SUMMARY
Cells keep their energy balance and avoid oxidative stress by regulating mitochondrial movement, distribution, and clearance. We report here that two Parkinson’s disease proteins, the Ser/Thr-kinase PINK1 and ubiquitin-ligase Parkin, participate in this regulation by arresting mitochondrial movement. PINK1 phosphorylates Miro, a component of the primary motor/adaptor complex that anchors kinesin to the mitochondrial surface. The phosphorylation of Miro activates proteasomal degradation of Miro in a Parkin-dependent manner. Removal of Miro from the mitochondrion also detaches kinesin from its surface. By preventing mitochondrial movement, the PINK1/Parkin pathway may quarantine damaged mitochondria prior to their clearance. PINK1 has been shown to act upstream of Parkin but the mechanism corresponding to this relationship has not been known. We propose that PINK1 phosphorylation of substrates triggers the subsequent action of Parkin and the proteasome.
INTRODUCTION
Parkinson’s disease (PD) is one of the most common neurodegenerative disorders. The causes of neuronal death in PD remain elusive, but mitochondrial malfunction is likely to be an important component (Whitworth and Pallanck, 2009; Youle and Narendra, 2010). Two hereditary forms of early onset, recessive PD arise from mutations in PINK1, a Ser/Thr kinase, and Parkin, an E3 ubiquitin ligase (Kitada et al., 1998; Valente et al., 2004). These genes affect mitochondrial function and morphology (Clark et al., 2006; Exner et al., 2007; Park et al., 2006; Poole et al., 2008) and participate in a pathway for mitochondrial quality control (Geisler et al., 2010; Narendra et al., 2008, 2010; Whitworth and Pallanck, 2009; Youle and Narendra, 2010).
Genetic studies in Drosophila established that PINK1 is upstream of Parkin (Clark et al., 2006; Exner et al., 2007; Park et al., 2006; Poole at al., 2008). PINK1 resides partially on the outer mitochondrial membrane (OMM) with its C-terminal kinase domain facing the cytosol (Zhou et al., 2008), though it is also imported into mitochondria. The presence of PINK1 on the mitochondrial surface is enhanced by mitochondrial damage and depolarization, which prevent mitochondrial import of PINK1 and its PARL-dependent cleavage (Deas et al., 2010; Jin et al., 2010). No substrates of PINK1 on the mitochondrial surface have presently been identified, but, by an unknown mechanism, PINK1 recruits Parkin to depolarized mitochondria. Activation of the PINK1/Parkin pathway is required to remove the damaged mitochondria (Geisler et al., 2010; Narendra et al., 2008, 2010). Consequently, neurons lacking PINK1 or Parkin may be impaired in mitochondrial clearance and therefore, in response to other stresses, be prone to degeneration (Gandhi et al., 2009; Surmeier et al., 2010). It is unknown how PINK1 and Parkin promote the clearance of the damaged mitochondria, nor how PINK1 recruits Parkin to mitochondria. Little is known about the substrates of these enzymes, although recent data indicate that mitofusin is a Parkin substrate (Poole et al., 2010; Karbowski and Youle, 2011; Ziviani et al., 2010). Parkin causes degradation of mitofusin (Clark et al., 2006; Exner et al., 2007; Park et al., 2006; Poole et al., 2008) and the zinc-finger protein PARIS/ZNF746 (Shin et al., 2011). Additionally, several mitochondrial proteins are down-regulated upon Parkin expression in HeLa cells and may therefore also be substrates of Parkin (Chan et al., 2011). Thus the Parkin pathway may regulate multiple aspects of mitochondrial biology.
Recently, PINK1 immunoprecipitates were found to include components of the motor/adaptor complex mediating axonal transport of mitochondria (Weihofen et al., 2009). This complex contains kinesin-1 heavy chain (KHC) and two essential adaptor proteins, Miro and milton (Glater et al., 2006; Stowers et al., 2002). Miro (also called RhoT) resides in the OMM (Fransson et al., 2006) and milton (also called OIP98/106, and TRAK1/2), by binding both Miro and KHC, recruits KHC to the mitochondrial surface.
The Miro/Milton/KHC complex is conserved from flies to mammals (Brickley et al., 2005; Fransson et al., 2006; Glater et al., 2006; Stowers et al., 2002), and is also the means by which mitochondrial movement is regulated by cytoplasmic Ca2+ (MacAskill et al., 2009; Saotome et al., 2008; Wang and Schwarz, 2009a).
The association of PINK1 with this motor/adaptor complex suggested that PINK1 might also be a regulator of mitochondrial motility. We show that both PINK1 and Parkin halt mitochondrial movement; PINK1 phosphorylates Miro and thereby initiates the rapid degradation of Miro through a Parkin- and proteasome-dependent pathway. The resulting release of kinesin from mitochondria may be an initial quarantining step prior to mitophagy or a mechanism to spatially restrict their deleterious effects.
RESULTS
Overexpression of PINK1 or Parkin in Rat Hippocampal Axons Decreases Mitochondrial Movement
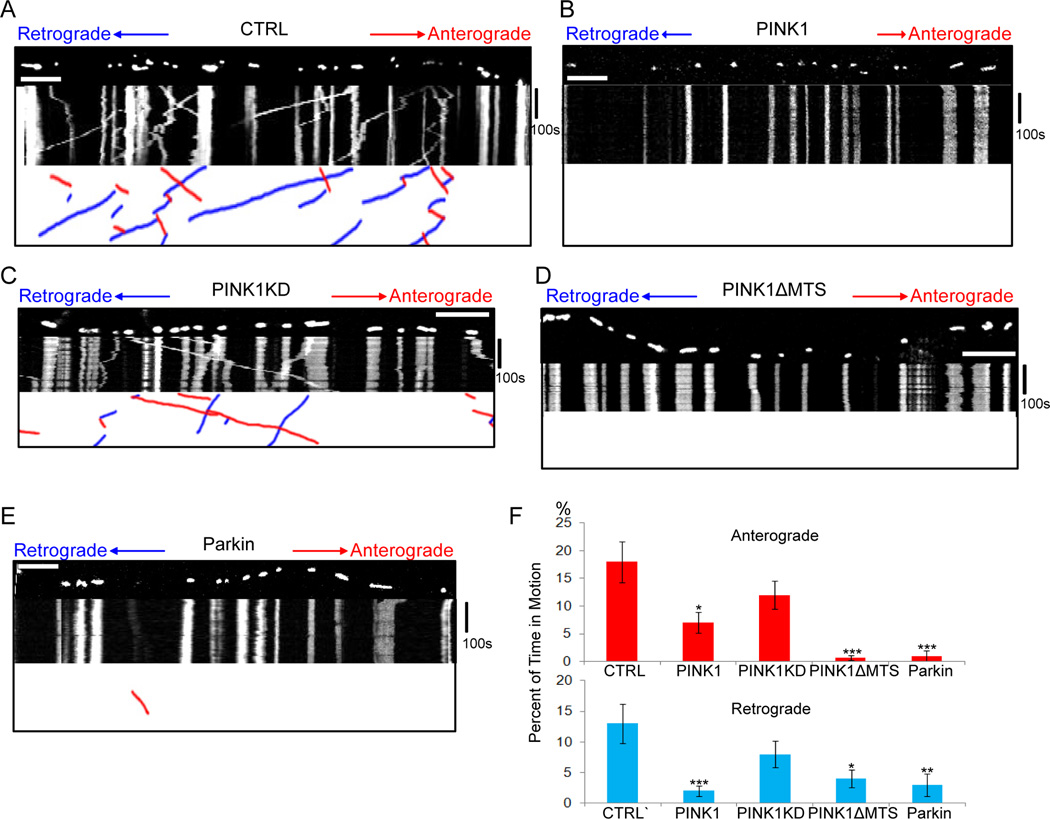
We examined the effects of PINK1 on axonal transport of mitochondria by live imaging in cultured rat hippocampal neurons. We co-expressed mito-dsRed to label mitochondria, synaptophysin-YFP (SYP-YFP) to label axons (Wang and Schwarz, 2009a, b), and PINK1-Flag (Weihofen et al., 2009). By retrospective staining, 96.7±2.27% of neurons that were mito-dsRed and SYP-YFP positive were also PINK1-Flag positive (Figure S1). We determined the percent of time mitochondria spent in motion, their average velocity, frequency of stopping and reversing direction, density and length (Table S1A). Percent of time in motion was selected as the primary parameter for quantifying motility, rather than percent of mitochondria that move, because the former distinguishes mitochondria that move for only a fraction of the observation period from those that move continuously. Expression of PINK1 significantly inhibited mitochondrial movement in both directions and, as expected from previous studies, shortened their length (Deng et al., 2008; Poole et al., 2008), but did not significantly alter their average velocities (Figure 1A–B, F, Movies S1A–B, Table S1A). Expression of a kinase-dead mutant (PINK1KD) did not arrest mitochondria, but expression of a mutant lacking the mitochondrial targeting sequence did (PINK1ΔMTS) (Figure 1A–D, F, Table S1A). The targeting sequence has previously been shown to be dispensable for PINK1 binding to mitochondria via the milton/Miro complex (Weihofen et al., 2009).
Figure 1. PINK1 or Parkin Overexpression Arrests Mitochondria in Rat Hippocampal Axons.
(A–E) Mitochondrial movement in representative axons transfected with mito-dsRed. The first frame of each live-imaging series is shown above a kymograph generated from the movie. The x axis of each represents mitochondrial position and the y axis is time (moving from top to bottom). Vertical white lines correspond to stationary mitochondria and diagonal lines are moving mitochondria. To illustrate the interpretation of kymographs, the lower panels extract only the moving mitochondria, which are shown as red (anterograde) or blue (retrograde) diagonal lines. Axons were tranfected with mito-dsRed alone (A), or together with PINK1-Flag (B), kinase-dead PINK1 (PINK1KD-Flag) (C), PINK1ΔMTS-Flag (D), or YFP-Parkin (E). (F) From kymographs as in (A–E), the percent of time each mitochondrion was in motion was determined and averaged. n=81–136 mitochondria from 8 axons and 4 separate transfections per genotype. * P<0.05, ** P<0.01, *** P<0.001, and error bars represent mean±S.E.M. here and for all figures unless otherwise stated. Scale bars, 10 µm. See also Figure S1, Movies S1, and Tables S1.
To examine the effects of Parkin, we co-expressed mito-dsRed and YFP-Parkin (Narendra et al., 2008) and selected axons by morphological criteria (Wang and Schwarz, 2009a) in YFP-positive cells. Expression of Parkin also significantly decreased the amount of mitochondrial movement, but not velocity, in both directions (Figure 1E–F, Movie S1C, Table S1A). The inhibition was specific to mitochondria, because movement of synaptic vesicles labeled with SYP-YFP was not affected by either PINK1 or Parkin expression (Table S1B). The density of axonal mitochondria was also significantly decreased by PINK1 or Parkin expression (Table S1A), as might be expected from either a diminished movement into the axon or increased mitophagy.
PINK1 Functions Upstream of Parkin to Inhibit Mitochondrial Motility
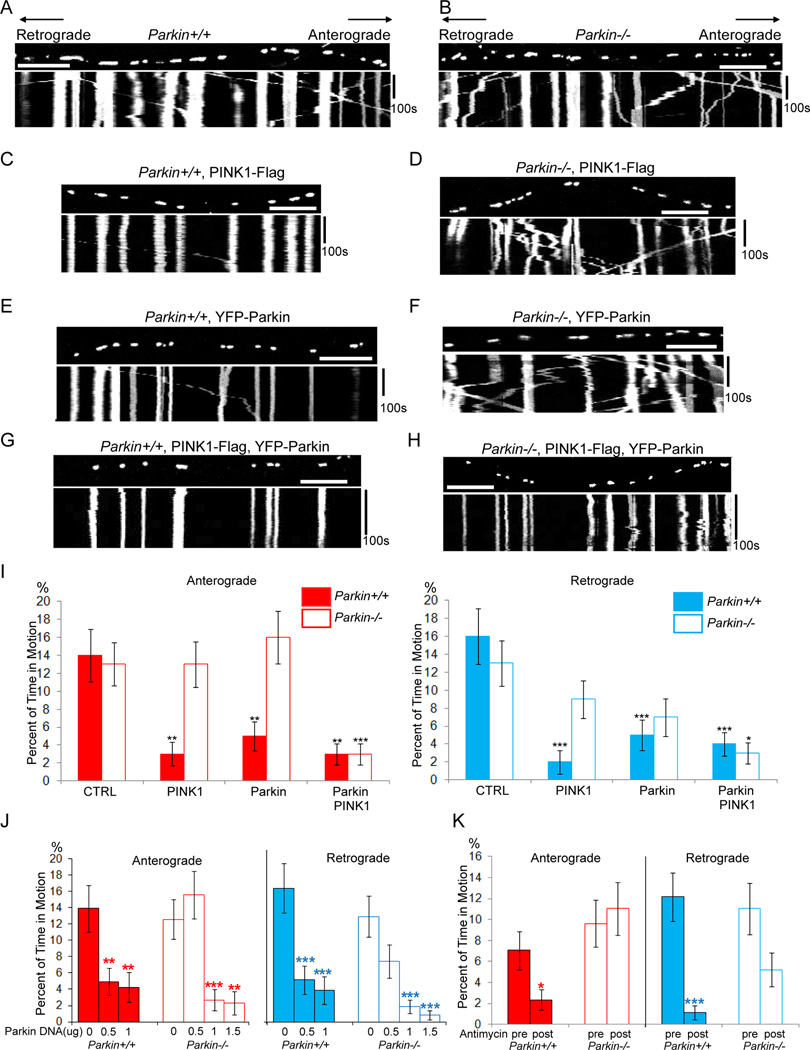
Because PINK1 requires Parkin for control of mitochondrial morphology and clearance (Clark et al., 2006; Exner et al., 2007; Narendra et al., 2010; Park et al., 2006), we asked whether PINK1 also required Parkin for regulating mitochondrial motility. We did not detect any differences in mitochondrial dynamics, density or length between cultured embryonic hippocampal neurons from wildtype and Parkin−/− mice (Goldberg et al., 2003) in the absence of PINK1 overexpression (Table S2A). However, whereas expression of PINK1 halted mitochondria in wildtype axons, it failed to do so in the Parkin−/− background (Figure 2A–D, I; Movies S2; Table S2A), even when cells were transfected with three-fold higher concentrations of DNA than were needed to arrest mitochondria in wildtype axons (Figure S2A). The efficacy of PINK1 expression in Parkin−/− neurons could be recovered if Parkin was co-expressed (Figure 2H, I, Table S2A). Thus PINK1-dependent mitochondrial arrest requires Parkin. The arrest of mitochondrial motility by Parkin in the absence of overexpressed PINK1 was dependent on the level of Parkin expression. In wildtype neurons, transfection with 0.5µg Parkin DNA/well was sufficient to arrest both directions, but in Parkin−/− neurons 1µg DNA/well was necessary (Figure 2E, F, I, J). Presumably the higher level of transfection was necessary to compensate for the lack of endogenous Parkin.
Figure 2. PINK1-dependent Mitochondrial Arrest Requires Parkin.
Mouse hippocampal axons were transfected with mito-dsRed and analyzed with in kymographs, as in Figure 1. (A) A wildtype(Parkin+/+) axon. (B) A Parkin−/− axon in which mitochondrial movement appears normal. (C, D) Expression of PINK1-Flag arrested mitochondria in a wildtype axon (C) but not in a Parkin−/− axon (D). (E, F) Tranfection with 0.5 µg YFP-Parkin DNA per well arrested mitochondria in wildtype (E), but not Parkin−/− (F) axons. (G, H) Tranfection with 0.5 µg YFP-Parkin DNA allowed PINK1 expression to arrest mitochondria in both wildtype (G) or Parkin−/− (H) axons. (I) From kymographs as in (A–H), the percent of time each mitochondrion was in motion was determined and averaged. n=108–157 mitochondria from 8 axons and 4 separate transfections per genotype. (J) Mitochondrial motility as a function of the amount of transfected YFP-Parkin DNA/well of a 24-well plate. n=97–157 mitochondria from 8 axons and 4 separate transfections per genotype. (K) Movies were taken before and after 15 min incubation with 80 µM Antimycin A and analyzed by kymograph (see Figure S2C–D). n=140–174 mitochondria from 8 axons and 8 separate transfections per genotype. Before Antimycin A, Parkin+/+ and Parkin−/− axons did not significantly differ. P values were calculated by comparing a given genotype to its control in (I), to 0 µg Parkin DNA in (J), and by comparing before and after Antimycin A in (K). Scale bars, 10 µm. See also Figure S2, Movies S2 and Table S2.
Physiologically, the PINK1/Parkin pathway can be activated by mitochondrial depolarization (Geisler et al., 2010; Narendra et al., 2008, 2010). We therefore asked whether depolarization regulated motility via this pathway. When neurons were treated with 80 µM Antimycin A, a complex III inhibitor, YFP-Parkin accumulated on axonal mitochondria (Figure S2B), as has been observed on depolarized non-neuronal mitochondria (Narendra et al., 2008), and bidirectional motility decreased. This effect was only partially dependent on Parkin: mitochondria stopped after 15 min in wildtype but not Parkin−/− axons (Figure 2K, S2C, D, Table S2B); however by 30 min little movement was observed in either genotype (data not shown). Antimycin A also decreased mitochondrial size in both genotypes (Table S2B).
PINK1 Functions Upstream of Parkin to Inhibit Mitochondrial Motility in Drosophila Larval Axons
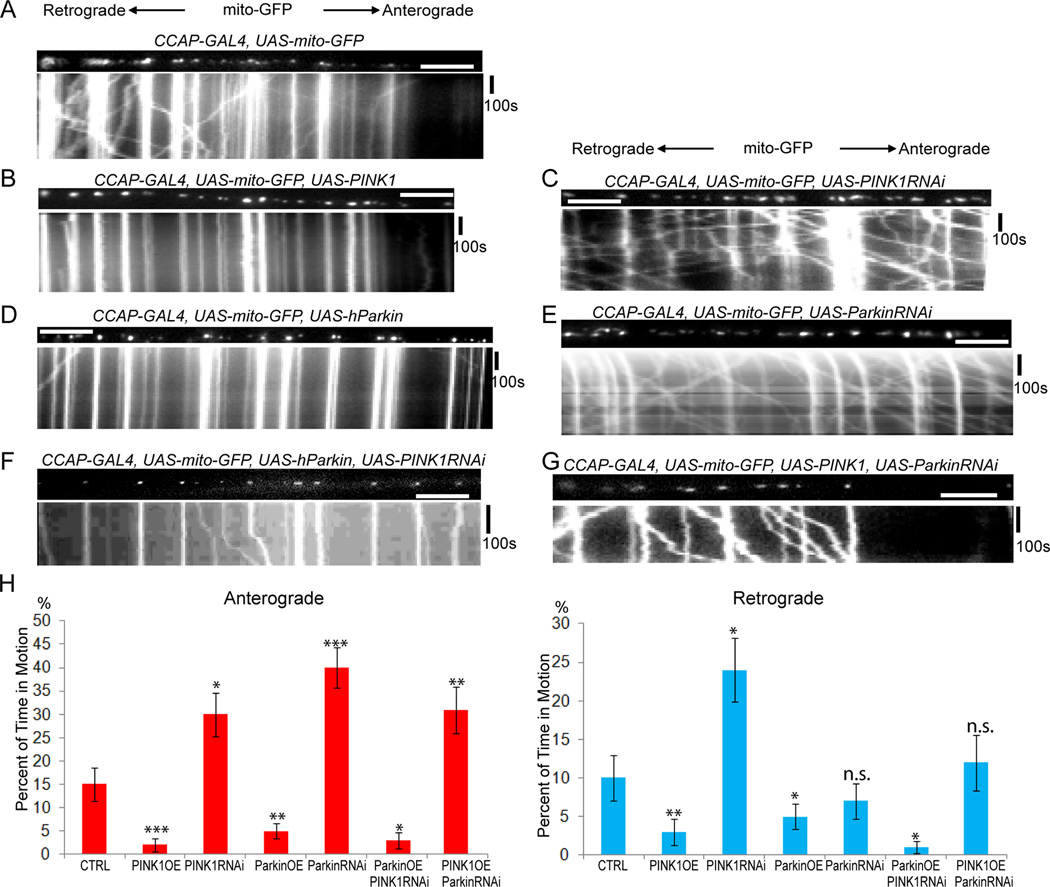
We took advantage of the mutations and transgenes available in Drosophila to examine axonal transport of mitochondria in dissected third-instar larvae (Wang and Schwarz, 2009b). As in mammalian neurons, overexpression of PINK1 or Parkin significantly decreased bidirectional motility. Knock-down of PINK1 or Parkin by RNAi significantly increased anterograde mitochondrial motility and PINK1 RNAi also increased retrograde movement. The effect of Parkin-RNAi did not reach statistical significance for the retrograde direction for reasons not presently understood (Figure 3A–E, H, Movies S3, Table S3). PINK1 or Parkin overexpression also significantly shortened mitochondria (Table S3). Expression of Parkin RNAi together with PINK1 prevented PINK1-mediated arrest of mitochondrial motility but expression of PINK1 RNAi did not interfere with the ability of Parkin expression to stop mitochondria (Figure 3F–H, Table S3). Thus PINK1-dependent mitochondrial arrest required Parkin, but Parkin-dependent mitochondrial arrest did not require normal levels of PINK1 expression. These findings indicate that PINK1 and Parkin function in a common pathway to inhibit mitochondrial motility with Parkin acting downstream of PINK1 in both Drosophila and mice.
Figure 3. PINK1 Functions Upstream of Parkin to Inhibit Mitochondrial Movement in Drosophila CCAP Axons.
For each genotype, UAS-mito-GFP was expressed in a single axon within the segmental nerve by CCAP-GAL4. The first frame of the live-imaging series appears above the kymograph. (A) Mitochondrial movement in a control larva. (B–E) PINK1 or hParkin decreased, and PINK1-RNAi or Parkin-RNAi increased mitochondrial movement when expressed in that axon. Expression of hParkin together with PINK1-RNAi arrested mitochondria (F), but expression of PINK1 together with Parkin-RNAi did not (G). (H) From kymographs as in (A–G), the percent of time each mitochondrion was in motion was determined, averaged, and compared with control. n=66–162 mitochondria from 8 axons and 4 animals per genotype. “n.s.”, not significant. Scale bars, 10 µm. See also Movies S3 and Table S3.
The PINK1/Parkin Pathway Induces Miro Degradation and Releases Kinesin from Mitochondria
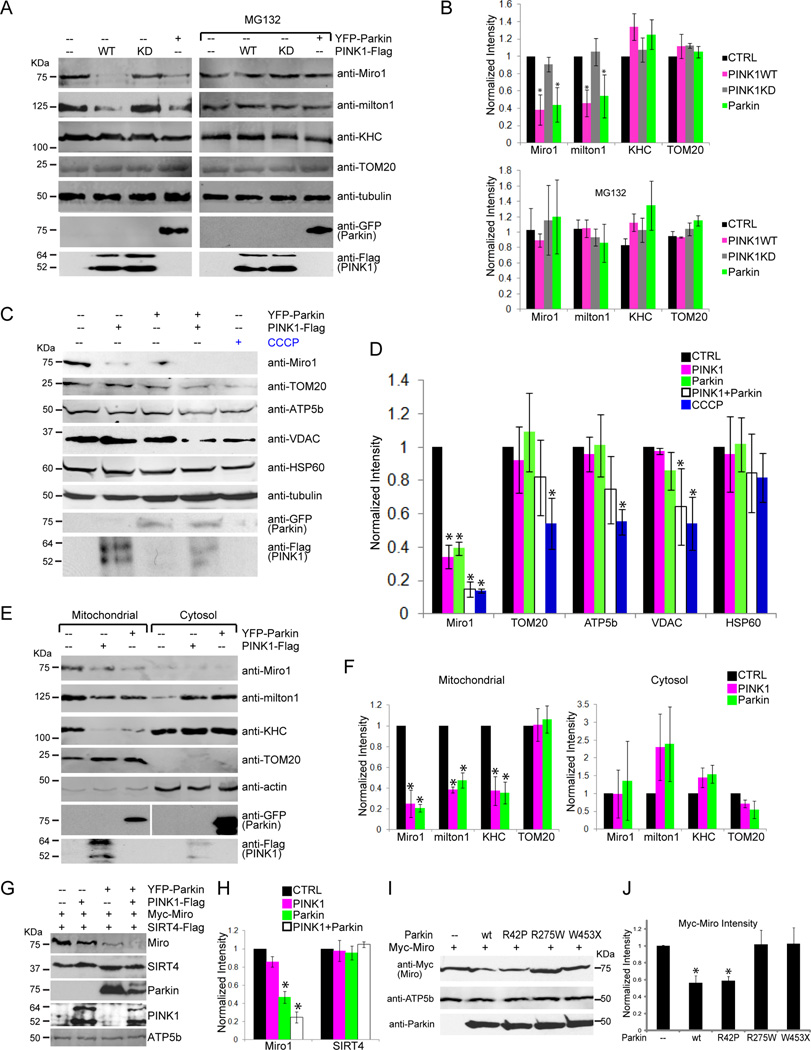
Because Parkin is an E3 ubiquitin ligase, we hypothesized that mitochondrial arrest could involve the degradation of components of the motor/adaptor complex. We expressed either PINK1 or Parkin in HEK293T cells with approximately 60% transfection efficiency (Figure S3A). Levels of endogenous Miro and milton were significantly decreased compared to control when either wildtype Parkin or PINK1 but not PINK1KD was overexpressed. Loss of Miro and milton could be prevented with the proteasome inhibitor MG132 (Figure 4A–B). The decrease in Miro and milton was not due to a uniform destruction of mitochondria because neither the OMM proteins TOM20 and VDAC, nor the matrix proteins ATP5β and HSP60 were decreased (Figure 4A–D). Expression of PINK and Parkin together significantly degraded both Miro and VDAC, and levels of TOM20, HSP60 and ATP5β also tended to decrease; this broader loss of mitochondrial proteins upon coexpression of PINK1 and Parkin might represent an early stage of mitophagy (Figure 4C–D). In contrast to the selective loss of Miro upon PINK1 or Parkin expression, the explicit induction of mitophagy by a 24 h incubation with CCCP (Geisler et al., 2010; Narendra et al., 2010) significantly degraded all of the mitochondrial markers except HSP60 (Figure 4C–D). HSP60 induction by mitochondrial stress may cause its levels to decline more slowly (Chan et al., 2011). Expressed human Miro or Drosophila Miro was also degraded upon expression of human PINK1 or human Parkin (Figure S3B, C). The likely conservation of the degradation pathway is consistent with the ability of PINK1 and Parkin to arrest mitochondrial motility in both mammalian and Drosophila neurons.
Figure 4. The PINK1/Parkin Pathway Causes Miro to be Degraded.
(A, B) Endogenous Miro1 and h-milton1 were degraded by either PINK1 or Parkin expression in a proteasome-dependent manner. HEK293T cells were transfected with PINK1-Flag, PINK1KD-Flag, or YFP-Parkin and grown in the presence or absence of MG132. Cell lysates were analyzed by immunoblot as indicated. n=3 transfections. (C, D) PINK1 and Parkin selectively decrease Miro1 levels and not other mitochondrial proteins. Cells were transfected with PINK1-Flag, YFP-Parkin, or both, or treated with CCCP 20µM for 24 h to promote mitophagy. Lysates were analyzed by immunoblot as indicated. n=3 transfections. In (A–D) quantification was with a fluorescence scanner in the linear range. The intensity of each band was normalized to that of tubulin and expressed relative to normalized levels in control cells. (E, F) PINK1 or Parkin expression in HEK293T cells decreased Miro1, h-milton1, and KHC levels on mitochondria and increased cytosolic h-milton1. For quantification, the intensity of each band was normalized to that of actin and expressed as a fraction of control levels. n=3 transfections. (G, H) PINK1-dependent degradation of Miro requires Parkin. HeLa cells were transfected with PINK1-Flag, or YFP-Parkin, or YFP-Parkin and PINK1-Flag, together with Myc-Miro and SIRT4-Flag (a mitochondrial matrix marker), and cell lysates were analyzed with antibodies to the tagged proteins. (H) The intensity of each Miro1 and SIRT4 band was normalized to the intensity of ATP5β (a mitochondrial matrix loading control) and expressed as a fraction of levels in control cells. n=3 transfections. (I, J) HEK293T cells transfected with Myc-Miro, together with wildtype (wt) Parkin, ParkinR42P, ParkinR275W, or ParkinW453X were lysed and immunoblotted with anti-Myc and detected with a linear range fluorescence scanner. The expression levels of the Parkin constructs were indistinguishable by quantitative immunoblot and one-way Anova: P=0.996, n=4 transfections. (J) Quantifications of Myc-Miro. The intensity of each Myc-Miro band was normalized to the intensity of ATP5β, and the control band was set as 1. n=4 transfections. See also Figure S3.
Expression of either PINK1 or Parkin in HEK293T cells significantly lowered the levels of Miro, milton and KHC on mitochondria. In the cytosol, levels of milton and KHC tended to increase, though these changes were highly variable and did not reach statistical significance (Figure 4E–F). The presence of KHC on other organelles may have obscured detection of KHC released from mitochondria. Miro is a requisite receptor for Milton and KHC on the mitochondrial surface (Glater et al., 2006). Therefore, if Miro is a direct target of proteasome-dependent degradation, it would likely cause loss of the entire complex from the mitochondrial surface and thereby explain mitochondrial arrest, although degradation of milton and KHC is also possible. The proteasomal degradation of membrane proteins such as Miro typically involves the intermediate step of removing the protein from the membrane by proteins such as p97 prior to its degradation and p97 has been shown to be activated by the PINK1/Parkin pathway (Karbowski and Youle, 2011). Loss of mitochondrial motility might therefore be achieved when Miro is removed from the membrane, even before its eventual degradation.
Because PINK1 acts via Parkin in arresting mitochondria, PINK1 was predicted to require Parkin to promote Miro degradation. This relationship was tested in HeLa cells, because they lack endogenous Parkin (Narendra et al., 2008). In HeLa cells, PINK1 overexpression did not significantly decrease cotransfected Myc-Miro levels unless Parkin was also co-transfected. As seen in HEK293T cells, Myc-Miro was selectively degraded when both PINK1 and Parkin were expressed; levels of the co-transfected mitochondrial marker (SIRT4) were unaffected (Figure 4G–H). Thus Parkin is required for PINK1-induced loss of Miro. Although expression of Parkin without co-expression of PINK1 could also cause loss of Myc-Miro in HeLa cells, it required a higher ratio of Parkin to Miro DNA in the transfection (1:1) than that which was needed for HEK293T cells (1:2) (Figure 4G, S3D–E). As was seen in Parkin−/− neurons, higher levels of Parkin transfection are likely required in HeLa cells due to the lack of endogenous Parkin.
We also tested pathogenic point mutations found in human Parkin (Berger et al., 2009) (Figure 4I–J). R42P disrupts the ubiquitin-like domain and R275W disrupts the RING1 domain, while W453X truncates the protein after the RING2 domain. The phenotypes associated with these alleles suggest that they have attenuated functions. We found that expression in HEK293T cells of either ParkinR275W or ParkinW453X failed to decrease co-expressed Myc-Miro levels, but Parkin R42P did. The latter allele has previously been shown to retain ligase activity and an affinity for some but not all targets (Sriram et al., 2005).
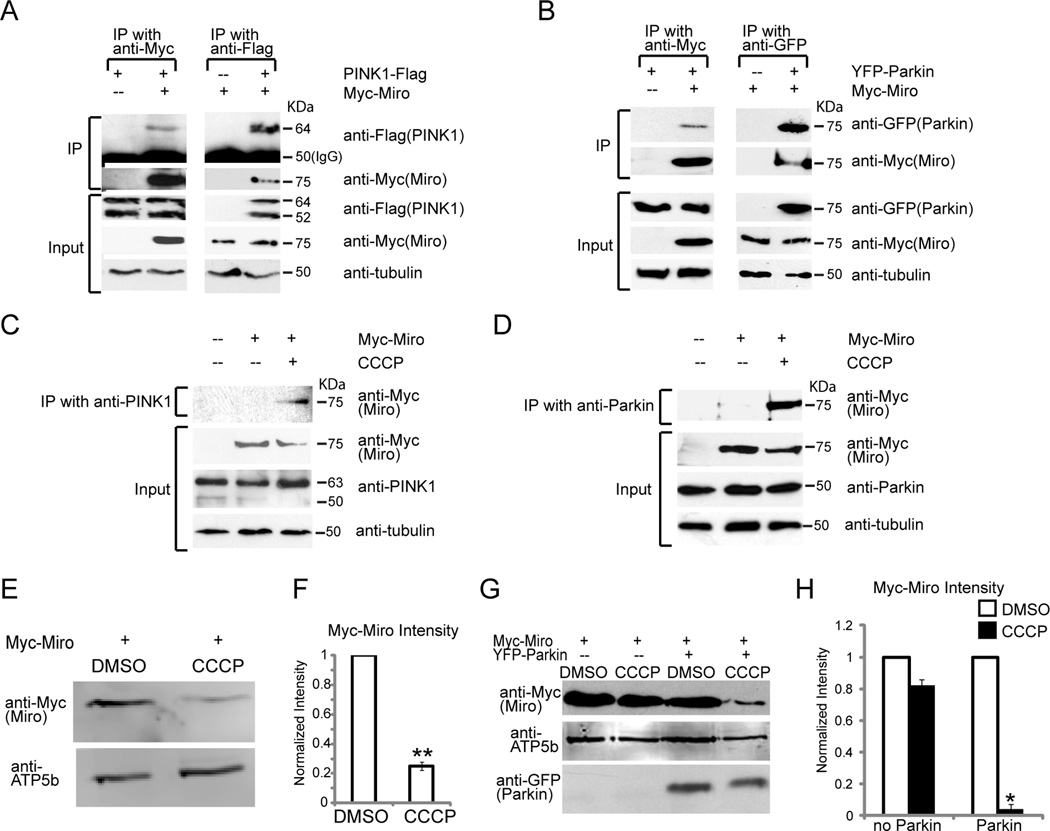
Mitochondrial Depolarization Causes PINK1 and Parkin to Associate with Miro and Causes Miro Degradation
To examine the mechanism that targets Miro for degradation, we analyzed its interactions with PINK1 and Parkin. Both PINK1-Flag and YFP-Parkin coprecipitated with Myc-Miro when expressed (Figure 5A–B). Without exogenous expression, however, very little interaction was observed for either endogenous PINK1 or endogenous Parkin in HEK293T cells with expressed Myc-Miro. Their coprecipitation was greatly enhanced, however, if cells were treated with CCCP (Figure 5C, D). Thus, the interaction of Miro with PINK1 and Parkin is triggered by mitochondrial depolarization, consistent with findings that PINK1 is not imported into damaged mitochondria (Jin et al., 2011) and accumulates, along with Parkin, on the OMM (Narendra et al., 2008).
Figure 5. PINK1 and Parkin Interact with Miro upon Mitochondrial Depolarization.
The interactions of PINK1, Parkin, and Miro were examined in HEK293T cells transfected as indicated. For each assay, 50% of the precipitate and 10% of the input were loaded. (A, B) Immunoprecipitations using anti-Myc, anti-Flag or anti-GFP. PINK1-Flag (A) and YFP-Parkin (B) were detected in Myc-Miro immunoprecipitates, and Myc-Miro was detected in PINK1-Flag (A) or YFP-Parkin immunoprecipitates (B). MG132 was used to prevent Myc-Miro degradation. (C, D) Immunoprecipitation of endogenous PINK1 (C) or Parkin (D). Myc-Miro coprecipitated efficiently with anti-PINK1 or anti-Parkin when cells were treated with 40µM CCCP for 10 min. (E, F) HEK293T cells transfected with Myc-Miro were incubated with 10µM CCCP in DMSO or DMSO alone for 3 h prior to lysing the cells. Immunoblots of lysates were probed with anti-Myc and detected with a fluorescence scanner for quantification (F) after normalization to the mitochondrial loading control ATP5β and expressed as a fraction of the control value. n=6 transfections. (G) Cell lysates from HeLa cells transfected as indicated and exposed to CCCP or DMSO as in (E). (H) Quantification of Myc-Miro levels, normalized to ATP5β and expressed as a fraction of the DMSO control. n=4 transfections. See also Figure S4.
Parkin facilitated the arrest of mitochondrial movement upon the loss of mitochondrial membrane potential (Figure 2K). To ask whether this effect correlated with Miro degradation, we applied 10 µM CCCP to HEK293T cells and examined Miro levels after 3 h, a time prior to the onset of mitophagy, and normalized levels of Miro immunoreactivity to ATP5β levels to control for any general loss of mitochondrial proteins due to early mitophagy or cell death. CCCP selectively reduced Miro levels to 25% of control (Figure 5E–F). Thus the degradation of Miro can be evoked not only by overexpression of PINK1 or Parkin, but also by mitochondrial depolarization.
Parallel experiments were conducted in HeLa cells transfected with Myc-Miro, with or without co-transfection of Parkin at a 1:4 ratio of Parkin to Miro DNA. At this ratio, Miro expression persists in Hela cells (Figure S3D–E, 5G), presumably because Miro synthesis outpaces its degradation. Under these conditions, but not in the absence of Parkin, 10 µM CCCP caused the loss of Miro (Figure 5G–H). Thus, as with PINK1 overexpression, CCCP-induced degradation of Miro cannot proceed in HeLa cells unless Parkin expression is restored. Similarly, Miro was degraded in neurons upon mitochondrial depolarization with 80 µM Antimycin A. The decrease in Miro could be prevented by MG132 (Figure S4). However, stabilization of Miro levels by MG132 could not rescue the mitochondrial arrest caused by Antimycin A (Figure 2K and data not shown). As discussed below, although mitochondrial depolarization causes Miro degradation in both cell lines and neurons, the pathway may not need to proceed all the way to proteasomal degradation of Miro in order to stop mitochondria,
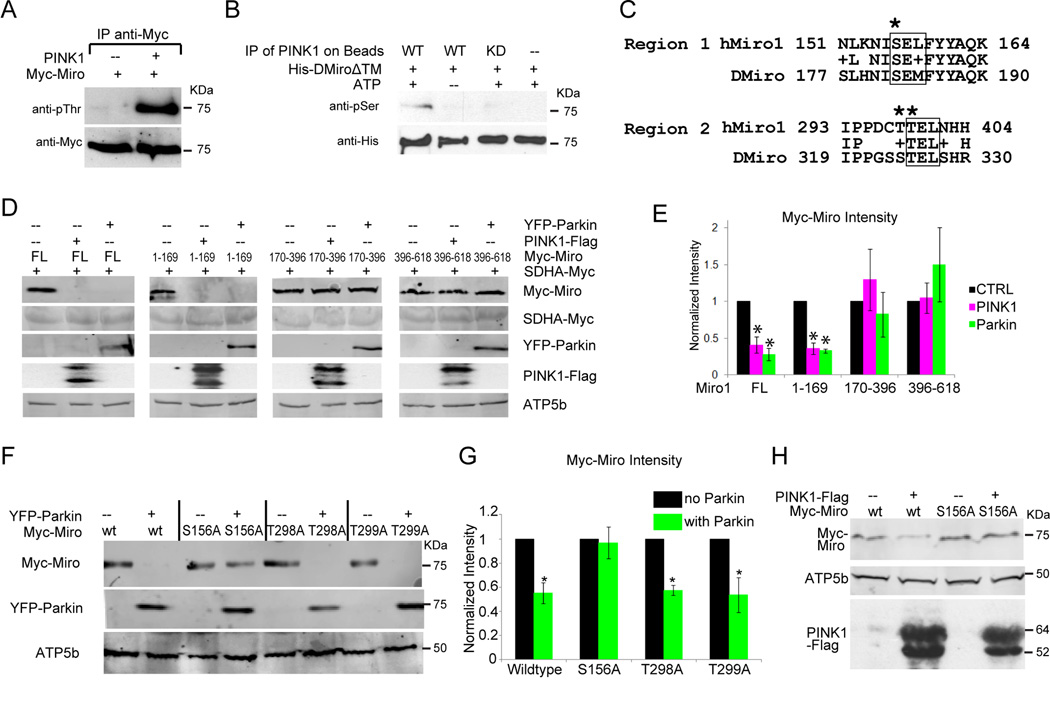
PINK1 Phosphorylates Miro
To determine if Myc-Miro is a substrate of PINK1, they were co-transfected in HEK293T cells. In an anti-Myc immunoprecipate, anti-phospho-Threonine detected a band that comigrated with Miro (Figure 6A). To look for a direct phosphorylation, bacterially expressed Drosophila Miro1-617, a construct lacking the C-terminal transmembrane domain, was incubated with Flag-tagged PINK1 or PINK1 KD. The in vitro phosphorylated Miro was then analyzed by immunoblotting. A band of the size of Miro was detected with anti-phospho-Serine when ATP and PINK1 but not PINK1 KD were added (Figure 6B). The immunoprecipitate was also analyzed by mass spectrometry. Two phosphopeptides were identified corresponding to amino acids 177–190 and 319–330 of Drosophila Miro, although the spectra could not determine which serine or threonine residues within the peptides were modified (Figures 6C and S5). When either ATP or PINK1 was omitted from the reaction, mass spectrometry did not detect the phosphorylated peptides, although the peaks corresponding to unphosphorylated Miro peptides were equivalent in the samples. In human Miro1 and Miro2, three of the potentially phosphorylated serine and threonine residues within the Drosophila phosphopeptides had clear homologs: in Miro1, Ser156 (homologous to Ser182 in Drosophila) and Thr298, 299 (homologous to Ser324, 325 in Drosophila, Figure 6C).
Figure 6. PINK1 Phosphorylation of Miro is Necessary for PINK1/Parkin-dependent Miro Degradation.
(A) Immunoprecipitated Myc-Miro was recognized by anti-phospho-Threonine only when PINK1-Flag was co-expressed in HEK293T cells. MG132 was present to prevent Miro degradation. (B) PINK1 or PINK1 KD was immunoprecipitated from HEK293T cell lysates and added to bacterially expressed His-tagged Drosophila Miro for an in vitro kinase assay and then probed with anti-phospho-Serine and anti-His. (C) The two phosphopeptides identified in Drosophila Miro by mass spectrometry are aligned with hMiro1. Potential phosphorylation sites that are conserved between the species are marked (*) and a box indicates a potential consensus for a PINK1 target. (D) Miro1-169 can be degraded by PINK1 or Parkin overexpression. Lysates from cells transfected as indicated were immunoblotted with anti-Myc, anti-GFP, anti-Flag and anti-ATP5β. FL, full length. Succinyldehydrogenase (SDHA) is a mitochondrial matrix marker. (E) Quantifications of MiroFL, Miro1-169, Miro170-396 and Miro396-618. The intensity of Myc-Miro bands were normalized to SDHA-Myc levels with the control level set as 1. n=3 transfections. (F) After transfection with wildtype or mutated forms of Myc-Miro and YFP-Parkin, lysates were prepared and immunoprobed with anti-Myc, anti-ATP5β and anti-GFP. (G) Quantifications of Myc-Miro immunoreactivity in (F). The intensity of each Myc-Miro band was normalized to the intensity of ATP5β and the control band was set as 1. n=4 transfections. (H) Cells transfected as indicated were blotted with anti-Myc, anti-ATP5β and anti-Flag. All quantifications were with a fluorescence scanner in the linear range. See also Figure S5.
Ser156 of Human Miro Mediates the Effects of PINK1 and Parkin Expression
Identification of phosphorylation sites allowed us to investigate their significance in triggering Miro degradation. To identify the necessary regions, we first split the coding region of human Miro into three pieces: Miro1-169, which contains the first GTPase domain and the predicted first phosphorylation site at Ser 156; Miro170-396, which contains the two EF hand motifs and the second set of predicted phosphorylation sites; and Miro396-618, which contains the second GTPase motif and the transmembrane domain. We predicted that only regions of the protein containing the necessary sequences for PINK1 and Parkin action would be degraded upon their coexpression in HEK293T cells. Levels of full-length Miro and Miro1-169 were significantly lower in response to either PINK1 or Parkin expression, but levels of Miro170-396 and 396–618 were not diminished (Figure 6D–E). We also individually mutated to Alanine each of the three potential PINK1 phosphorylation sites in human Miro (Ser156, Thr298, and Thr299), and determined whether those alterations influenced the effect of Parkin expression. Only the Ser156Ala mutation (MiroS156A) significantly prevented the loss of Miro in response to overexpressed Parkin (Figure 6F–G). Ser156Ala also prevented the decrease in Miro levels caused by overexpressed PINK1 (Figure 6H). Levels of ATP5β were unaffected by PINK1 or Parkin expression, confirming that their effect was not due to mitophagy. Ser156 resides within the N-terminal portion of Miro and its significance for PINK1- and Parkin-dependent degradation is thus consistent with the sensitivity of this domain to degradation (Figure 6D–E).
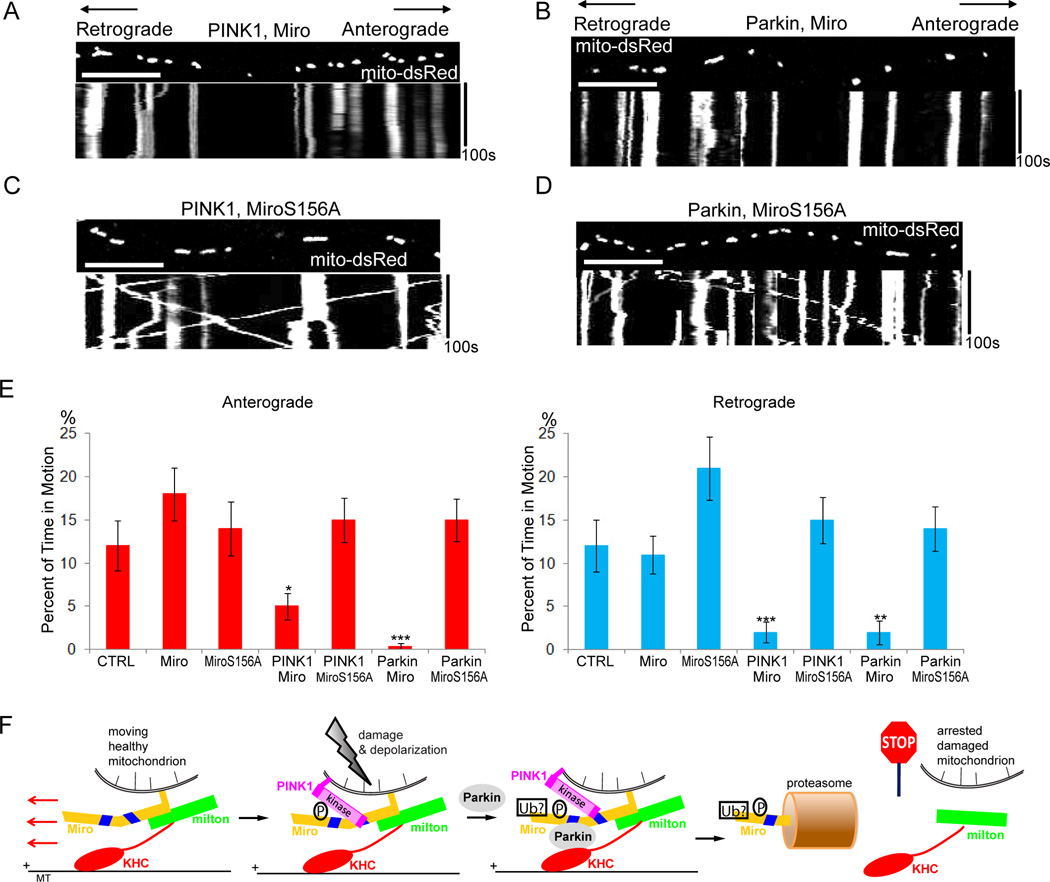
The preceding experiments suggested a model in which Ser156 of Miro is phosphorylated by PINK1, which triggers Parkin-dependent degradation of Miro by the proteasome. This model predicts that MiroS156A expression should permit axonal transport of mitochondria to persist when PINK1 or Parkin is expressed. We found, however, that overexpressed wildtype Miro was sufficient to resist the arrest of mitochondria if transfected at a 1:1 DNA ratio with PINK1 or Parkin (Table S4). Increased Miro synthesis probably compensated for Miro degradation. To examine the importance of the phosphorylation site, therefore, we reduced expression of the Miro transgene to a level where wildtype Miro synthesis did not outpace its degradation: a 3:1 DNA transfection ratio of PINK1 or Parkin to Miro. At this ratio, transfection of PINK1 or Parkin with wildtype Miro stopped mitochondria but movement persisted when MiroS156A was used. Overexpression of Miro or MiroS156A alone at this level did not affect mitochondrial motility (Figure 7A–E, Table S4), and protein levels of overexpressed Miro or MiroS156A on mitochondria were indistinguishable (Figure S6). Ser156 therefore has a key role in PINK1- and Parkin-driven degradation of Miro and mitochondrial arrest.
Figure 7. MiroS156A Prevents Mitochondrial Arrest by PINK1/Parkin Overexpression in Rat Hippocampal Axons.
(A,B) Expression of PINK1-Flag (A) or YFP-Parkin (B) at a 3:1 ratio to wild type Miro arrested mitochondria. (C,D) Expression of PINK1-Flag (C) or YFP-Parkin (D) at a 3:1 ratio to MiroS156A did not arrest mitochondria. (E) From kymographs as in (A–D), the percent of time each mitochondrion was in motion was determined and averaged (n=85–153 mitochondria from 8 axons and 4 separate transfections per genotype). (F) Schematic representation of the proposed mechanism of PINK1/Parkin-dependent mitochondrial arrest. Mitochondrial depolarization stabilizes PINK1 on the surface of the mitochondrion, promotes its interaction with Miro, and causes PINK1 to phosphorylate Ser156 of Miro. Subsequent interaction of Parkin with Miro and likely ubiquitination causes Miro to be removed from the membrane and degraded by the proteasome, releasing milton and kinesin from the organelle. Scale bars, 10 µm. See also Figure S6 and Table S4.
DISCUSSION
Although genetic and cell biological data have placed PINK1 upstream of Parkin in a pathway that regulates mitochondrial morphology and degradation, the relationship of the two enzymes has been obscure. One model proposes that Parkin is a PINK1 substrate activated by phosphorylation, but others have failed to find this phosphorylation (Vives-Bauza and Przedborski, 2011). Our findings indicate an alternative model: PINK1 and Parkin bind to the same target (Figure 5A–D) and its phosphorylation by PINK1 (Figure 6) allows Parkin, presumably acting as an ubiquitin-ligase, to designate that protein for removal from the mitochondrial membrane and proteasomal degradation. Indeed, hMiro1 and hMiro2 recently appeared among a list of proteins down-regulated by Parkin overexpression and CCCP (Chan et al., 2011). The ability of Parkin to bring about Miro degradation is consistent with its ability to ubiquitinate mitofusin and thereby to cause its degradation through the sequential action of p97/VPC and the proteasome (Karbowski and Youle, 2011). Interestingly, Miro and mitofusin interact with one another (Misko et al., 2010) and their shared interaction with Parkin suggests coordinated regulation. We have identified two Miro peptides phosphorylated by PINK1 and at least one phosphorylation site, Serine 156, is important for the subsequent action of Parkin and the proteasome (Figures 6, 7). We have not directly examined the function of p97/VPC, but it is likely also to be required to remove Miro from the OMM so that Miro can be accessed by the proteasome. This may be the stage of the pathway at which milton and kinesin are released from the mitochondrial surface and mitochondria stop. Our genetic and biochemical data indicate that arrest by PINK1 expression occurs downstream of Parkin, but the inability of MG132 to prevent this arrest suggests that the degradation of Miro occurs after the complex has been removed from the surface and motility has ceased.
Mitochondrial motility is especially critical to neurons where it may take days for a mitochondrion to move between the cell body and a distant axonal or dendritic ending. The need for mitochondria to undergo turnover, as well as their redistribution to balance changes in local energy demand, make mitochondrial movement an important on-going and regulated process. The mitochondrion-specific adaptor proteins, Miro and milton, are control points for this motility as illustrated by the pathway we have examined (Figure 7F).
Arrest-by-removal via this pathway provides an interesting contrast with the previously reported mechanism of Ca2+-dependent arrest (Wang and Schwarz, 2009a). The latter is accomplished by a conformational change within the complex: when Ca2+-binds to the EF hands of Miro, the motor domain of kinesin binds directly to Miro and is thereby prevented from interacting with microtubules. Kinesin remains bound to the mitochondrion and therefore the mechanism is easily reversible once Ca2+ is removed. Although an alternative model was proposed in which Ca2+ dissociated the motor/adaptor complex (MacAskill et al., 2009), this has not been observed by others (Wang and Schwarz, 2009a; Brickley et al, 2010). The Ser/Thr-kinase and ubiquitin-ligase cascade that constitutes the PINK1/Parkin pathway, however, brings about an irreversible change via the degradation of Miro (Figures 4 and 7F).
This distinction likely reflects the different cellular purposes of the pathways. Whereas Ca2+ likely triggers a transient local response to a temporary need for additional mitochondria, PINK1 and Parkin initiate an irreversible pathway for clearing damaged mitochondria. Consistent with this, depolarization of mitochondria with CCCP greatly increased the association of PINK1 and Parkin with Miro (Figure 5C, D) and depolarization was sufficient to trigger the loss of Miro from HEK293T cells, HeLa cells, and neurons, provided Parkin was present (Figures 5E–H, S4).
Damaged mitochondria in cell lines selectively recruit Parkin and are in turn targeted for mitophagy (Narendra et al., 2008). In contrast to an earlier report (Van Laar et al, 2011), we find that this recruitment also occurs in axons; when highly expressed, YFP-Parkin was observed on mitochondria without depolarization (consistent with its ability to arrest mitochondrial motility upon overexpression), but with lower expression levels it was recruited to mitochondria by treatment with Antimycin A (Figure S2B). Parkin recruitment is initiated by the depolarization-induced stabilization of PINK1 on the mitochondrial surface (Geisler et al., 2010; Jin et al., 2010; Narendra et al., 2010) and PINK1 is also upstream of Parkin in regulating mitochondrial morphology (Clark et al., 2006; Exner et al., 2007; Park et al., 2006; Poole at al., 2008). This relationship also holds for mitochondrial motility. PINK1 arrested mitochondrial motility in wildtype but not Parkin−/− mice or Parkin RNAi flies. Mitochondrial depolarization with CCCP caused the degradation of Miro in a Parkin-dependent manner. Similarly, PINK1 expression brought about the degradation of Miro in Parkin expressing cells, but not in Parkin-lacking HeLa cells (Figures 2–5).
Why is Miro targeted as part of a mitophagy pathway? One possibility is that the arrest of mitochondrial motility, like the prevention of mitochondrial fusion by mitofusin degradation, helps to quarantine a depolarized mitochondrion or mitochondrial fragment prior to its engulfment by an autophagosome. Stationary mitochondria are less likely to undergo fusion with other mitochondria (Twig et al., 2010) and, by immobilizing unhealthy mitochondria, damage from released reactive oxygen species would be confined to a smaller region. The fate of depolarized axonal mitochondria is not known and they may be degraded primarily in the soma. The neuron may, however, need first to sequester the damaged organelle locally within an autophagosome. Subsequent autophagosomal transport to the soma would be independent of milton and Miro, which are selectively on mitochondria.
In previous genetic studies of PINK1 and Parkin differences were noted between mice and Drosophila. Drosophila loss of function mutants had profound defects in mitochondrial morphology (Deng et al., 2008; Poole et al., 2008; Yang et al., 2006) that were only seen in knockout mice when neurons were additionally stressed (Exner et al., 2007; Gautier et al., 2008; Palacino et al., 2004). We also observed differences between Drosophila and murine models. In both, PINK1 or Parkin overexpression arrested mitochondria and in both Parkin was required downstream of PINK1. However, in Drosophila neurons RNAi knockdown of PINK1 or Parkin increased mitochondrial motility whereas we observed no statistically significant differences of motility in murine Parkin−/− neurons. These differences may reflect a difference in how the species employ the pathway: in mammals, it may be strictly reserved for the response to mitochondrial depolarization (Figure 2K) whereas in the fly, whose short lifespan may make mitochondrial damage less critical, it may contribute to the ongoing turnover of proteins that participate in mitochondrial dynamics.
The ability of Parkin overexpression to alter mitochondrial motility in the presence of PINK1 RNAi (Figure 3) or mitochondrial morphology in a PINK1 null background (Clark et al., 2006) indicates that, although PINK1 can stimulate Parkin function, Parkin can act independently as well. In the current study, we do not know if Parkin is effective because of residual PINK1 in the RNAi-expressing cells, because other kinases can also activate Miro as a Parkin substrate, or because elevated levels of Parkin can lead to Miro degradation even in the absence of a phosphorylation. PINK1 is likely to enhance Parkin function but probably is not required.
Our observation that two PD-associated genes encode regulators of mitochondrial motility is consistent with other findings linking misregulation of mitochondrial dynamics to neurodegeneration. Changes in mitochondrial distribution, transport, and dynamics are implicated in Charcot-Marie-Tooth, Amyotrophic Lateral Sclerosis, Alzheimer’s and Huntington’s diseases (Schon and Przedborski, 2011). These findings underscore the importance of mitochondrial dynamics for supplying distal regions with sufficient energy and Ca2+-buffering capacity, compensating for changes in energy demand, refreshing older mitochondria through fusion with newly-synthesized mitochondria (Frederick and Shaw, 2007), and clearing damaged mitochondria (Whitworth and Pallanck, 2009; Youle and Narendra, 2010).
Miro joins a growing list of reported Parkin substrates (Chan et al., 2011; Geisler et al., 2010; Poole et al., 2010; Shin et al., 2011; Karbowski and Youle, 2011; Ziviani et al., 2010). It is presently unknown whether any single substrate is critical for subsequent mitophagy or if ubiquitination or proteasomal degradation of multiple mitochondrial surface proteins is needed. It will also be important to determine if PINK1 serves as a priming kinase for each of the Parkin substrates and whether substrate phosphorylation is sufficient to promote the action of Parkin. The phosphorylation sites recognized by PINK1 fall in two regions of Miro: 1) at the C-terminal end of the first GTPase domain of Miro and 2) between the two EF hands (Figure 6C). Some sequence similarity between these sites suggests a partial consensus for the enzyme (S/T E L/M). Interestingly, a similar sequence can be found in mitofusin, suggesting that PINK1 may phosphorylate mitofusin and thereby regulate its Parkin-dependent ubiquitination (Poole et al., 2010; Karbowski and Youle, 2011; Ziviani et al., 2010).
Clarification of the relationship of PINK1 and Parkin supports the view that PD is a mitochondrial disorder (Whitworth and Pallanck, 2009; Youle and Narendra, 2010). In the etiology of PD, the regulation of Miro levels may be significant. Either through a specific sorting pathway or as a consequence of the random reassortment of mitochondrial proteins that occur with repeated fusion and fission, some organelles or fragments of the organelle will arise in which the burden of dysfunctional proteins is sufficient to compromise the membrane potential. The resulting stabilization of PINK1 on the surface (Deas et al., 2010; Jin et al., 2010) and targeting of Miro, mitofusin, and other proteins for Parkin action and degradation, will bring about the sequestration and eventual engulfment of that dysfunctional organelle. Sequestration and mitophagy thereby prevent further cellular damage due to reactive oxygen species and enable the cellular complement of mitochondria to be replenished by healthier organelles. The greater the stresses on mitochondria, the more acute the need for this clearance pathway. The heightened sensitivity of the dopaminergic neurons in the substantia nigra to disruption of this ubiquitous pathway may therefore reflect exceptional challenges for mitochondria in these cells. Those stresses may include the susceptibility of dopamine to oxidation and high rates of Ca2+ influx (Surmeier et al., 2010). When this quality control mechanism is defective in patients carrying mutations in either gene, damaged mitochondria will retain Miro and mitofusin, and therefore may move about in the neuron and, through fusion reactions, reintroduce damaged components to otherwise healthy organelles rather than undergo mitophagy.
EXPERIMENTAL PROCEDURES
Fly stocks
The following fly stocks were used: CCAP-GAL4 (Park et al., 2003), UAS-mito-GFP (Pilling et al., 2006), UAS-PINK1(Clark et al., 2006), UAS-hParkin (Yang et al., 2006), UAS-PINK1RNAi (Yang et al., 2006), UAS-ParkinRNAi (Venderova et al., 2009, VDRC ID 47636).
Constructs
The following constructs were used: mito-dsRed (Clontech); Synaptophysin-YFP (Wang and Schwarz, 2009a); PINK1-Flag, PINK1KD-Flag, and PINK1ΔMTS-Flag (Weihofen et al., 2009); YFP-Parkin, mCherry-Parkin (Narendra et al., 2008); pcDH-Parkin, pcDH-ParkinR275W, pcDH-ParkinR42P, and pcDH-ParkinW453X (Berger et al., 2009); pA1T7-DMiro (Glater et al., 2006); pRK5Myc-Miro1 (Fransson et al., 2006). pRK5Myc-Miro1(1–169), pRK5Myc-Miro1(170–396), and pRK5Myc-Miro1(396–618) were generated by PCR amplification from pRK5Myc-Miro1 with BamHI/EcoRI engineered primers and then ligated into BamHI/EcoRI digested pRK5Myc-Miro1. Residues 1–5 of Miro1 were thereby retained in all three constructs but no Ser or Thr are present in this region. pRK5Myc-Miro1S156A, pRK5Myc-Miro1T298A, and pRK5Myc-Miro1T299A were generated by site-directed mutagenesis (Stratagene). Transfections of multiple constructs were done with 1:1 ratios unless otherwise stated.
Cell Culture
Hippocampal cells were dissociated from day 18 (rat) or day 17 (mouse) embryos, cultured 5–7 days, transfected with Lipofectamine 2000 (Invitrogen), and imaged 2 days later. Parkin−/− mice, lacking exon3 (Goldberg et al., 2003), were generated by intercrosses of C57BL/6 and 129/Sv hybrid parkin+/− mice. Wild-type mice were used as controls (Goldberg et al., 2003). HEK293T cells were transfected with calcium phosphate (Wang and Schwarz, 2009a). When used, MG132 (Sigma) was applied at 5µM for 12–20 h before lysis of the cells.
Live Image Acquisition and Quantification
For neuronal cultures, confocal images were captured every 2–8 s as previously described (Wang and Schwarz, 2009a). Dissected third-instar larvae were imaged in a chamber on a glass slide and maintained in Schneider’s medium with 5mM EGTA at 22 °C for less than 1 h (Wang and Schwarz, 2009b). Images were captured every 2 s and kymographs were generated for an interval of 100 s. Details of quantification are in Supplementary Methods and Wang and Schwarz, 2009a.
Immunoprecipitation and Western Blotting
HEK293T or HeLa cells lysis and immunoprecipitations were performed as in Wang and Schwarz, 2009a. Lysates were incubated with mouse anti-Myc (9E10, Santa Cruz Biotechnology), rabbit anti-GFP (Invitrogen), rabbit anti-Flag (Sigma), rabbit anti-PINK1 (BC100-494, Novus), mouse anti-Parkin (sc32282, Santa Cruz), mouse anti-His (Invitrogen), and protein A-Sepharose beads (GE Healthcare, Piscataway, NJ) for 3 h at 4°C. See Supplementary Methods for use of antibodies.
In Vitro PINK1 Kinase Assay
See Supplementary Methods for more details. The His-DMiro1-617 coding sequence was PCR amplified from a full-length cDNA construct (Drosophila Genomics Resource Center clone RE22983) and inserted into a pET17b(+) vector (Novagen). The plasmid was expressed in E. coli BL21(DE3)RP cells. PINK1-Flag was immunoprecipitated from HEK293T cells, added to the expressed His-DMiro1-617, with ATP (Sigma) and phosphatase inhibitor cocktails I and II (Sigma) in CK1 buffer (NEB). The reaction was at 30 °C for 2 h and analyzed by immunoblotting.
Mass Spectrometry Analysis
Coomassie-stained gel bands were excised and minced. Details of the procedures can be found in Supplementary Methods.
Statistical Analysis
Throughout the paper, the distribution of data points is expressed as mean ± standard error of the mean. The Mann-Whitney U tests were used for statistical analysis unless otherwise indicated.
Supplementary Material
ACKNOWLEDGEMENT
We thank Drs. P. Aspenström, A. M. Craig, M. Guo, B. Lu, R. Youle, M., Haigis, and G. Hajnóczky for reagents; S. Vasquez and Dr. M. Sahin for assistance with hippocampal cultures. This work was supported by the Ellison Medical Foundation (T.L.S.), Hartman Foundation for Parkinson’s Research (T.L.S.), NIH GM069808 (T.L.S.), NIH NS065013 (M.J.L.), NIH K99NS067066 (X.W.) and LSRF Novartis Fellowship (X.W.) and the proteomics, genetics, and imaging cores of the IDDRC (grant number P30HD18655).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Berger AK, Cortese GP, Amodeo KD, Weihofen A, Letai A, LaVoie MJ. Parkin selectively alters the intrinsic threshold for mitochondrial cytochrome c release. Hum. Mol. Genet. 2009;18:4317–4328. doi: 10.1093/hmg/ddp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickley K, Smith MJ, Beck M, Stephenson FA. GRIF-1 and OIP106, members of a novel gene family of coiled-coil domain proteins: association in vivo and in vitro with kinesin. J. Biol. Chem. 2005;280:14723–14732. doi: 10.1074/jbc.M409095200. [DOI] [PubMed] [Google Scholar]
- Brickley K, Pozo K, Stephenson FA. N-acetylglucosamine transferase is an integral component of a kinesin-directed mitochondrial trafficking complex. Biochim. Biophys. Acta. 2010;1813:269–281. doi: 10.1016/j.bbamcr.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011 Feb 18; doi: 10.1093/hmg/ddr048. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins L, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 2010 doi: 10.1093/hmg/ddq526. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M .The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner N, Treske B, Paquet D, Holmström K, Schiesling C, Gispert S, Carballo-Carbajal I, Berg D, Hoepken HH, Gasser T, et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson S, Ruusala A, Aspenstrom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun. 2006;344:500–510. doi: 10.1016/j.bbrc.2006.03.163. [DOI] [PubMed] [Google Scholar]
- Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S, Wood-Kaczmar A, Yao Z, Plun-Favreau H, Deas E, Klupsch K, Downward J, Latchman DS, Tabrizi SJ, Wood NW, et al. PINK1-associated Parkinson's disease is caused by neuronal vulnerability to calcium-induced cell death. Mol. Cell. 2009;33:627–638. doi: 10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl. Acad. Sci. U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, Meloni EG, Wu N, Ackerson LC, Klapstein GJ, et al. Parkindeficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Youle RJ. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr. Opin. Cell Biol. 2011 Jun 24; doi: 10.1016/j.ceb.2011.05.007. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- MacAskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler JT. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009;61:541–555. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J. Neurosci. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J. Biol. Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- Park JH, Schroeder AJ, Helfrich-Forster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development. 2003;130:2645–2656. doi: 10.1242/dev.00503. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck LJ. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, Rizzuto R, Hajnóczky G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc. Natl. Acad. Sci. U S A. 2008;105:20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) Repression of PGC-1α Contributes to Neurodegeneration in Parkinson's Disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram SR, Li X, Ko HS, Chung KK, Wong E, Lim KL, Dawson VL, Dawson TM. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum. Mol. Genet. 2005;14:2571–2586. doi: 10.1093/hmg/ddi292. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Surmeier DJ, Guzman JN, Sanchez-Padilla J, Goldberg JA. What causes the death of dopaminergic neurons in Parkinson's disease? Prog. Brain Res. 2010;183:59–77. doi: 10.1016/S0079-6123(10)83004-3. [DOI] [PubMed] [Google Scholar]
- Twig G, Liu X, Liesa M, Wikstrom JD, Molina AJ, Las G, Yaniv G, Hajnóczky G, Shirihai OS. Biophysical properties of mitochondrial fusion events in pancreatic beta-cells and cardiac cells unravel potential control mechanisms of its selectivity. Am. J. Physiol. Cell Physiol. 2009;299:C477–C487. doi: 10.1152/ajpcell.00427.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary earlyonset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Van Laar VS, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum. Mol. Genet. 2011;20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venderova K, Kabbach G, Abdel-Messih E, Zhang Y, Parks RJ, Imai Y, Gehrke S, Ngsee J, Lavoie MJ, Slack RS, et al. Leucine-Rich Repeat Kinase 2 interacts with Parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson's disease. Hum. Mol. Genet. 2009;18:4390–4404. doi: 10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Przedborski S. Mitophagy: the latest problem for Parkinson’s disease. Trends in Molecular Medicine. 2011;17:158–165. doi: 10.1016/j.molmed.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. The mechanism of Ca2+ regulation of kinesin-mediated mitochondrial motility. Cell. 2009a;136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. Imaging axonal transport of mitochondria. Methods in Enzymology. 2009b;457:319–333. doi: 10.1016/S0076-6879(09)05018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen A, Thomas KJ, Ostaszewsk,i B, Cookson M, Selkoe DJ. Pink1 forms a multi-protein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry. 2009;48:2045–2052. doi: 10.1021/bi8019178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway: a mitochondrial quality control system? J. Bioenerg. Biomembr. 2009;41:499–503. doi: 10.1007/s10863-009-9253-3. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. U S A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. U S A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Huang Y, Shao Y, May Z, Prou D, Perier C, Dauer W, Schon E, Przedborski E. Proc. Natl. Acad. Sci. U S A. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.