with shRNAs CtIP-1 and CtIP-2 or control Luc shRNA (a) and complementation by expressing human CtIP (hCtIP; b). The times relative to the first shRNA transfection are indicated. Black line indicates lanes merged from separate gels. (c) Translocation frequencies of Ku70−/− pCr15 cells treated with the indicated shRNAs alone (blue bars), with an empty expression vector (green bars) and with an hCtIP expression vector (orange bars) as compared to that of mock-treated Ku70−/− cells (black bars). All translocation frequencies are normalized for colony survival after shRNA transfection (Supplementary Fig. 1c). Data represent the mean ± 1 s.d. from three or more independent experiments. (d) Distribution of microhomology lengths at der(17) breakpoint junctions from Ku70−/− pCr15 cells treated with the control Luc shRNA (black bars), CtIP-2 shRNA (blue bars) or CtIP-2 shRNA rescued by an hCtIP expression vector (orange bars) or an empty vector (green bars). (e) Distribution of deletion lengths for der(17) breakpoint junctions from Ku70−/− pCr15 cells treated as in d. P values were calculated by a two-tailed Mann-Whitney test.
CtIP loss does not affect microhomologies in Ku70−/− cells
In both wild-type and Ku70−/− cells, the mean length of microhomology (2.59 bp versus 2.56 bp, respectively; P = 0.798) and the fraction of junctions with >2 bp microhomology (56% versus 49%) were similar (Table 1), as previously reported15. These results are consistent with translocations arising in wild-type cells primarily by alt-NHEJ rather than the canonical pathway. Unlike in wild-type cells, however, microhomology was only marginally reduced upon CtIP depletion in Ku70−/− cells (2.33 bp for shRNA CtIP-1, P = 0.459; 2.39 bp for shRNA CtIP-2, P = 0.475; Fig. 2d and Table 1), and the fraction of junctions with >2 bp microhomology was also similar (49% versus 48% and 44%, respectively; Table 1). That CtIP depletion differentially affects the remaining breakpoint junctions in wild-type and Ku-deficient cells firmly supports a role, albeit a minor one, for the canonical NHEJ pathway in translocation formation.
As seen in wild-type cells, CtIP knockdown in Ku70−/− cells led to a shift toward shorter deletions, with reduced mean and median lengths, that were restored by complementation with human CtIP (Fig. 2e, Table 1 and Supplementary Fig. 2b). Thus, in both wild-type and Ku70−/− cells, a lower translocation frequency with CtIP depletion is associated with smaller deletions, suggesting that there is a link between translocation efficiency and CtIP-mediated end resection. Consistent with this idea, the Mre11 complex, a partner with CtIP in resection, has recently been characterized as a player in mammalian alt-NHEJ25,26. Our preliminary results also indicate that chemical inhibition of Mre11 nuclease activity reduces translocation frequency (Y.Z. and M.J., unpublished results).
DISCUSSION
Oncogenic translocations represent the initiating lesion for many types of tumors, most recently even some carcinomas. A paradox has existed in that these translocations arise almost exclusively by NHEJ, yet model systems have shown that translocations do not require the canonical NHEJ pathway for their formation, implicating alt-NHEJ without providing definitive evidence for its involvement. Here we directly demonstrate that alt-NHEJ is required for efficient translocation formation in mammalian cells through CtIP, thereby providing the first direct role for a specific DNA repair component in promoting translocations while addressing a longstanding question about the genetic requirements for translocation formation.
Our experiments point to a primary role for microhomology-prone alt-NHEJ in translocation formation (Fig. 3). Although the small difference in microhomology usage from alt-NHEJ is likely inconsequential on a biological level, it provides a clear indication of a distinct mechanism in translocation formation, one that is more prone to genomic rearrangement than the canonical NHEJ pathway. In most oncogenic translocations junctions, as well as the junctions reported here, microhomology lengths are short9, requiring only limited resection of DNA ends. For example, in a random sequence, only 64 bp would need to be resected for a 3-bp microhomology to be exposed for annealing. In yeast, the CtIP ortholog Sae2, together with the Mre11 complex, is involved in the initial step of end resection, removing about 50–100 nucleotides from the 5′ end of the DSB27,28, and this function of Sae2 has been implicated in promoting alt-NHEJ by exposing microhomologies at DNA ends for annealing19,22. Our data suggest that CtIP has a similar role in translocation formation in mammalian cells. In this model, longer microhomologies would be infrequently used in joining, even if they would be more energetically favorable, because the length of resection would need to be enormous, larger than many of the introns in which joining occurs in oncogenic translocations. For instance, repeats of 7 or 8 bp, such as those placed close to DNA ends in other studies of alt-NHEJ23,29, would require 16,000 or 64,000 bp of resection, respectively, in order for microhomology to be exposed for annealing in a random sequence. Deletions of this size are not typically found in translocation junctions9.
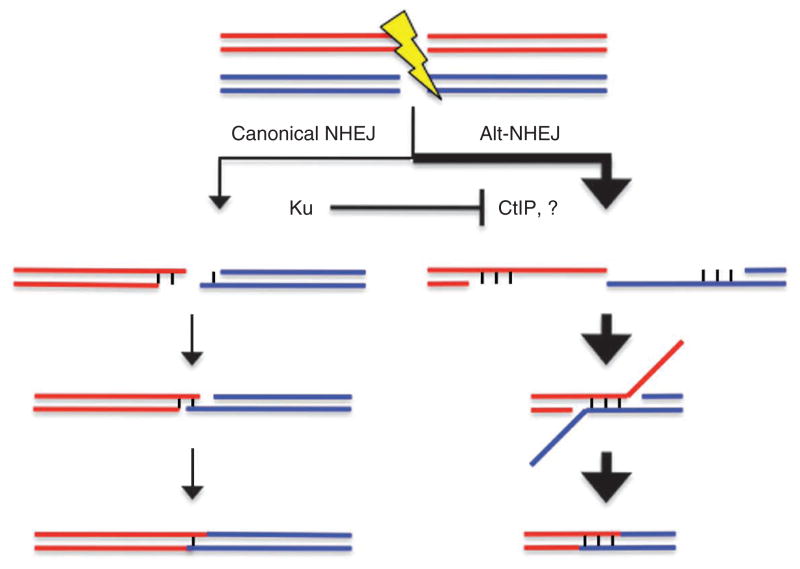
Figure 3.
Model for pathways involved in chromosomal translocations. Translocations primarily arise from an alt-NHEJ pathway that is largely dependent on CtIP. CtIP promotes the resection of DNA ends to uncover microhomologies that anneal for end joining. In the absence of Ku, resection factors such as CtIP (and possibly other unknown ones, represented by the question mark) have greater access to DNA ends, such that translocations increase. In wild-type cells, a minor portion of translocations may arise through the canonical NHEJ pathway, which can efficiently join ends without microhomology.
Notably, canonical NHEJ keeps translocations in check by inhibiting the resection step of alt-NHEJ (Fig. 3). Physical assays for resection in yeast have directly demonstrated that resection increases in both Ku and DNA ligase IV mutants30,31. Moreover, in mammalian cells, homologous recombination, which also relies on resection, is increased in Ku and ligase IV mutants32,33. At the same time, canonical NHEJ appears to have a minor role in forming translocations in wild-type cells (Fig. 3). That the canonical pathway contributes at all is supported by the reduced microhomology at junctions upon CtIP depletion when Ku is present compared to when it is absent. Notably, translocation formation is not abolished in CtIP-depleted, Ku-deficient cells. There are several nonexclusive possibilities to account for these remaining events, a trivial one being that CtIP depletion is not complete. In addition, loss of both CtIP and Ku may result in a high level of unrepaired breaks. Finally, the existence of a CtIP-independent alt-NHEJ pathway suppressed by Ku cannot be ruled out (Fig. 3). Our model for multiple NHEJ pathways in translocation formation is buttressed by a systematic survey of genomic rearrangements in cancer cells10.
Although NHEJ with microhomology preference34,35 and NHEJ in the absence of canonical NHEJ factors17,18,35 were documented more than two decades ago, the significance of alt-NHEJ was largely unappreciated until recently, mainly because of the uncertainty about its functional components and biological relevance. The establishment of a crucial role for CtIP in alt-NHEJ and the definition of a primary role for alt-NHEJ in chromosomal translocation formation have now substantially illuminated the nature of this pathway. In concordance with our findings, a role for CtIP-dependent alt-NHEJ in CSR has also been recently uncovered24. In contrast to its clear benefits in the context of CSR, it is unlikely that alt-NHEJ is inherently advantageous in the context of chromosomal translocation formation. Whether such a function is an unintended consequence of its other cellular functions or whether it staves off cell death in the face of multiple DSBs remains to be determined. In any case, targeting of components of the pathway, such as CtIP, has therapeutic potential to reduce translocations, especially when individuals are at high risk of these, as from cancer treatment.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
Acknowledgments
We thank J. Chaudhuri and colleagues (Memorial Sloan-Kettering Cancer Center) for sharing unpublished results, R. Baer (Columbia University) for reagents, and E. Brunet, D. Simsek, P. Sung and other members of the Jasin laboratory for helpful discussions. This work was supported by a Leukemia and Lymphoma Society fellowship (Y.Z.), a Dorothy Rodbell Cohen Cancer Research Program Grant and US National Institutes of Health grant R01 NIHGM54668 (M.J.).
Footnotes
Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
Y.Z. performed the experiments. Y.Z. and M.J. designed the research and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst) 2006;5:1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 3.Brandt VL, Roth DB. Recent insights into the formation of RAG-induced chromosomal translocations. Adv Exp Med Biol. 2009;650:32–45. doi: 10.1007/978-1-4419-0296-2_3. [DOI] [PubMed] [Google Scholar]
- 4.Wang JH, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbiani DF, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 7.Boboila C, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA. 2010;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 13.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 14.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4–ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Difilippantonio MJ, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 18.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 19.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 22.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end-joining during class-switch recombination. Nat Struct Mol Biol. 2010 Dec 5; doi: 10.1038/nsmb.1942. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rass E, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 26.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double- strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SE, et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 32.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinstock DM, Jasin M. Alternative pathways for the repair of RAG-induced DNA breaks. Mol Cell Biol. 2006;26:131–139. doi: 10.1128/MCB.26.1.131-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackwell TK, et al. Isolation of scid pre-B cells that rearrange kappa light chain genes: formation of normal signal and abnormal coding joins. EMBO J. 1989;8:735–742. doi: 10.1002/j.1460-2075.1989.tb03433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.