Abstract
Bacillus anthracis, the causative agent of anthrax, is considered a serious threat as a bioweapon. The drugs most commonly used to treat anthrax are quinolones, which act by increasing DNA cleavage mediated by topoisomerase IV and gyrase. Quinolone resistance most often is associated with specific serine mutations in these enzymes. Therefore, to determine the basis for quinolone action and resistance, we characterized wild-type B. anthracis topoisomerase IV, the GrlAS81F and GrlAS81Y quinolone-resistant mutants, and the effects of quinolones and a related quinazolinedione on these enzymes. Ser81 is believed to anchor a water-Mg2+ bridge that coordinates quinolones to the enzyme through the C3/C4 keto acid. Consistent with this hypothesized bridge, ciprofloxacin required increased Mg2+ concentrations to support DNA cleavage by GrlAS81F topoisomerase IV. The three enzymes displayed similar catalytic activities in the absence of drugs. However, the resistance mutations decreased the affinity of topoisomerase IV for ciprofloxacin and other quinolones, diminished quinolone-induced inhibition of DNA religation, and reduced the stability of the ternary enzyme-quinolone-DNA complex. Wild-type DNA cleavage levels were generated by mutant enzymes at high quinolone concentrations, suggesting that increased drug potency could overcome resistance. 8-Methyl-quinazoline-2,4-dione, which lacks the quinolone keto acid (and presumably does not require the water-Mg2+ bridge to mediate protein interactions), was more potent than quinolones against wild-type topoisomerase IV and was equally efficacious. Moreover, it maintained high potency and efficacy against the mutant enzymes, effectively inhibited DNA religation, and formed stable ternary complexes. Our findings provide an underlying biochemical basis for the ability of quinazolinediones to overcome clinically-relevant quinolone resistance mutations in bacterial type II topoisomerases.
Bacillus anthracis, the causative agent of anthrax, is a highly pathogenic Gram-positive soil bacterium that is considered a serious threat as a weapon of mass destruction and an agent of bioterrorism (1-5). B. anthracis is used as a biological weapon in large part because it forms durable spores (1, 2, 4, 5). These spores can enter the body through multiple routes and then germinate and grow as vegetative cells. Toxic factors secreted by the vegetative cells accumulate and usually cause the death of the host within several days, particularly if spores enter via the lungs or gut (1, 2, 4, 5). Mortality rates for respiratory anthrax, the most lethal form of the disease, approach 100% if untreated.
Natural strains of B. anthracis are sensitive to several antibacterial agents, which can be used to treat anthrax (4, 5). The most effective and commonly used drug for the treatment of anthrax is ciprofloxacin (4, 5), a broad-spectrum quinolone antibacterial (6-10). Following the mailings of letters containing lethal B. anthracis spores in the autumn of 2001 in the United States, it is estimated that as much as one billion dollars worth of ciprofloxacin was prescribed to treat individuals who potentially were exposed to the spores (11).
Ciprofloxacin and other quinolones kill bacteria by increasing levels of DNA strand breaks generated by enzymes known as type II topoisomerases (6-10). Nearly all bacteria encode two type II topoisomerases, gyrase and topoisomerase IV (7, 12-19). Both enzymes are comprised of two protomer subunits and have an A2B2 quaternary structure (7, 12-14, 16, 17, 20). Gyrase is made up of two GyrA subunits (that contain the active site tyrosines involved in DNA cleavage and ligation) and two GyrB subunits (that bind ATP, which is required for overall catalytic activity). Topoisomerase IV is made up of two GrlA (named as gyrase-like) and two GrlB subunits that are homologous to GyrA and GyrB, respectively (12-14, 17). Gyrase and topoisomerase IV alter DNA topology by passing an intact double helix through a transient break that they generate in a separate segment of DNA (12-14, 16-18, 20). Although these enzymes share a common mechanism, they appear to have different physiological functions. Specific interactions between DNA substrates and the C-terminus of GyrA enable gyrase to introduce negative supercoils into relaxed molecules. As a result, gyrase plays critical roles in maintaining the superhelical density of the bacterial genome and is primarily responsible for removing positive supercoils that accumulate ahead of replication forks and transcription complexes (12-14, 17). In contrast, topoisomerase IV is a far more efficient decatenase than is gyrase. It is primarily responsible for removing knots and tangles that form in the bacterial chromosome during recombination and replication (13, 14, 17). Both type II enzymes are essential for cell survival (12-14, 16-18) and appear to be physiological targets for quinolone antibacterials in B. anthracis (21-24).
Recently, structures have been reported for covalent complexes formed between topoisomerase IV or gyrase and cleaved DNA (i.e., cleavage complexes) in the presence of quinolones (25-28). As predicted from studies on anticancer drugs that target human type II topoisomerases (29-32), quinolones were found at the cleaved scissile bonds in the active site of the bacterial enzymes (25-28). However, there is disagreement within the field as to how quinolones are positioned within the cleavage complex. In some cases, the orientation of the drug is rotated as much as 180° (25-28). Furthermore, relatively little is understood regarding the mechanism by which quinolones increase levels of topoisomerase IV- or gyrase-mediated DNA cleavage or how mutations in either enzyme lead to drug resistance.
Quinolone resistance is most often associated with specific mutations in topoisomerase IV and/or gyrase (6-8, 10, 21-24, 33). Generally, mutation of one type II enzyme confers ≤10-fold drug resistance. Selection for higher levels of resistance (~10- to 100-fold) usually yields strains with mutations in both enzymes (6-8, 10, 21-24, 33). Considering the emergence of quinolone resistance in many infectious bacterial strains (34-36) and the potential use of quinolone-resistant B. anthracis as a bioweapon (1-5), more effective drugs that display activity against these strains need to be developed.
Therefore, as an important step towards this goal, we characterized wild-type B. anthracis topoisomerase IV and the corresponding GrlAS81F and GrlAS81Y quinolone-resistant mutants. We also examined the effects of clinically-relevant quinolones and an associated quinazolinedione on the DNA cleavage and religation activities of these enzymes. Our results shed light on the biochemical mechanism of quinolone action against the bacterial type II enzyme. Furthermore, they provide a mechanistic basis for drug resistance induced by mutations at the amino acid most commonly associated with decreased quinolone sensitivity and a rationale for overcoming this resistance in B. anthracis topoisomerase IV. These findings may have broad applicability to quinolone-resistant type II topoisomerases from other bacterial species.
EXPERIMENTAL PROCEDURES
Enzymes and Materials
Genes encoding wild-type B. anthracis GrlA and GrlB and drug-resistant GrlAS81F and GrlAS81Y (generated by site-directed mutagenesis) were individually cloned, N-terminally His-tagged, and expressed in Escherichia coli. The resulting proteins were purified by affinity chromatography (37), dialyzed into 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 20% glycerol, and stored at −20 °C. In all assays, topoisomerase IV was used as a 1:1 mixture of GrlA:GrlB.
Negatively supercoiled pBR322 plasmid DNA was prepared from E. coli using a Plasmid Mega Kit (Qiagen) as described by the manufacturer. Kinetoplast DNA (kDNA) was isolated from Crithidia fasiculata as described previously (3). [γ-32P]ATP (~6000Ci/mmol) was obtained from Perkin-Elmer.
Ciprofloxacin was obtained from LKT Laboratories, stored at −20 °C as a 40 mM stock solution in 0.1 N NaOH, and diluted five-fold with 10 mM Tris-HCl (pH 7.9) immediately prior to use. Moxifloxacin was obtained from LKT Laboratories, and levofloxacin, sparfloxacin, and norfloxacin were obtained from Sigma Aldrich. CP-115,953 was synthesized as described (39). 3-Amino-7-[(3S)-3-(aminomethyl)-1-pyrrolidinyl]-1-cyclopropyl-6-fluoro-8-methyl-2,4(1H,3H) -quinazolinedione was synthesized using established methods as previously reported (40). For simplicity, this compound will be referred to as 8-methyl-quinazoline-2,4-dione. All drugs other than ciprofloxacin were stored at 4 °C as 20 mM stock solutions in 100% DMSO. All other chemicals were analytical reagent grade.
DNA Relaxation
DNA relaxation assays were based on the protocol of Fortune and Osheroff (41). Reaction mixtures (20 μL) contained 50 nM wild-type or mutant topoisomerase IV and 5 nM negatively supercoiled pBR322 in relaxation buffer [40 mM HEPES (pH 7.6), 100 mM potassium glutamate, 10 mM Mg(OAc)2, 50 mM NaCl, and 1 mM ATP] and were incubated at 37 °C. Relaxation was stopped at times ranging from 0-30 min by the addition of 3 μL of stop solution [0.77% SDS and 77.5 mM EDTA]. Samples were mixed with 2 μL of agarose gel loading buffer [60% sucrose, 10 mM Tris-HCl (pH 7.9), 0.5% bromophenol blue, and 0.5% xylene cyanol FF], heated at 45 °C for 5 min, and subjected to electrophoresis in 1% agarose gels in 100 mM Tris-borate (pH 8.3) and 2 mM EDTA. Gels were stained with 0.75 μg/mL ethidium bromide for 30 min. DNA bands were visualized with medium-range ultraviolet light and quantified using an Alpha Innotech digital imaging system. The percent relaxed DNA was determined by the loss of supercoiled DNA substrate.
Kinetoplast DNA Decatenation
Decatenation assays were carried out by the procedure of Anderson et al. (42). Reaction mixtures (20 μL) contained 50 nM wild-type or mutant topoisomerase IV and 0.3 μg kinetoplast DNA (kDNA) in relaxation buffer and were incubated at 37 °C. Decatenation was stopped at times ranging from 0-30 min by the addition of 3 μL of stop solution. Samples were mixed with 2 μL of agarose gel loading buffer, heated at 45 °C for 5 min, and subjected to electrophoresis in 1% agarose gels in 100 mM Tris-borate (pH 8.3) and 2 mM EDTA containing 0.5 μg/mL ethidium bromide. DNA bands were visualized and quantified as described above. The percent decatenated DNA was determined by the appearance of monomeric circular DNA molecules.
Plasmid DNA Cleavage
DNA cleavage reactions were carried out using the procedure of Fortune and Osheroff (41). Reactions contained 200 nM wild-type or mutant topoisomerase IV and 10 nM negatively supercoiled pBR322 in a total of 20 μL of cleavage buffer [40 mM Tris–HCl (pH 7.9), 10 mM MgCl2, 50 mM NaCl, and 2.5% (v/v) glycerol]. In some reactions, the concentration dependence of MgCl2 was examined or the divalent metal ion was replaced with either CaCl2 or MnCl2. Reaction mixtures were incubated at 37 °C for 10 min, and enzyme-DNA cleavage complexes were trapped by the addition of 2 μL of 5% SDS followed by 1 μL of 250 mM EDTA (pH 8.0). Proteinase K (2 μL of a 0.8 mg/mL solution) was added, and samples were incubated at 45 °C for 45 min to digest the enzyme. Samples were mixed with 2 μL of agarose gel loading buffer, heated at 45 °C for 5 min, and subjected to electrophoresis in 1% agarose gels in 40 mM Tris-acetate (pH 8.3) and 2 mM EDTA containing 0.5 μg/mL ethidium bromide. DNA bands were visualized and quantified as described above. DNA cleavage was monitored by the conversion of supercoiled plasmid to linear molecules.
Assays that monitored the DNA cleavage activities of wild-type and mutant B. anthracis topoisomerase IV in the absence of drugs substituted 1 mM CaCl2 for 10 mM MgCl2 in the cleavage buffer. Assays that assessed the DNA cleavage activities of the wild-type and mutant enzymes in the presence of drugs contained 0-30 μM compound for the wild-type enzyme and 0-500 μM compound for the mutant enzymes.
For assays that monitored competition between ciprofloxacin (0-150 μM) and 8-methyl-quinazoline-2,4-dione (20 μM), the level of cleavage seen with the corresponding concentration of ciprofloxacin in the absence of the quinazolinedione was used as a baseline and was subtracted from the cleavage level seen in the presence of both compounds. Ciprofloxacin and 8-methyl-quinazoline-2,4-dione were added simultaneously to reaction mixtures.
DNA Cleavage Site Utilization
DNA cleavage sites were mapped using a modification (43) of the procedure of O’Reilly and Kreuzer (44). The pBR322 DNA substrate was linearized by treatment with HindIII. Terminal 5′-phosphates were removed by treatment with calf intestinal alkaline phosphatase and replaced with [32P]phosphate using T4 polynucleotide kinase and [γ-32P]ATP. The DNA was treated with EcoRI, and the 4332 bp singly-end-labeled fragment was purified from the small EcoRI-HindIII fragment by passage through a CHROMA SPIN+TE-100 column (Clontech). Reaction mixtures contained 200 nM wild-type or mutant topoisomerase IV and 1 nM labeled pBR322 DNA substrate in 50 μL of DNA cleavage buffer in the absence or presence of compounds. Reaction mixtures were incubated at 37 °C for 10 min, and enzyme-DNA cleavage complexes were trapped by the addition of 5 μL of 5% SDS followed by 3 μL of 250 mM EDTA (pH 8.0). Proteinase K (5 μL of a 0.8 mg/mL solution) was added, and samples were incubated at 45 °C for 45 min to digest the enzyme. DNA products were precipitated with ethanol and resuspended in 5 μL of polyacrylamide gel loading buffer [10% agarose gel loading buffer, 80% formamide, 100 mM Tris-borate (pH 8.3), and 2 mM EDTA]. Samples were subjected to electrophoresis in denaturing 6% polyacrylamide sequencing gels. Gels were dried in vacuo, and DNA cleavage products were visualized with a Bio-Rad Molecular Imager FX.
DNA Religation
DNA religation assays were carried out by the procedure of Robinson and Osheroff (45). Reaction mixtures (20 μL) contained 200 nM wild-type or mutant topoisomerase IV and 10 nM negatively supercoiled pBR322 in cleavage buffer containing 5 mM MgCl2. Initial DNA cleavage/religation equilibria were established at 37 °C for 10 min. Religation was initiated by rapidly shifting the temperature from 37 to 75 °C. Reactions were stopped at times ranging from 0-135 s by the addition of 2 μL of 5% SDS followed by 1 μL of 250 mM EDTA (pH 8.0). Samples were digested with proteinase K and processed as described above for plasmid cleavage assays. Levels of DNA cleavage were set to 100% at time zero, and religation was determined by the loss of linear reaction product over time.
Persistence of Topoisomerase IV-DNA Cleavage Complexes
The persistence of topoisomerase IV-DNA cleavage complexes established in the presence of drugs was determined using the procedure of Gentry et al. (46). Initial reaction mixtures contained 1 μM wild-type or mutant topoisomerase IV, 50 nM DNA, and 20 μM (for the wild-type enzyme) or 200 μM (for the mutant enzymes) ciprofloxacin or 20 μM 8-methyl-quinazoline-2,4-dione in a total of 20 μL of DNA cleavage buffer. Reactions were incubated at 37 °C for 10 min and then diluted 20-fold with DNA cleavage buffer warmed to 37 °C. Samples (20 μL) were removed at times ranging from 0-300 min, and DNA cleavage was stopped with 2 μL of 5% SDS followed by 1 μL of 250 mM EDTA (pH 8.0). Samples were digested with proteinase K and processed as described above for plasmid cleavage assays. Levels of DNA cleavage were set to 100% at time zero, and the persistence of cleavage complexes was determined by the decay of the linear reaction product over time.
RESULTS
Characterization of Wild-Type, GrlAS81F, and GrlAS81Y B. anthracis Topoisomerase IV
Quinolone-based antibacterials are the main prophylactic drugs used to treat potential exposure to B. anthracis and are the most common treatment for individuals who have contracted anthrax (4, 5). Therefore, as a first step towards defining interactions between quinolones and B. anthracis type II topoisomerases, we purified and characterized topoisomerase IV.
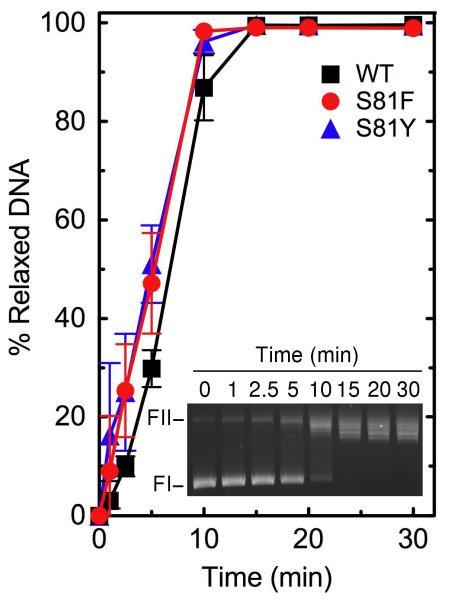
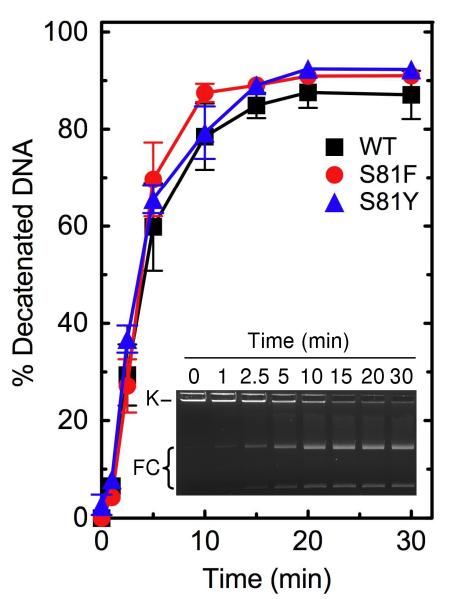
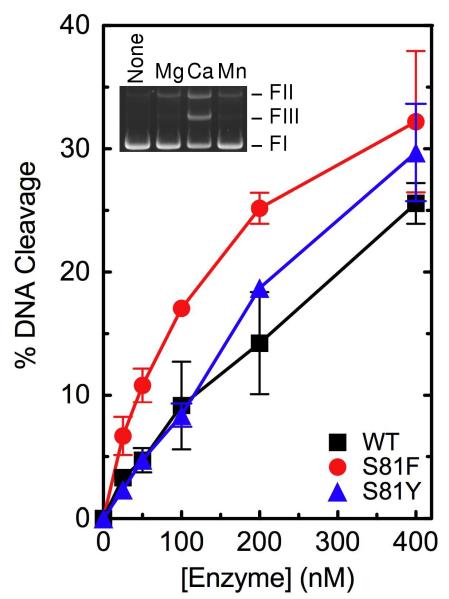
As seen in Figures 1 and 2, wild-type topoisomerase IV readily relaxes negatively supercoiled plasmid DNA and decatenates kDNA. In addition, the enzyme requires a divalent metal ion for DNA cleavage, and the reaction is supported by Mg2+, Ca2+, and Mn2+ (Figure 3, inset). As observed for other prokaryotic and eukaryotic type II topoisomerases (47-50), high levels of DNA scission were seen in reactions that contained Ca2+. Therefore, an enzyme titration was carried out in the presence of this divalent metal ion (Figure 3). Under “standard reaction conditions” that included 200 nM topoisomerase IV, the enzyme cut nearly 15% of the negatively supercoiled substrate. Using equivalent conditions in reactions that replaced Ca2+ with Mg2+, baseline levels of DNA cleavage were ~2%.
Figure 1.
DNA relaxation activities of wild-type, GrlAS81F, and GrlAS81Y B. anthracis topoisomerase IV. The ability of wild-type (WT, black squares), GrlAS81F (S81F, red circles), and GrlAS81Y (S81Y, blue triangles) topoisomerase IV to relax negatively supercoiled pBR322 plasmid DNA is shown. Error bars represent the standard deviation of three or more independent experiments. The inset shows an agarose gel of a typical 30 min relaxation time course catalyzed by wild-type topoisomerase IV. The positions of negatively supercoiled (replicative form I, FI) and nicked (replicative form II, FII) plasmids are indicated.
Figure 2.
DNA decatenation activities of wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV. The ability of wild-type (WT, black squares), GrlAS81F (S81F, red circles), and GrlAS81Y (S81Y, blue triangles) topoisomerase IV to decatenate kinetoplast DNA is shown. Error bars represent the standard deviation of three or more independent experiments. The inset shows an agarose gel of a typical 30 min decatenation time course catalyzed by wild-type topoisomerase IV. The positions of kDNA (K) and monomeric free circles (FC) resulting from decatenation of kDNA are indicated.
Figure 3.
DNA cleavage activities of wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV. The ability of wild-type (WT, black squares), GrlAS81F (S81F, red circles), and GrlAS81Y (S81Y, blue triangles) topoisomerase IV to cleave negatively supercoiled pBR322 plasmid DNA is shown. Assays were carried out in the presence of 1 mM CaCl2. Error bars represent the standard deviation of three or more independent experiments. The inset shows an agarose gel of a typical DNA cleavage assay mediated by wild-type topoisomerase IV in the absence of divalent metal ion (None) or in the presence of Mg2+, Ca2+, or Mn2+. The positions of negatively supercoiled (replicative form I, FI), nicked (replicative form II, FII), and linear (replicative form III, FIII) plasmids are indicated.
Quinolone resistance in clinical isolates of pathogenic bacteria is most often associated with specific mutations in topoisomerase IV and/or gyrase (6, 8, 36). Although the primary target of quinolones (topoisomerase IV or gyrase) is species- and drug-dependent, both enzymes contribute to drug resistance (6, 33, 51-55). The most common mutations occur at a specific serine residue in the A (or equivalent) subunit of either enzyme (34). This residue originally was described as Ser83 in E. coli gyrase. The homologous residue in B. anthracis GrlA is Ser81. Indeed, in laboratory studies of B. anthracis that selected for drug resistance, GrlAS81F and GrlAS81Y were frequently observed in quinolone-resistant strains (22-24). Therefore, GrlAS81F and GrlAS81Y topoisomerase IV were isolated and characterized.
As seen in Figures 1 and 2, both mutant enzymes displayed DNA relaxation and decatenation activities that were similar to those of wild-type B. anthracis topoisomerase IV. In addition, baseline levels of DNA cleavage mediated by GrlAS81F and GrlAS81Y topoisomerase IV in the presence of Ca2+ appeared to be somewhat higher than that seen with the wild-type enzyme (Figure 3). Similar results were observed in the presence of Mg2+ (see the 0 μM drug points in Figure 5). Thus, quinolone resistance in GrlAS81F and GrlAS81Y topoisomerase IV is not due to a general loss of enzyme activity. This finding has important implications for the potential treatment of B. anthracis infections that carry these common mutations. Because quinolone-induced cell death is triggered by the generation of topoisomerase IV- and gyrase-mediated DNA strand breaks, it would be extremely difficult to overcome quinolone resistance that results from decreased activity of one or both type II enzymes.
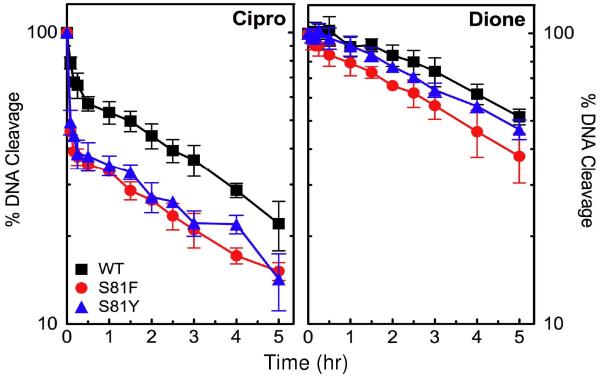
Figure 5.
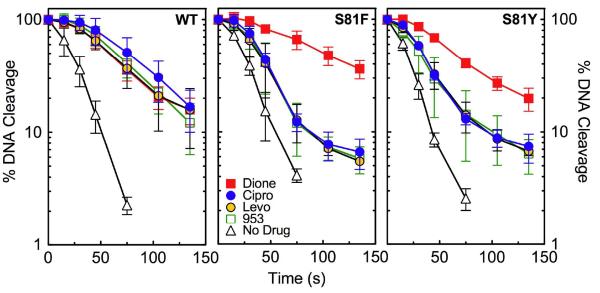
Effects of quinolones and 8-methyl-quinazoline-2,4-dione on the DNA cleavage activities of wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV. DNA cleavage mediated by wild-type (WT), GrlAS81F (S81F), and GrlAS81Y (S81Y) topoisomerase IV in the presence of drugs are shown in the left, center, and right panels, respectively. Results with ciprofloxacin (Cipro, blue circles), levofloxacin (Levo, yellow circles), CP-115,953 (953, open green squares), norfloxacin (Nor, open diamonds), moxifloxacin (Moxi, black diamonds), sparfloxacin (Spar, purple triangles), and 8-methyl-quinazoline-2,4-dione (Dione, red squares) are shown for wild-type topoisomerase IV. Data with ciprofloxacin, levofloxacin, CP-115,953, and 8-methyl-quinazoline-2,4-dione are shown for the mutant enzymes. Error bars represent the standard deviation of three or more independent experiments.
Effects of Quinolones and 8-Methyl-quinazoline-2,4-dione on DNA Cleavage Mediated by Wild-Type, GrlAS81F, and GrlAS81Y Topoisomerase IV
In order to explore the mechanism of quinolone action and resistance, initial experiments examined the effects of quinolones on DNA cleavage mediated by wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV. A number of clinically-relevant quinolones were used for these experiments, including ciprofloxacin, levofloxacin, moxifloxacin, norfloxacin, and sparfloxacin (10) (Figure 4). The experimental quinolone CP-115,953 also was used. It is the only topoisomerase II poison that displays high activity against bacterial and eukaryotic type II enzymes (42, 56, 57). All of the quinolones were examined with wild-type B. anthracis topoisomerase IV. While some differences in drug potency and efficacy were noted among the compounds, all of the quinolones fundamentally had the same effect on the enzyme. Therefore, only data for ciprofloxacin, levofloxacin, and CP-115,953 are shown for the mutant enzymes.
Figure 4.
Structures of the quinolones and quinazolinedione utilized in this study. Ciprofloxacin, levofloxacin, moxifloxacin, norfloxacin, and sparfloxacin are clinically-relevant fluoroquinolones (10). CP-115,953 is an experimental fluoroquinolone that displays high activity against both prokaryotic and eukaryotic type II enzymes (42, 56, 57). 8-Methyl-quinazoline-2,4-dione is a quinolone-related compound that previously has been shown to overcome quinolone-resistance mutations, including the GyrAS83W mutation, in E. coli gyrase (60).
Results of DNA cleavage assays are shown in Figure 5. All of the quinolones enhanced DNA cleavage mediated by wild-type topoisomerase IV (left panel). Ciprofloxacin was the most potent quinolone and, together with levofloxacin, was the most efficacious. Although sparfloxacin increased DNA cleavage several-fold, it was the least potent and efficacious quinolone examined.
As predicted from quinolone resistance studies with B. anthracis cultures (22-24) and previous studies with gyrase and topoisomerase IV from other bacterial species that carry mutations at the “Ser83 equivalent” residue (34, 48, 58-60), ciprofloxacin, levofloxacin, and CP-115,953 displayed decreased activity against GrlAS81F and GrlAS81Y topoisomerase IV (Figure 5, center and right panels, respectively). The quinolone concentration required to triple levels of enzyme-mediated DNA cleavage (CC3) rose ~27– to 42-fold with GrlAS81F topoisomerase IV (as compared to the wild-type enzyme) and ~10– to 17-fold with GrlAS81Y topoisomerase IV (Table 1). In order to determine the effects of the Ser81 mutations on the efficacy of quinolone action, maximal levels of DNA cleavage for wild-type, GrlAS81F, and GrlAS81Y B. anthracis topoisomerase IV were obtained by extending the drug concentration range. As seen in Figure 6, levels of DNA cleavage observed with the mutant enzymes at 300 μM ciprofloxacin, levofloxacin, or CP-115,953 approached those seen at lower drug concentrations with wild-type topoisomerase IV. Quinolone efficacy (as determined by the maximal level of DNA cleavage) ranged from 0.90 to 1.06 for GrlAS81F topoisomerase IV (compared to that seen with the wild-type enzyme) and 0.77 to 0.95 for GrlAS81Y topoisomerase IV (Table 1).
Table 1.
Potencya and efficacy of drugs against wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV from B. anthracis.
| WT |
GrlAS81F |
GrlAS81Y |
||||
|---|---|---|---|---|---|---|
| Drug | CC3 μM |
Max. DNA cleavage % |
CC3 μM |
Max. DNA cleavage % |
CC3 μM |
Max. DNA cleavage % |
| Ciprofloxacin | 0.53 | 30.0 | 22.3 (42.1)b | 26.9 (0.90)b | 9.1 (17.2)b | 23.4 (0.78)b |
| Levofloxacin | 1.45 | 30.4 | 38.8 (26.8) | 29.3 (0.96) | 15.0 (10.3) | 23.5 (0.77) |
| CP-115,953 | 0.85 | 23.5 | 30.8 (36.2) | 25.0 (1.06) | 8.2 (9.65) | 22.4 (0.95) |
| 8-Methyl-quinazoline- 2,4-dione |
0.20 | 29.0 | 0.30 (1.50) | 39.5 (1.36) | 0.18 (0.90) | 30.3 (1.04) |
CC3, the concentration of drug required to triple the percent DNA cleavage observed in the absence of drug, is used as an indicator of potency.
Values in parentheses represent a relative comparison of mutant values to wild-type values, as calculated by dividing the mutant value by the corresponding wild-type value.
Figure 6.
DNA cleavage induced by GrlAS81F and GrlAS81Y topoisomerase IV at high quinolone concentrations. A titration is shown for ciprofloxacin and GrlAS81F (S81F, red circles) or GrlAS81Y (S81Y, blue triangles) topoisomerase IV. Results with low concentrations of ciprofloxacin (up to 30 μM, the concentration that generated maximal DNA cleavage) and the wild-type enzyme (WT, black squares) are shown for reference. The inset shows results for levofloxacin (Levo, solid bars) and CP-115,953 (953, stippled bars) with wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV (black, red, and blue, respectively). Quinolone concentrations that generated maximal levels of DNA cleavage (20 μM and 300 μM for the wild-type and mutant enzymes, respectively) were used. Error bars represent the standard deviation of three or more independent experiments.
As above, these results have important implications for the potential treatment of quinolone-resistant strains of B. anthracis. They suggest that it should be possible to overcome drug resistance in B. anthracis if quinolones or quinolone-like drugs with higher affinities for the most common mutant type II enzymes can be developed.
Although the underlying basis for quinolone resistance generated by “Ser83” mutations has yet to be determined, it has been suggested that it is related to the ability of quinolones to bind Mg2+ ions. Quinolones require divalent metal ions in order to unwind DNA (59, 61, 62), and it has long been assumed that Mg2+ is required to coordinate the interactions of quinolones in the enzyme-DNA complex (10, 63, 64). Based on the ability of ciprofloxacin to alter conformational equlibria in E. coli GyrA (but not GyrAS83W) in the presence of Mg2+ ions, Sissi et al. (64) suggested that “Ser83” was involved in mediating quinolone-Mg2+-protein interactions. A recent crystallographic structure of a DNA cleavage complex formed with Acinetobacter baumannii topoisomerase IV in the presence of moxifloxacin supports this conclusion (28). The structure shows a Mg2+ ion interacting with the C3/C4 keto acid of moxifloxacin and with the enzyme through four water molecules. Two of these water molecules appear to be coordinated by Ser84 (which is homologous to Ser81 in B. anthracis GrlA and Ser83 in E. coli GyrA) (28). This Mg2+-mediated interaction appears to be one of the primary points of contact between the quinolone and the type II enzyme. Based on these findings, the authors proposed that mutation of the serine residue causes drug resistance by disrupting the proper coordination of the water-Mg2+ interaction that bridges the quinolone to the enzyme (28).
If the above hypothesis is correct, then quinolone-like drugs that do not require the water-Mg2+ bridge might be relatively unaffected by “Ser83” mutations. In this regard, quinazolinediones lack the C3/C4 keto acid that is characteristic of quinolones (see Figure 4). Although it has not been demonstrated directly, it is likely that quinazolinediones do not bind the Mg2+ ion that bridges quinolones to the protein.
Quinazolinediones display similar or better activity than quinolones against a variety of Gram-positive and Gram-negative bacterial species in vitro (35, 59, 65, 66). Furthermore, the efficacy of PD 0305970 (a quinazolinedione that is similar to the 8-methyl-quinazoline-2,4-dione used in the present work) in a murine acute lethal Gram-positive infection model was greater than that of either ciprofloxacin or levofloxacin (35). Previous studies have shown that cultures of quinolone-resistant E. coli (66) and Streptococcus pneumoniae (59) harboring “Ser83” mutations in topoisomerase IV and/or gyrase retain sensitivity to quinazolinediones. In addition, a limited number of quinazolinediones have been shown to induce high levels of DNA cleavage mediated by E. coli gyrase (60) and Streptococcus pneumoniae gyrase and topoisomerase IV that contain “Ser83” mutations (59).
Therefore, the effects of 8-methyl-quinazoline-2,4-dione on DNA cleavage mediated by wild-type, GrlAS81F, and GrlAS81Y B. anthracis topoisomerase IV were determined (Figure 5 and Table 1). The quinazolinedione-induced increase in DNA cleavage mediated by wild-type topoisomerase IV was similar to that seen with the quinolones. However, this drug was ~2.5 times more potent (as reflected by the CC3 value) than ciprofloxacin. Dramatic differences were seen for 8-methyl-quinazoline-2,4-dione with the GrlAS81F and GrlAS81Y mutant enzymes. In contrast to the quinolones, little to no resistance was observed with the quinazolinedione. With GrlAS81F topoisomerase IV, the CC3 concentration for the quinazolinedione rose only 1.5-fold and the level of maximal DNA cleavage was actually 36% higher than observed with the wild-type enzyme. With GrlAS81Y topoisomerase IV, potency and efficacy both were slightly better than those seen with the wild-type enzyme.
The striking sensitivity of GrlAS81F and GrlAS81Y topoisomerase IV to 8-methyl-quinazoline-2,4-dione is consistent with the hypothesis of Wohlkonig et al. (28) and suggests that the proposed water-Mg2+ bridge that coordinates the bacterial type II enzyme with quinolones is a major determinant for drug action. Furthermore, it provides a unique opportunity to address the proposed mechanism for quinolone resistance caused by mutations in “Ser83.” Therefore, the Mg2+-dependence of DNA cleavage mediated by wild-type and GrlAS81F topoisomerase IV in the presence of ciprofloxacin was compared to that seen with 8-methyl-quinazoline-2,4-dione. Results are shown in Figure 7. In order to facilitate direct comparisons between each different combination of drug and enzyme, levels of DNA cleavage generated in the presence of 10 mM Mg2+ were normalized to 100% for each drug-enzyme pair.
Figure 7.
Effects of Mg2+ on DNA cleavage mediated by wild-type and GrlAS81F topoisomerase IV in the presence of ciprofloxacin and 8-methyl-quinazoline-2,4-dione. Results are shown for 50 μM ciprofloxacin (Cipro, left panel) and 10 μM 8-methyl-quinazoline-2,4-dione (Dione, right panel) with the wild-type (WT, black squares) and GrlAS81F (S81F, red circles) enzymes. DNA cleavage for each drug-enzyme pair was normalized to 100% at 10 mM Mg2+ to facilitate direct comparisons. Error bars represent the standard deviation of three or more independent experiments.
The quinolone and quinazolinedione displayed similar requirements for Mg2+ when wild-type topoisomerase IV was employed; half-maximal and maximal DNA cleavage were observed at ~0.35 mM and ~1 mM Mg2+, respectively. In contrast, the two drugs displayed markedly different Mg2+ requirements for DNA cleavage mediated by GrlAS81F. While the metal ion utilization for 8-methyl-quinazoline-2,4-dione closely resembled that seen with the wild-type enzyme (p > 0.05), ciprofloxacin required higher levels of Mg2+ to support DNA cleavage. Levels of Mg2+ required to generate half-maximal and maximal DNA cleavage rose to ~0.75 mM and ~2.5 mM Mg2+, respectively, in the presence of the quinolone. Both of these values are significantly higher than those seen with wild-type topoisomerase IV (p = 0.002 and 0.01, respectively).
The above findings strongly suggest that the GrlA S81->F mutation in B. anthracis topoisomerase IV disrupts the water-Mg2+ bridge that mediates quinolone-protein binding in the cleavage complex. This conclusion supports the hypothesis that quinolone resistance results from the disruption of this critical quinolone-protein interaction (28) and also provides an explanation for the lack of quinazolinedione resistance seen with GrlAS81F.
Biochemical Mechanism for Quinolone Resistance in GrlAS81F and GrlAS81Y Topoisomerase IV
The differential requirement for Mg2+ by quinolones and 8-methyl-quinazoline-2,4-dione has the potential to alter a number of drug actions against wild-type and mutant topoisomerase IV. Therefore, several experiments were carried out to further characterize the biochemical mechanism of quinolone resistance in B. anthracis topoisomerase IV and the differences between quinolones and quinazolinediones. First, sites of DNA cleaved by wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV were examined in the presence of ciprofloxacin, levofloxacin, CP-115,953, and 8-methyl-quinazoline-2,4-dione (Figure 8). Some differences in site utilization were seen, and overall levels of quinolone-induced DNA cleavage mediated by the mutant enzymes dropped. However, no major differences in cleavage site specificity were observed for any drug-enzyme combination. Although some DNA cleavage bands became very faint in the presence of quinolones and the resistant enzymes, these bands were visible at higher drug concentrations (data not shown).
Figure 8.
Effects of quinolones and 8-methyl-quinazoline-2,4-dione on sites of DNA cleavage mediated by wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV. An autoradiogram of a polyacrylamide gel identifying DNA sites cleaved by the wild-type (WT), GrlAS81F (S81F), and GrlAS81Y (S81Y) enzymes is shown. Reactions contained no enzyme (No topo), or topoisomerase IV in the presence of the indicated concentrations of Mg2+, Ca2+, or ciprofloxacin (Cipro), levofloxacin (Levo), CP-115,953 (953), or 8-methyl-quinazoline-2,4-dione (Dione). Mg2+ (10 mM) was used in all drug-containing reactions. The autoradiogram is representative of three or more independent experiments.
These findings indicate that the resistance of GrlAS81F and GrlAS81Y topoisomerase IV to quinolones and the ability of 8-methyl-quinazoline-2,4-dione to overcome the resistance are not related to the selection of DNA cleavage sites by B. anthracis topoisomerase IV. They further suggest that drug-DNA interactions formed in the presence of the mutant enzymes are essentially those formed in the presence of wild-type topoisomerase IV (albeit weaker with quinolones) and that quinazolinedione-DNA interactions are similar to those established by quinolones in the cleavage complex.
The increased quinolone CC3 values for GrlAS81F and GrlAS81Y topoisomerase IV suggest that these drugs bind less tightly to the mutant enzymes than to wild-type topoisomerase IV. This finding is consistent with previous binding and kinetic studies with “Ser83” mutants of E. coli gyrase and Staphylococcus aureus topoisomerase IV (48, 58, 60, 67). In contrast, the high affinity of 8-methyl-quinazoline-2,4-dione for the mutant enzymes appears to be maintained. Therefore, the second experiment explored the basis underlying these differences by examining the ability of ciprofloxacin to compete with the quinazolinedione.
Competition assays took advantage of the fact that lower levels of DNA cleavage were observed in the presence of the quinolone. Therefore, the relative contributions of ciprofloxacin and 8-methyl-quinazoline-2,4-dione to DNA cleavage were determined by comparing levels of scission generated in the presence of both drugs to the cleavage observed in the presence of either drug alone.
Results are shown for GrlAS81F topoisomerase IV in Figure 9. Similar results were observed with GrlAS81Y topoisomerase IV (data not shown). A concentration of 20 μM 8-methyl-quinazoline-2,4-dione, which is saturating, was used for these experiments. Ciprofloxacin concentrations ranged from 0-150 μM; greater concentrations of the quinolone produced high levels of cleavage that interfered with the analysis of the assays.
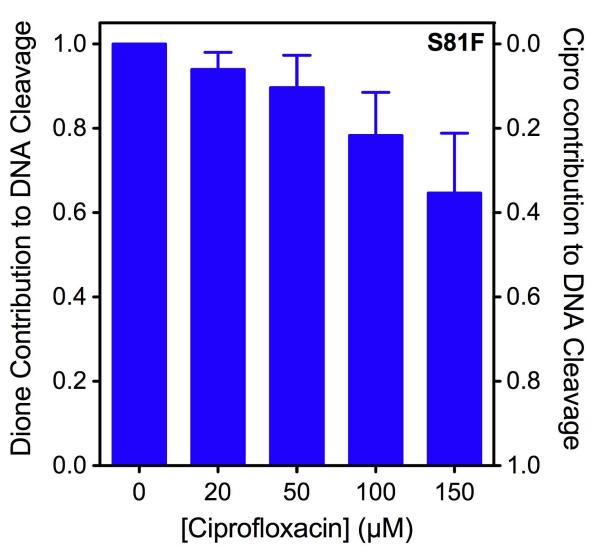
Figure 9.
Competition between ciprofloxacin and 8-methyl-quinazoline-2,4-dione. The ability of 0-150 μM ciprofloxacin to compete with 20 μM 8-methyl-quinazoline-2,4-dione for GrlAS81F topoisomerase IV was determined using DNA cleavage assays. Both drugs were added to reaction mixtures simultaneously. The relative contribution of the quinazolinedione (Dione, left axis) to the total level of DNA cleavage was calculated as follows: (% DNA cleavage with both drugs – % DNA cleavage with ciprofloxacin only) ÷ (% DNA cleavage with quinazolinedione only – % DNA cleavage with ciprofloxacin only). The relative contribution of ciprofloxacin (Cipro) to the total level of DNA cleavage (1 minus the above equation) can be read from the right axis. Error bars represent the standard deviation of three or more independent experiments.
Even at quinolone concentrations that were 7.5 times higher than that of the quinazolinedione, ciprofloxacin contributed less than half of the drug-induced DNA cleavage (Figure 9). This finding suggests that ciprofloxacin is unable to compete effectively with 8-methyl-quinazoline-2,4-dione for binding to the GrlAS81F topoisomerase IV-DNA complex and provides further evidence that the mutation at Ser81 of B. anthracis topoisomerase IV disproportionately affects the affinity of the quinolone as compared to the quinazolinedione.
The third experiment assessed the ability of drugs to inhibit topoisomerase IV-mediated DNA religation. Although quinolones impair DNA religation mediated by type II topoisomerases, their effects range from modest (<2-fold) to strong (~10-fold), depending on the enzyme species and drug employed (42, 56-58). As seen in Figure 10, quinolones and the quinazolinedione (20 μM) had a similar moderate effect on DNA religation mediated by wild-type B. anthracis topoisomerase IV, increasing t1/2 values by ~3-fold (as compared to reactions carried out in the absence of drug, left panel). However, the two drug classes had markedly different effects on religation mediated by GrlAS81F and GrlAS81Y topoisomerase IV (middle and right panels, respectively). Whereas 8-methyl-quinazoline-2,4-dione (20 μM) maintained (or even increased) its ability to inhibit religation, very little inhibition was seen in the presence of 200 μM ciprofloxacin, levofloxacin, or CP-115,953. The abilities of quinolones and the quinazolinedione to inhibit religation mediated by GrlAS81F and GrlAS81Y differ significantly (p = 0.002 and 0.001, respectively) and imply that the resistance mutations impair quinolone function as well as binding.
Figure 10.
Effects of quinolones and 8-methyl-quinazoline-2,4-dione on the DNA religation activities of wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV. Results for assays carried out in the absence of drugs (No Drug, open triangle) or in the presence of ciprofloxacin (Cipro, blue circles), levofloxacin (Levo, yellow circles), CP-115,953 (953, open green squares), or 8-methyl-quinazoline-2,4-dione (Dione, red squares) are shown. DNA religation mediated by wild-type (WT), GrlAS81F (S81F), and GrlAS81Y (S81Y) topoisomerase IV are shown in the left, center, and right panels, respectively. Religation was assessed by monitoring the loss of double-stranded DNA breaks (linear product) over time. Cleavage at time zero was set to 100%. Quinolone concentrations were 20 μM in assays that examined wild-type topoisomerase IV and were increased to 200 μM in assays that examined the GrlAS81F and GrlAS81Y enzymes. The concentration of 8-methyl-quinazoline-2,4-dione was 20 μM in all assays. Reactions carried out in the absence of drugs replaced Mg2+ with Ca2+ in order to achieve readily quantifiable levels of DNA cleavage. Error bars represent the standard deviation of three or more independent experiments.
The fourth experiment examined the effects of resistance mutations on the persistence of drug-induced cleavage complexes. This was accomplished by establishing DNA cleavage-religation equilibria with wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV in the presence of ciprofloxacin (20 μM with the wild-type or 200 μM with the mutant enzymes) or 8-methyl-quinazoline-2,4-dione (20 μM), diluting reaction mixtures 20-fold, and monitoring the decay of cleavage complexes over time (Figure 11). The t1/2 for DNA cleavage induced by ciprofloxacin dropped ~20-fold with the mutant enzymes (p = 0.007 for both enzymes), while the drop seen with 8-methyl-quinazoline-2,4-dione was (at most) 30% (p > 0.05 for both enzymes). These findings indicate that quinolone resistance correlates with a decreased stability of the topoisomerase IV-drug-DNA ternary complex, while the high sensitivity of GrlAS81F and GrlAS81Y topoisomerase IV towards the quinazolinedione correlates with the maintenance of a stable ternary complex.
Figure 11.
Effects of ciprofloxacin and 8-methyl-quinazoline-2,4-dione on the persistence of ternary enzyme-drug-DNA cleavage complexes formed with wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV. Results are shown for ciprofloxacin (Cipro, left panel) and 8-methyl-quinazoline-2,4-dione (Dione, right panel) with the wild-type (WT, black squares), GrlAS81F (S81F, red circles), and GrlAS81Y (S81Y, blue triangles) enzymes. Initial DNA cleavage-religation reactions were allowed to come to equilibrium and were then diluted 20-fold with DNA cleavage buffer. The persistence of cleavage complexes was assessed by monitoring the loss of double-stranded DNA breaks (linear product) over time. Cleavage at time zero was set to 100%. The concentration of ciprofloxacin was 20 μM in assays that examined wild-type topoisomerase IV and was increased to 200 μM in assays that examined the GrlAS81F and GrlAS81Y enzymes. The concentration of 8-methyl-quinazoline-2,4-dione was 20 μM in all assays. Error bars represent the standard deviation of three or more independent experiments.
DISCUSSION
As described above, the most common quinolone-resistance mutations in B. anthracis topoisomerase IV (GrlAS81F and GrlAS81Y) (22-24) have little effect on the catalytic activity and DNA cleavage specificity of the enzyme, but significantly decrease the affinity of topoisomerase IV for several commonly prescribed quinolones. However, wild-type levels of DNA cleavage generated by GrlAS81F and GrlAS81Y topoisomerase IV could be achieved with high concentrations of quinolones (15 to 20 times higher than required to reach maximal levels of DNA cleavage with the wild-type enzyme), suggesting that it should be possible to overcome resistance if a more potent quinolone or related drug could be developed.
It has been proposed that “Ser83” mutations lead to quinolone resistance because they alter an amino acid residue that plays an important role in coordinating quinolones to type II topoisomerases through a water-Mg2+ bridge (28, 64). Therefore, we examined the effects of 8-methyl-quinazoline-2,4-dione, which lacks the keto acid used by quinolones to bind the Mg2+ ion, on DNA cleavage and religation mediated by wild-type, GrlAS81F, and GrlAS81Y topoisomerase IV. As predicted by the hypothesis, the quinazolinedione displayed high activity against all three enzymes. 8-Methyl-quinazoline-2,4-dione was more potent than the quinolones against the wild-type enzyme and was equally efficacious. Moreover, in marked contrast to the quinolones, the quinazolinedione maintained its potency and efficacy against the mutant enzymes.
In order to directly test the importance of the proposed water-Mg2+ bridge in coordinating quinolone-topoisomerase IV interactions, we analyzed the Mg2+ concentration dependence for drug-induced DNA cleavage mediated by wild-type and GrlAS81F B. anthracis topoisomerase IV. The requirement for higher Mg2+ concentrations to support quinolone-induced, but not quinazolinedione-induced, DNA cleavage by the mutant enzyme provides the first functional evidence supporting the importance of the water-Mg2+ bridge in coordinating quinolone-topoisomerase IV binding and the role of “Ser83” in this interaction.
Type II topoisomerases require two divalent metal ions per active site to support DNA cleavage (27, 49, 50, 68-71). One of the metal ions binds with high affinity to site A and mediates the chemistry of DNA scission and religation. The other binds with lower affinity to site B and appears to play a structural role aligning the double helix during these processes. The KD values for Mg2+ binding to metal ion sites A and B are ~0.1 mM and ~1 mM, respectively, for E. coli topoisomerase IV (50). Because half-maximal DNA cleavage was observed at ~0.35 mM Mg2+ for both the quinolone and the quinazolinedione with wild-type B. anthracis topoisomerase IV, it is likely that this value reflects the affinity of metal ion B rather than the metal ion used to mediate quinolone-protein binding. This supposition implies that the affinity of the metal ion involved in the protein-water-Mg2+-quinolone interaction is higher than that of active site metal ion B, and that the Mg2+-quinolone interaction only becomes limiting in the presence of “Ser83” (or potentially other) mutants that disrupt the coordination of the water-Mg2+ bridge. Consequently, it is probable that the GrlA S81->F mutation in B. anthracis topoisomerase IV actually lowers the affinity of the quinolone-bridging Mg2+ by more than the two-fold effect seen in Figure 7.
The increased Mg2+ concentration required to support ciprofloxacin-induced DNA cleavage by GrlAS81F topoisomerase IV suggests that the mutant protein can still bind the quinolone-bridging Mg2+, but does so with diminished affinity. Furthermore, since DNA cleavage assays utilize 10 mM Mg2+, which is significantly above the saturating metal ion concentration seen for the GrlAS81F enzyme (~2.5 mM), the decreased potency of quinolones against the resistant mutant enzymes cannot be attributed solely to decreased Mg2+ binding. Thus, it is likely that quinolones still interact with GrlAS81F topoisomerase IV through a water-Mg2+ bridge. However, because “Ser83” appears to coordinate two of the four water molecules that contact the Mg2+ ion, we propose that the water-Mg2+ bridge formed with the mutant enzymes is altered and less stable. As a result, quinolones display a decreased potency for DNA cleavage, exhibit a reduced ability to inhibit DNA religation, and form less stable cleavage complexes with the mutant enzymes. In contrast, because quinazolinediones presumably do not require the water-Mg2+ bridge to mediate their interactions with bacterial type II topoisomerases, mutation of the serine residue has little effect on the ability of 8-methyl-quinazoline-2,4-dione to inhibit enzyme-mediated DNA religation and form a stable ternary enzyme-drug-DNA complex.
In the present study, the DNA cleavage potency of 8-methyl-quinazoline-2,4-dione never decreased more than 1.5-fold with GrlAS81F or GrlAS81Y topoisomerase IV from B. anthracis. This is as compared to decreases in potencies of ~27– to 42-fold and ~10– to 17-fold, respectively, seen with a variety of quinolones. Similar results have been reported for other enzymes. Previous work that examined drug resistance accompanying a GyrAS83W mutation in E. coli gyrase reported that the DNA cleavage potency of 8-methyl-quinazoline-2,4-dione dropped ~5.6-fold as compared to a drop of ~400-fold seen with ciprofloxacin (60). Moreover, in a study that examined similar S. pneumoniae gyrase and topoisomerase IV resistance mutations, the potency of a related quinazolinedione (PD 0305970) dropped only ~2-fold and ~2.5-fold, respectively, as compared to drops of ~8-fold and >16-fold seen with ciprofloxacin (59). Thus, quinazolinediones represent quinolone-like drugs that retain high activity against quinolone-resistance mutations commonly found in clinical bacterial isolates.
The potential use of quinolone-resistant B. anthracis as a future bioweapon remains high (2, 5). Furthermore, quinolone resistance is rising in a wide variety of bacterial pathogens that infect humans (34-36). The present study provides a rational basis for the development of quinolone-like agents that overcome the major resistance mutations in bacterial type II topoisomerases. It also represents a first step towards identifying a drug that overcomes this resistance and could be an effective therapy for anthrax or other clinically-relevant quinolone-resistant bacterial infections.
ACKNOWLEDGEMENTS
We are grateful to Adam C. Ketron, MaryJean Campbell, and R. Hunter Lindsey for critical reading of the manuscript.
Footnotes
K.J.A. was a trainee under grant T32 CA09582 from the National Institutes of Health. This work was supported by National Institutes of Health research grant AI81775 (to C.L.T), National Institutes of Health grant AI87671 (to R.J.K.), Department of Defense Breast Cancer Research Program grant BC095831P1 (to D.E.G.), and National Institutes of Health research grant GM33944 (to N.O.)
REFERENCES
- 1.Mock M, Fouet A. Anthrax. Annu. Rev. Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- 2.Bossi P, Tegnell A, Baka A, Van Loock F, Hendriks J, Werner A, Maidhof H, Gouvras G. Bichat guidelines for the clinical management of anthrax and bioterrorism-related anthrax. Eurosurveillance. 2004;9:1–7. doi: 10.2807/esm.09.12.00500-en. [DOI] [PubMed] [Google Scholar]
- 3.Danzig R. Proliferation of biological weapons into terrorist hands. In: Campbell KM, editor. The Challenge of Proliferation: A Report from the Aspen Strategy Group. The Aspen Institute; Washington, D.C: 2005. pp. 65–84. [Google Scholar]
- 4.Schwartz M. Dr. Jekyll and Mr. Hyde: a short history of anthrax. Mol. Aspects Med. 2009;30:347–355. doi: 10.1016/j.mam.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Waterer GW, Robertson H. Bioterrorism for the respiratory physician. Respirology. 2009;14:5–11. doi: 10.1111/j.1440-1843.2008.01446.x. [DOI] [PubMed] [Google Scholar]
- 6.Hooper DC. Mode of action of fluoroquinolones. Drugs. 1999;58(Suppl. 2):6–10. doi: 10.2165/00003495-199958002-00002. [DOI] [PubMed] [Google Scholar]
- 7.Anderson VE, Osheroff N. Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Curr. Pharm. Des. 2001;7:337–353. doi: 10.2174/1381612013398013. [DOI] [PubMed] [Google Scholar]
- 8.Hooper DC. Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin. Infect. Dis. 2001;32(Suppl. 1):S9–S15. doi: 10.1086/319370. [DOI] [PubMed] [Google Scholar]
- 9.Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. Quinolones: action and resistance updated. Curr. Top. Med. Chem. 2009;9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson KM, Armstrong RE, Thompson D. Bayes, bugs, and bioterrorists: Lessons learned from the anthrax attacks. Center for Technology and National Security Policy, National Defense University Press; Washington, D.C: 2005. [Google Scholar]
- 12.Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 13.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 14.Velez-Cruz R, Osheroff N. Encyclopedia of Biological Chemistry. Elsevier Inc; 2004. DNA topoisomerases: type II; pp. 806–811. [Google Scholar]
- 15.Aubry A, Fisher LM, Jarlier V, Cambau E. First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2006;348:158–165. doi: 10.1016/j.bbrc.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 17.Deweese JE, Osheroff MA, Osheroff N. DNA topology and topoisomerases: teaching a “knotty” subject. Biochem. Molec. Biol. Educ. 2009;37:2–10. doi: 10.1002/bmb.20244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Deibler RW, Chan HS, Zechiedrich L. The why and how of DNA unlinking. Nuc. Acids Res. 2009;37:661–671. doi: 10.1093/nar/gkp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tretter EM, Lerman JC, Berger JM. A naturally chimeric type IIA topoisomerase in Aquifex aeolicus highlights an evolutionary path for the emergence of functional paralogs. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22055–22059. doi: 10.1073/pnas.1012938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nuc. Acids Res. 2009;37:738–749. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brook I, Elliott TB, Pryor HI, 2nd, Sautter TE, Gnade BT, Thakar JH, Knudson GB. In vitro resistance of Bacillus anthracis Sterne to doxycycline, macrolides and quinolones. Int. J. Antimicrob. Agents. 2001;18:559–562. doi: 10.1016/s0924-8579(01)00464-2. [DOI] [PubMed] [Google Scholar]
- 22.Price LB, Vogler A, Pearson T, Busch JD, Schupp JM, Keim P. In vitro selection and characterization of Bacillus anthracis mutants with high-level resistance to ciprofloxacin. Antimicrob. Agents Chemother. 2003;47:2362–2365. doi: 10.1128/AAC.47.7.2362-2365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grohs P, Podglajen I, Gutmann L. Activities of different fluoroquinolones against Bacillus anthracis mutants selected in vitro and harboring topoisomerase mutations. Antimicrob. Agents Chemother. 2004;48:3024–3027. doi: 10.1128/AAC.48.8.3024-3027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bast DJ, Athamna A, Duncan CL, de Azavedo JC, Low DE, Rahav G, Farrell D, Rubinstein E. Type II topoisomerase mutations in Bacillus anthracis associated with high-level fluoroquinolone resistance. J. Antimicrob. Chemother. 2004;54:90–94. doi: 10.1093/jac/dkh294. [DOI] [PubMed] [Google Scholar]
- 25.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 26.Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010;5:e11338. doi: 10.1371/journal.pone.0011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, Giordano I, Hann MM, Hennessy A, Hibbs M, Huang J, Jones E, Jones J, Brown KK, Lewis CJ, May EW, Saunders MR, Singh O, Spitzfaden CE, Shen C, Shillings A, Theobald AJ, Wohlkonig A, Pearson ND, Gwynn MN. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 28.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, Leydon VR, Miles TJ, Pearson ND, Perera RL, Shillings AJ, Gwynn MN, Bax BD. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 2010;17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 29.Freudenreich CH, Kreuzer KN. Localization of an aminoacridine antitumor agent in a type II topoisomerase-DNA complex. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11007–11011. doi: 10.1073/pnas.91.23.11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capranico G, Binaschi M. DNA sequence selectivity of topoisomerases and topoisomerase poisons. Biochim. Biophys. Acta. 1998;1400:185–194. doi: 10.1016/s0167-4781(98)00135-3. [DOI] [PubMed] [Google Scholar]
- 31.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 32.Pommier Y, Marchand C. Interfacial inhibitors of protein-nucleic acid interactions. Curr. Med. Chem. Anticancer Agents. 2005;5:421–429. doi: 10.2174/1568011054222337. [DOI] [PubMed] [Google Scholar]
- 33.Fournier B, Zhao X, Lu T, Drlica K, Hooper DC. Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 2000;44:2160–2165. doi: 10.1128/aac.44.8.2160-2165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drlica K, Zhao X. DNA Gyrase, Topoisomerase IV, and the 4-Quinolones. Microbiol. Mol. Biol. Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huband MD, Cohen MA, Zurack M, Hanna DL, Skerlos LA, Sulavik MC, Gibson GW, Gage JW, Ellsworth E, Stier MA, Gracheck SJ. In vitro and in vivo activities of PD 0305970 and PD 0326448, new bacterial gyrase/topoisomerase inhibitors with potent antibacterial activities versus multidrug-resistant gram-positive and fastidious organism groups. Antimicrob. Agents Chemother. 2007;51:1191–1201. doi: 10.1128/AAC.01321-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan-Linnell SK, Becnel Boyd L, Steffen D, Zechiedrich L. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob. Agents Chemother. 2009;53:235–241. doi: 10.1128/AAC.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong S, McPherson SA, Wang Y, Li M, Wang P, Turnbough CL, Jr., Pritchard DG. Characterization of the enzymes encoded by the anthrose biosynthetic operon of Bacillus anthracis. J. Bacteriol. 2010;192:5053–5062. doi: 10.1128/JB.00568-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Englund PT. The replication of kinetoplast DNA networks in Crithidia fasciculata. Cell. 1978;14:157–168. doi: 10.1016/0092-8674(78)90310-0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Mu F, Robinson B, Wang P. Concise route to the key intermediate for divergent synthesis of C7-substituted fluoroquinolone derivatives. Tetrahedron Lett. 2010;51:600–601. [Google Scholar]
- 40.Malik M, Marks KR, Mustaev A, Zhao X, Chavda K, Kerns RJ, Drlica K. Fluoroquinolone and quinazolinedione activities against wild-type and gyrase mutant strains of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 2011;55:2335–2343. doi: 10.1128/AAC.00033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fortune JM, Osheroff N. Merbarone inhibits the catalytic activity of human topoisomerase IIα by blocking DNA cleavage. J. Biol. Chem. 1998;273:17643–17650. doi: 10.1074/jbc.273.28.17643. [DOI] [PubMed] [Google Scholar]
- 42.Anderson VE, Gootz TD, Osheroff N. Topoisomerase IV catalysis and the mechanism of quinolone action. J. Biol. Chem. 1998;273:17879–17885. doi: 10.1074/jbc.273.28.17879. [DOI] [PubMed] [Google Scholar]
- 43.Baldwin EL, Byl JA, Osheroff N. Cobalt enhances DNA cleavage mediated by human topoisomerase IIα in vitro and in cultured cells. Biochemistry. 2004;43:728–735. doi: 10.1021/bi035472f. [DOI] [PubMed] [Google Scholar]
- 44.O’Reilly EK, Kreuzer KN. A unique type II topoisomerase mutant that is hypersensitive to a broad range of cleavage-inducing antitumor agents. Biochemistry. 2002;41:7989–7997. doi: 10.1021/bi025897m. [DOI] [PubMed] [Google Scholar]
- 45.Robinson MJ, Osheroff N. Effects of antineoplastic drugs on the post-strand-passage DNA cleavage/religation equilibrium of topoisomerase II. Biochemistry. 1991;30:1807–1813. doi: 10.1021/bi00221a012. [DOI] [PubMed] [Google Scholar]
- 46.Gentry AC, Pitts SL, Jablonsky MJ, Bailly C, Graves DE, Osheroff N. Interactions between the etoposide derivative F14512 and human type II topoisomerases: implications for the C4 spermine moiety in promoting enzyme-mediated DNA cleavage. Biochemistry. 2011;50:3240–3249. doi: 10.1021/bi200094z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osheroff N, Zechiedrich EL. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: trapping the covalent enzyme-DNA complex in an active form. Biochemistry. 1987;26:4303–4309. doi: 10.1021/bi00388a018. [DOI] [PubMed] [Google Scholar]
- 48.Barnard FM, Maxwell A. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87) Antimicrob. Agents Chemother. 2001;45:1994–2000. doi: 10.1128/AAC.45.7.1994-2000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deweese JE, Burgin AB, Osheroff N. Human topoisomerase IIα uses a two-metal-ion mechanism for DNA cleavage. Nuc. Acids Res. 2008;36:4883–4893. doi: 10.1093/nar/gkn466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pitts SL, Liou GF, Mitchenall LA, Burgin AB, Maxwell A, Neuman KC, Osheroff N. Use of divalent metal ions in the DNA cleavage reaction of topoisomerase IV. Nuc. Acids Res. 2011;39:4808–4817. doi: 10.1093/nar/gkr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munoz R, De la Campa AG. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan X-S, Ambler J, Mehtar S, Fisher LM. Involvement of Topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan X-S, Fisher LM. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan X-S, Fisher LM. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Higgins PG, Fluit AC, Schmitz FJ. Fluoroquinolones: structure and target sites. Curr. Drug Targets. 2003;4:181–190. doi: 10.2174/1389450033346920. [DOI] [PubMed] [Google Scholar]
- 56.Robinson MJ, Martin BA, Gootz TD, McGuirk PR, Moynihan M, Sutcliffe JA, Osheroff N. Effects of quinolone derivatives on eukaryotic topoisomerase II. A novel mechanism for enhancement of enzyme-mediated DNA cleavage. J. Biol. Chem. 1991;266:14585–14592. [PubMed] [Google Scholar]
- 57.Anderson VE, Zaniewski RP, Kaczmarek FS, Gootz TD, Osheroff N. Quinolones inhibit DNA religation mediated by Staphylococcus aureus topoisomerase IV: changes in drug mechanism across evolutionary boundaries. J. Biol. Chem. 1999;274:35927–35932. doi: 10.1074/jbc.274.50.35927. [DOI] [PubMed] [Google Scholar]
- 58.Anderson VE, Zaniewski RP, Kaczmarek FS, Gootz TD, Osheroff N. Action of quinolones against Staphylococcus aureus topoisomerase IV: basis for DNA cleavage enhancement. Biochemistry. 2000;39:2726–2732. doi: 10.1021/bi992302n. [DOI] [PubMed] [Google Scholar]
- 59.Pan XS, Gould KA, Fisher LM. Probing the differential interactions of quinazolinedione PD 0305970 and quinolones with gyrase and topoisomerase IV. Antimicrob. Agents Chemother. 2009;53:3822–3831. doi: 10.1128/AAC.00113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oppegard LM, Streck KR, Rosen JD, Schwanz HA, Drlica K, Kerns RJ, Hiasa H. Comparison of in vitro activities of fluoroquinolone-like 2,4- and 1,3-diones. Antimicrob. Agents Chemother. 2010;54:3011–3014. doi: 10.1128/AAC.00190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tornaletti S, Pedrini AM. Studies on the interaction of 4-quinolones with DNA by DNA unwinding experiments. Biochim. Biophys. Acta. 1988;949:279–287. doi: 10.1016/0167-4781(88)90153-4. [DOI] [PubMed] [Google Scholar]
- 62.Palu G, Valisena S, Ciarrocchi G, Gatto B, Palumbo M. Quinolone binding to DNA is mediated by magnesium ions. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9671–9675. doi: 10.1073/pnas.89.20.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan JY, Sun D, Yu H, Kerwin SM, Hurley LH. Self-assembly of a quinobenzoxazine-Mg2+ complex on DNA: a new paradigm for the structure of a drug-DNA complex and implications for the structure of the quinolone bacterial gyrase-DNA complex. J. Med. Chem. 1995;38:408–424. doi: 10.1021/jm00003a003. [DOI] [PubMed] [Google Scholar]
- 64.Sissi C, Perdona E, Domenici E, Feriani A, Howells AJ, Maxwell A, Palumbo M. Ciprofloxacin affects conformational equilibria of DNA gyrase A in the presence of magnesium ions. J. Mol. Biol. 2001;311:195–203. doi: 10.1006/jmbi.2001.4838. [DOI] [PubMed] [Google Scholar]
- 65.Tran TP, Ellsworth EL, Sanchez JP, Watson BM, Stier MA, Showalter HD, Domagala JM, Shapiro MA, Joannides ET, Gracheck SJ, Nguyen DQ, Bird P, Yip J, Sharadendu A, Ha C, Ramezani S, Wu X, Singh R. Structure-activity relationships of 3-aminoquinazolinediones, a new class of bacterial type-2 topoisomerase (DNA gyrase and topo IV) inhibitors. Bioorg. Med. Chem. Lett. 2007;17:1312–1320. doi: 10.1016/j.bmcl.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 66.German N, Malik M, Rosen JD, Drlica K, Kerns RJ. Use of gyrase resistance mutants to guide selection of 8-methoxy-quinazoline-2,4-diones. Antimicrob. Agents Chemother. 2008;52:3915–3921. doi: 10.1128/AAC.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Willmott CJ, Maxwell A. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 1993;37:126–127. doi: 10.1128/aac.37.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noble CG, Maxwell A. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J. Mol. Biol. 2002;318:361–371. doi: 10.1016/S0022-2836(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 69.Deweese JE, Burch AM, Burgin AB, Osheroff N. Use of divalent metal ions in the DNA cleavage reaction of human type II topoisomerases. Biochemistry. 2009;48:1862–1869. doi: 10.1021/bi8023256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deweese JE, Osheroff N. The Use of Divalent Metal Ions by Type II Topoisomerases. Metallomics. 2010;2:450–459. doi: 10.1039/c003759a. [DOI] [PMC free article] [PubMed] [Google Scholar]